Abstract
Candida albicans is a fungal pathogen that causes severe disseminated infections that can be lethal in immunocompromised patients. Genetic factors are known to alter the initial susceptibility to and severity of C. albicans infection. We developed a next-generation computational genetic mapping program with advanced features to identify genetic factors affecting survival in a murine genetic model of hematogenous C. albicans infection. This computational tool was used to analyze the median survival data after inbred mouse strains were infected with C. albicans, which provides a useful experimental model for identification of host susceptibility factors. The computational analysis indicated that genetic variation within early classical complement pathway components (C1q, C1r, and C1s) could affect survival. Consistent with the computational results, serum C1 binding to this pathogen was strongly affected by C1rs alleles, as was survival of chromosome substitution strains. These results led to a combinatorial, conditional genetic model, involving an interaction between C5 and C1r/s alleles, which accurately predicted survival after infection. Beyond applicability to infectious disease, this information could increase our understanding of the genetic factors affecting susceptibility to autoimmune and neurodegenerative diseases.
INTRODUCTION
Genetic factors are known to alter susceptibility to and severity of Candida albicans infection in mice (1, 3, 22) and humans (42). Therefore, characterizing genetic factors affecting host susceptibility to C. albicans infection is of great importance. Since systemic candidiasis in mice closely resembles the human disease, inbred mouse strains provide a useful experimental model for identification of host susceptibility factors. Although virtually all organs are infected, the kidney is the major target, and the histopathology of infected lesions is similar in mice and humans. Mutations in several immune response genes have been associated with susceptibility to chronic mucocutaneous candidiasis in human families (14, 17, 36, 48), and several have been verified in murine models. Differences in survival after hematogenous C. albicans infection among inbred mouse strains have been associated with complement factor 5 (Hc or C5) alleles (1, 2, 4, 34). A 2-bp deletion polymorphism at the 5′ end of the C5 transcript shifts its reading frame and causes ∼50% of inbred strains to be C5 protein deficient (54). Disseminated candidiasis is rapidly fatal in C5-deficient strains because of uncontrolled fungal proliferation in most organs (34). Although C5 alleles make an important contribution, several previous analyses indicated that there are other genetic factors that affect the severity of tissue damage or survival after C. albicans infection (2, 38). However, no one has yet been able to identify these other genetic factors.
Since its inception in 2004, haplotype-based computational genetic mapping (HBCGM) (30) has been used to identify the genetic basis for many biomedical trait differences among inbred mouse strains, including differences in gene expression (30), pharmacogenetic factors (19, 20, 58), susceptibility to invasive aspergillosis (56) and respiratory syncytial virus infections (47), analgesic medication (43) and inflammatory pain responses (26, 27), incisional wound biology (23, 24), and narcotic drug responses (12, 28, 29, 43). In a mapping experiment, a property of interest is measured in ≥10 inbred mouse strains; genetic factors are then predicted computationally by identifying genomic regions where the pattern of genetic variation correlates with the distribution of trait values among the inbred strains (30). Despite multiple successes, this genetic mapping method has been unable to identify the underlying genetic differences in other, more complex biologic systems (59). The paucity of genomic regions covered by the genetic map was a significant contributor to these failures. The previous haplotype map covered only ∼15% of the genes in the mouse genome (30), and gene families were selected to enable analyses of specific phenotypes (i.e., drug metabolism). Also, the existing haplotype block construction algorithm (30) rewarded the inclusion of more single-nucleotide polymorphisms (SNPs), penalized the generation of more haplotypes in a block, and did not allow for overlapping blocks within a region. As a consequence, a causative block could easily be missed (producing false-negative results) if another block in a region with fewer haplotypes and fewer SNPs was selected. A new HBCGM method with whole-genome coverage and an improved method for haplotype block construction were needed to enable a wider range of biomedical phenotypes (including infectious disease) to be evaluated. Therefore, we produced a next-generation version of the HBCGM method and used it to analyze survival after hematogenous C. albicans infection in a panel of inbred mouse strains. The results led us to produce a novel combinatorial, conditional genetic model, involving an interaction between C5 and C1s alleles, that accurately predicted survival after infection.
MATERIALS AND METHODS
Survival after Candida albicans infection.
All mouse experiments were approved by the Los Angeles Biomedical Research Institute Animal Care and Use Committee and were performed according to the Guide for the Care and Use of Laboratory Animals (35a). Male mice were obtained from Jackson Laboratories and were used in survival studies at approximately 6 weeks of age. C. albicans strain SC5314 was grown in yeast extract-peptone-dextrose (YPD) broth at 30°C. The yeast-phase organisms were washed twice in phosphate-buffered saline (PBS) and enumerated with a hemacytometer. To induce disseminated candidiasis, 10 mice of each strain were inoculated via the lateral tail vein with 104 C. albicans cells per gram of body weight. The inoculum was confirmed by quantitative culture. In the survival experiments, the mice were monitored at least 3 times daily, and moribund animals were euthanized humanely. The kidney fungal burden and myeloperoxidase activity were determined with a separate group of infected mice that were sacrificed 1 day after inoculation. The kidneys obtained from these mice were harvested and homogenized in ice-cold PBS containing protease inhibitor cocktail (Sigma-Aldrich), and an aliquot was cultured quantitatively to measure the kidney fungal burden. The myeloperoxidase content of the kidney homogenates was measured to assess renal phagocyte accumulation as previously described (13). The kidney homogenates were clarified by centrifugation at 16,000 × g, and the myeloperoxidase content was determined using an enzyme immunoassay kit (Cell Sciences, Canton, MA). The log rank test was used to compare the survival curves for different C57BL/6 and B6.CSS6 mice after C. albicans infection (21).
Computational genetic mapping.
The methods for producing the genetic map and the new mapping method and the characterization of the method are described in the supplemental material. Using the new genomewide haplotype map, genetic factors were identified computationally using our previously described methods (30, 51). In brief, the pattern of genetic variation within each block was correlated with the distribution of trait values among the strains analyzed by using analysis of variance (ANOVA)-based statistical modeling. P values from the ANOVA model and the corresponding genetic effect size were calculated for each block (30, 51). The blocks were then ranked by their P values, and those below an input threshold were used as candidate predictions. The SNPs within the blocks were annotated using Ensembl mouse genome annotation information (http://www.ensembl.org; NCBI mouse genome build 37), and our software automatically identified SNPs causing nonsynonymous coding changes. The haplotype pattern, chromosomal location, presence of nonsynonymous SNPs, and calculated P values and genetic effect size for each block meeting the input criteria were also outputs of this program. For analysis of C. albicans survival, the median survival was used as the phenotypic input, since the median is more resistant to occasional outliers than the average. NZW mice had an extremely long survival time (median, 13 days) relative to the other strains. This complicated the computational analysis, since all blocks with NZW-specific haplotypes were identified. Therefore, the survival data were censored at 7 days for this analysis, which indicated that the strains had prolonged survival. This approach is very similar to that used for nonparametric analysis; it enabled genetic mapping to produce more stable results that were unaffected by the extreme value for one strain.
Determination of complement alleles for inbred strains.
Genomic DNAs from the following strains were purchased from Jackson Laboratory (ME): DBA/1J, LG/J, MRL/MpJ, NZW/LacJ, PL/J, SJL/J, and SM/J. Next, 9 genomic regions were PCR amplified and sequenced using the following sets of primers: for C1rb on chromosome 6 (124,469,409 bp), GGAGGGAGTTGGGGGTCTAGT and AAGTGGAGGACACCTGTGCAA for amplification and AGTGGAAACAGGCAAGGGTCT and GCAAGACTTCAGTCGGGCAAT for sequencing; for C1s on chromosome 6 (124,490,473 bp), CAGTGGCACAAAGCTGGAGTC and AGCACAGGAGGGAGAGGGATG for amplification and GCACAAAGCTGGAGTCTTGGA and AGGGAGAGGGATGGGAGGAG for sequencing; for C1qb on chromosome 4 (136,436,623 bp), AGCTTCAAGACTACCCCACCTG and GGAGGCTCTAGGAGGCCCATT for amplification and CTACCCCACCTGTGGTCACCT and CTGAACCCAGAGAGGCACAAG for sequencing; for C1qc on chromosome 4 (136,446,076 bp), CTCATAAGGTATTGATAAATGGCCACA and AGGTGTGGAGGGCCGATACAA for amplification and AAATGGCCACAGGAATAATACCA and GGCCGATACAAACAGAAGCAC for sequencing; for C1qc on chromosome 4 (136,446,490 bp), TGGCCGTATGCGATGTGTAGT and TTCCTGGAAGAGGAAACTGGA for amplification and GTAGTAGAGGCCCGGCACTTC and AAGGGGAAGGAGAAAGATCATCA for sequencing; for C1qc on chromosome 4 (136,448,449 bp), GAGGCTCCTCTTTATTCCCTTCT and AGTTGTGGTTTCCCCTCAGGT for amplification and GCTGCAAGGTCACCAGAGTCA and CTCAGGTACCCTCCTGAACCA for sequencing; for C1qa on chromosome 4 (136,453,706 bp), GGAGGAGAAAGGGGAAAGAGG and GGGGTGGGTGCTAGGGTTAAG for amplification and GGAAAGAGAAGCTGGGGACAA and AGGGCTAGGGGTTAGGGTCAA for sequencing; for C1s on chromosome 6 (124,489,825 bp), CTGCCTCTGCTTCCTGAGTGA and TGCCTTGCCTTTGTGTGACTT for amplification and sequencing; and for C1s on chromosome 6 (124,484,450 bp), AGACAACTCTGTCCCGGCATT and GAAAATGTGAGAATGTCTGAAGATGC for amplification and sequencing.
All PCRs were performed using Novagen KOD Hot Start DNA polymerase under the following conditions: 95°C for 2 min; 40 cycles of 95°C for 20 s, 60°C for 10 s, and 70°C for 15 s; and a final step of 70°C for 2 min. The single amplified band for each genomic region was purified using a QIAquick PCR purification kit (Qiagen). The only exception was chr. 6 region 124484450, for which the PCR products were gel fractionated and the band at 594 bp was purified for sequencing. The alleles were determined by aligning the resulting sequence with the reference C57BL/6 sequence (52), using the ClustalW alignment program (46).
Measurement of serum anti-Candida antibodies and serum C1q binding activity.
C. albicans SC5314 yeast cells were grown in YNB broth (Difco) at 30°C overnight, washed in PBS, and then frozen at −80°C in PBS as single-use aliquots. The presence of anti-C. albicans antibodies in mouse sera was determined using a method previously used to detect human anti-C. albicans antibody (8). Serum C1q binding to C. albicans cells was also assessed using a modification of a method previously used to measure human C1q binding to cultured cells (15). C. albicans cells were washed once with Veronal-buffered saline (VBS) and incubated at 1 × 107/ml for 1 h at 1°C with mouse serum that was diluted in 0.1 ml VBS containing 0.1% gelatin, 1.5 mM CaCl2, and 1 mM MgCl2. The cells were washed twice with PBS and incubated for 1 h at room temperature either with fluorescein isothiocyanate (FITC)-labeled anti-mouse Ig(H+L) (SouthernBiotech, Birmingham, AL) for detection of cell-bound mouse antibody or with polyclonal rabbit anti-mouse C1q antibody (Santa Cruz Biotechnology, Santa Cruz, CA) followed by FITC-conjugated goat anti-rabbit antibody (SouthernBiotech) to detect cell-bound C1q. The detection antibodies were used at a 1:50 dilution in 1% bovine serum albumin (BSA)-PBS. After being washed twice with PBS, C. albicans cells were resuspended in 1% BSA-PBS containing LDS 751 (Invitrogen, Carlsbad, CA). The amount of cell-bound antibody or C1q was quantified by flow cytometry using a Quanta SC MPL flow cytometer at a 488-nm excitation wavelength (Beckman Coulter, Miami, FL) as described previously (9). C. albicans cells were identified with LDS 751, and single cells were then defined by cell size with Coulter electronic volume technology and side scatter. The amount of mouse antibody or C1q bound to 5,000 single yeast cells was measured quantitatively as the fluorescence intensity of cell-bound FITC-labeled antibody. A positive cell was defined as having a fluorescence intensity of >10 fluorescence units. As controls, cells treated with FITC-labeled antibody alone were used to measure the level of background binding, which was less than 10 fluorescence units.
RESULTS
Next-generation computational genetic mapping method.
A 2.7-million-SNP database was generated from analysis of data obtained from two different sources and was used to provide genomewide coverage (≥95% of the genes in the murine genome) for 16 inbred strains: 129S1/SvImJ, A/J, AKR/J, BALB/cJ, C3H/HeJ, C57BL/6J, CBA/J, DBA/2J, LP/J, NOD/ShiLtJ, NZO/HiLtJ, BALB/cByJ, FVB/NJ, BTBR T+tf/J, KK/HlJ, and NZW/LacJ. We also developed a new haplotype block construction method that reduced the possibility that a computational genetic mapping experiment would miss a true causative haplotype block (i.e., produce false-negative results). The new “maximal” haplotype block construction method identifies all patterns of genetic variation within a region by allowing the haplotype blocks to overlap (see Fig. S1 in the supplemental material). When the allelic data from all 16 strains were evaluated, the maximal method generated 6-fold more blocks (n = 580,565) than the prior method (n = 92,109) (see Table S1). Our previous method generated a single haplotype map by using all available allelic data for all strains (30), and this map was used for all analyses. However, phenotypic data are usually available only for a subset of the strains in a typical mapping experiment, and inclusion of irrelevant alleles can disrupt haplotypic patterns that are uniform among the strains of interest. The 30,000-fold improvement in the computational efficiency of this implementation enabled customized haplotype blocks to be produced dynamically in real time for the strains with available phenotypic data. We also found that the use of whole-genome sequencing data enabled the haplotype map to provide a more complete representation of the pattern of genetic variation for new strains than could be obtained using genotyping arrays that characterize only previously known SNPs (see Tables S2 and S3). As described in the supplemental material, this new genetic map and mapping method exhibit superior performances over those of our prior genetic mapping method (see Fig. S4) and other available methods for association mapping (see Tables S4 and S5).
Survival after fungal infection.
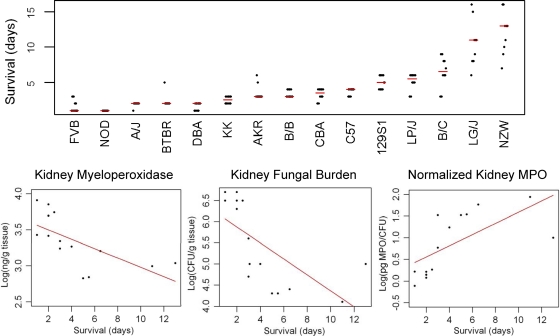
We examined the survival of 15 inbred mouse strains after hematogenous C. albicans infection. There was substantial variation in strain survival; the median survival for 7 strains was ≤3 days, while other strains survived for 10 to 13 days (Fig. 1). One day after infection, we also measured the kidney fungal burden and kidney myeloperoxidase activity (as a measure of phagocyte accumulation). Kidney fungal burden (P value, 0.007) and myeloperoxidase activity (P value, 0.03) were both inversely correlated with survival, while the normalized myeloperoxidase activity (relative to fungal burden) was directly correlated with survival (P value, 0.01). Since both measurements were made at a single time after infection, they provide an imperfect assessment of a dynamic and evolving response to infection. However, these correlations verify the expected result that the host's abilities to recruit phagocytes and limit fungal growth in the kidney are determinants of survival. C5 alleles could explain some, but not all, of the observed interstrain differences. All C5-deficient strains had a median survival of ≤3 days after infection, while the median survival among the C5-sufficient strains ranged from 3 to 13 days (Fig. 2). The large variation in survival among the C5-sufficient strains indicates that genetic factors other than C5 could affect survival.
Fig. 1.
Measured survival times, kidney myeloperoxidase activities, and kidney fungal burdens for a panel of inbred mouse strains. (Top) Survival time for each of 10 mice after hematogenous infection with C. albicans for each of the 15 inbred strains tested. The red bars represent median survival times. (Bottom) Scatter plots for kidney myeloperoxidase activity and kidney fungal burden versus survival time. The x axis represents the survival time (days), and the y axis represents the log10-transformed measured kidney myeloperoxidase activity or kidney fungal burden measured 1 day after infection. The red lines represent the fitted regression lines, and both have Pearson's correlation coefficients of −0.67. Myeloperoxidase activity and kidney fungal burden both show statistically significant inverse correlations with survival (P values of 0.03 and 0.007, respectively). On the right is a scatter plot for normalized myeloperoxidase activity, which is the myeloperoxidase activity divided by the kidney fungal burden (pg/CFU), relative to the survival time (days). The normalized myeloperoxidase activity was directly correlated with survival time (P value, 0.01).
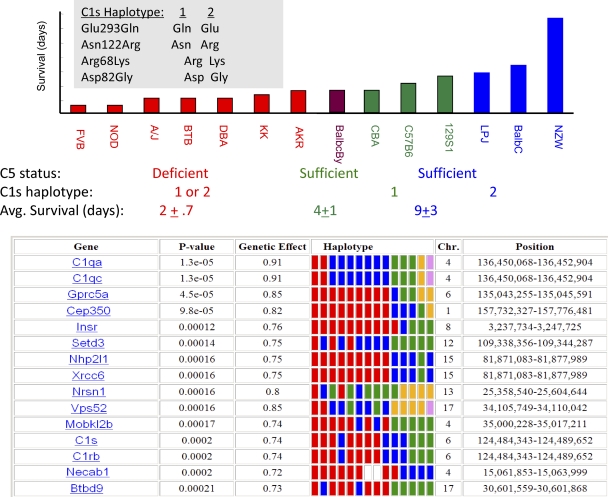
Fig. 2.
(Top) Median survival after hematogenous C. albicans infection for the 14 indicated strains. The C5-deficient strains are shown in red, and the C1s haplotypes of the C5-sufficient strains are indicated. The amino acids at the indicated positions for the two C1s haplotypes are also shown. All C5-deficient strains survived for ≤3 days after infection, while the C1s haplotype influenced the survival of the C5-sufficient strains. BALB/cByJ mice have a different C1s haplotype (due to a C allele at SNP NES16184810) from the other C5-sufficient strains, which is why the data for this strain are shown with a bar with a unique color. (Bottom) The survival data were analyzed by computational haplotype-based genetic mapping, and the 15 genes with the most correlation are shown. The columns show the gene symbol, calculated P value and genetic effect size, chromosome, and locations of the starting and ending positions for each correlated haplotype block. The haplotype pattern within each haplotypic block is also shown; each rectangle represents a single strain that appears in the same order as in the bar graph above. Each haplotype within a block is represented by a rectangle of a different color; strains with rectangles of the same color have the same haplotype.
The median survival data for the 14 strains with available allelic information were analyzed using the next-generation computational genetic mapping method. Two haplotype blocks encoding C1q components (C1qa and C1qc) had the highest correlation with median survival (P < 1 × 10−5). C1rb and C1s (P < 0.0002) were also among the 13 most highly correlated genes (Fig. 2). All three C1q component genes (C1qa, -b, and -c) are adjacent to each other on chromosome 4 (136.4 Mb), while C1rb and C1s are carried by opposite strands of the same genomic region on chromosome 6 (124.5 Mb) (see Fig. S2 in the supplemental material). There was high linkage disequilibrium among SNP alleles within each of these two genomic regions. SNPs within the four C1 genes gave the following alterations: Glu293Gln, Asn122Arg, Arg68Lys, and Asp82Gly substitutions within C1s; an Asn359His substitution within C1r; a Thr16Ile substitution in C1qa; and Val208Ala, Arg70Gln, and Pro10Gln substitutions in C1qc (see Fig. S2B). C1q binding to an antigen-antibody complex induces the Ca2+-dependent assembly of a C1s-C1r-C1s-C1r tetramer (5), which activates the classical complement pathway. C1s is a serine protease with a modular structure; all 4 nonsynonymous C1s SNPs are located within the NH2-terminal bone morphogenic protein and epidermal growth factor (EGF)-like modules (CUB1-EGF-CUB2) (reviewed in reference 5). This Ca2+ binding region of C1s mediates the interaction with C1r and C1q, which is essential for C1s activation (18). The haplotypic groupings created by the C1rs and C1q alleles are very similar (see Fig. S2). However, for the reasons discussed below, the C1s haplotype (Fig. 2) was used in the subsequent analyses. Nevertheless, it is possible that C1q haplotypes (which have a very similar pattern to those for C1rs) have an independent effect on survival, but analyzing this would require a much larger strain panel.
C1 allelic differences have functional consequences.
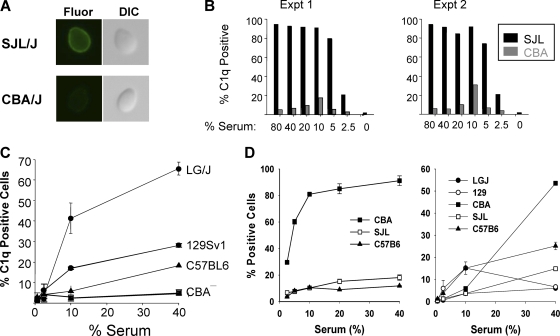
The complement system can be activated by three distinct mechanisms: C1 binding to the Fc region of an antigen-antibody complex activates the classical pathway, while the alternative and mannose-binding lectin pathways are activated by the direct binding of other complement proteins to the pathogen surface (49, 50). Of relevance here, the classical pathway plays an important role in host defense against many types of pathogens (55); it is the dominant pathway for protecting mice against Streptococcus pneumoniae infection (10). Bacteria can activate the classical pathway through direct binding of C1q to the bacterial surface (11) or via naturally occurring IgM antibodies bound to the bacterial surface (10). Similarly, naturally occurring mannan-specific human IgG antibodies can activate the classical complement pathway and induce C3 deposition on C. albicans (57). Since the allelic differences induced significant amino acid changes in C1 components, we compared C1 functional activities in sera obtained from 5 strains by measuring C1q binding to Candida albicans. In our initial experiment, the amount of C1q bound was >10-fold higher in SJL/J serum than in CBA/J serum (Fig. 3 A and B). Furthermore, the sera of all 3 strains with C1s haplotype 1 (C57BL/6, 129S1/SvImJ, and CBA/J) had low levels of C1q binding activity, while both strains (SJL/J and LG/J) with C1s haplotype 2 had high levels of C1q binding (Fig. 3B and C), and this strain-specific difference was reproducible in multiple independent experiments. Although LG/J and SJL/J sera had higher C1q binding activities, they had lower levels of anti-Candida antibody than CBA/J sera (Fig. 3D). Thus, the interstrain differences in the level of C1q binding to C. albicans were independent of the amount of naturally occurring anti-Candida antibody but were correlated with the C1 haplotype and with survival after C. albicans infection.
Fig. 3.
Candida albicans cells (1 × 107/ml) were incubated for 1 h at 1°C in 0.05 ml calcium-sufficient complement activation buffer containing the indicated amounts of mouse serum and then washed. Cell-bound C1q was detected using polyclonal rabbit anti-mouse C1q antibody and FITC–goat anti-rabbit antibody. C1q binding was either visualized by immunofluorescence microscopy (A) or quantified by flow cytometry (B and C), where ∼5,000 cells were analyzed and a positive cell was defined as having a fluorescence intensity of ≥10. Data for two independent experiments using dilutions of SJL/J and CBA/J sera are shown in panel B, while the averages ± standard deviations (SD) for 3 to 5 independent measurements for the indicated strain and serum concentrations are shown in panel C. (D) Amounts of naturally occurring anti-Candida antibody in sera obtained from five inbred strains. After the indicated dilution of serum was incubated with Candida albicans cells, the amount of bound antibody was measured by flow cytometry. Candida cells with ≥10 fluorescence units were scored as positive cells. Each data point indicates the average % positive cells for three independent measurements ± SD, and data from two different experiments are shown.
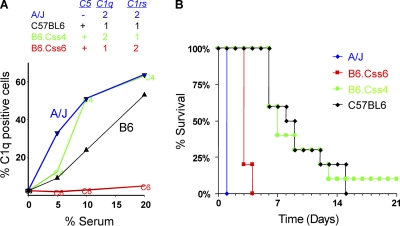
C57BL/6 and A/J mice have different C1q and C1r/s alleles. Therefore, C1q binding to C. albicans was measured using sera obtained from two chromosome substitution strains (CSS) that were derived from these 2 strains and had selectively altered C1q (chromosome 4) or C1rs (chromosome 6) alleles. Each CSS strain is homosomic for a single specified A/J chromosome on an otherwise C57BL/6 (C5-sufficient) genetic background (35). At all serum dilutions tested, A/J and B6.CSS4 (both C1q haplotype 2) sera had higher levels of C1q binding than C57BL/6 (C1q haplotype 1) sera (Fig. 4 A). The level of C1q binding in B6.CSS6 (C1rs haplotype 2 and C1q haplotype 1) sera was substantially lower than that in C57BL/6 sera. The C1qa-C1qb-C1qc and C1r-C1s proteins have different haplotypes in B6.CSS6 mice, and all 4 nonsynonymous C1s SNPs are located within the coding sequence for the Ca2+ binding region that mediates the interaction of C1s with C1r and C1q (5, 18). Thus, the interaction between C1 components with different haplotypes likely reduces the formation or stability of the C1 complex that is bound to the fungus, thereby decreasing C1q binding.
Fig. 4.
(A) C1q binding to C. albicans was quantified by flow cytometry as described in the legend to Fig. 3, using dilutions of sera obtained from C57BL/6, A/J, or the indicated CSS mice. The C5, C1q, and C1rs alleles for each strain are indicated. (B) Survival after hematogenous C. albicans infection for A/J, C57BL/6, or the indicated CSS strain mice (10 mice per group).
We also examined the survival of CSS mice after disseminated C. albicans infection. Consistent with their reduced level of serum C1q binding, B6.CSS6 mice exhibited remarkably poor survival after disseminated C. albicans infection, which was substantially shorter than that of C57BL/6 mice (Fig. 4B); the log rank test P value was 4 × 10−6 when their survival curves were compared. In contrast, the survival of B6.CSS4 mice was comparable to that of C57BL/6 mice. The CSS results and the other data indicate that C1rs alleles have a major effect on C1q binding to the fungal pathogen and on survival after C. albicans infection, which appears to also depend upon the C1q and C5 alleles.
A combinatorial genetic model predicts survival.
These results led us to propose a conditional, multiallelic model (involving 2 complement genes) for predicting inbred strain survival after hematogenous C. albicans infection. Four considerations formed the basis for this genetic model: (i) because complement pathway activation is essential for controlling fungal infection, the effect of a C5 deficiency will predominate over allelic differences within C1 components; (ii) among the C5-sufficient strains, allelic differences within C1 components that affect complement binding to the pathogen and the efficiency of classical complement pathway activation could alter host survival after infection; (iii) C1s alleles caused the largest number of (and the most significant) amino acid changes; and (iv) Although C1q alleles had the highest level of correlation with survival among all analyzed strains, C1s alleles exhibited the highest correlation with survival among the seven strains that were C5 sufficient. According to this model, all C5-deficient strains will have poor survival (≤3 days) after infection, while C5-sufficient strains will have a longer survival (>3 days) that is dependent upon the C1s haplotype, as indicated by the nonsynonymous amino acid changes shown in Fig. 2. According to this model, C5-sufficient strains with C1s haplotype 1 will have a median survival time of 4 ± 1 days after infection, while those with C1s haplotype 2 will have a longer median survival time (9 ± 3 days) (Fig. 2). In brief, this model predicts that genetic variation within the C1s and C5 genes will determine whether a strain will have short, medium, or long survival after C. albicans infection. This model was tested by measuring the survival after hematogenous C. albicans infection of four C5-sufficient strains whose C1s haplotypes were unknown. The C1s alleles for these 4 strains and for LG/J mice (for which we had survival but not allelic data) were then determined. The C1s and C5 allele-based conditional genetic model accurately predicted the measured survival for all 5 strains (Table 1). Note that the C1q haplotype was the same as the C1s haplotype in these strains (i.e., all C1q haplotype 2 strains were also C1s haplotype 2). Although the C1q haplotype could have an independent effect on survival after infection, its impact will have to be analyzed using recombinant strains with targeted substitutions in both C1q and C1rs alleles on a C5-sufficient genetic background.
Table 1.
Comparison of measured median survival rates after C. albicans infection among 5 strains (10 mice per strain) relative to those predicted using the C5 and C1s composite genetic modela
| Strain | C1s haplotype | Median (±SD) survival (days) |
|
|---|---|---|---|
| Predicted | Measured | ||
| SM/J | 1 | 4 + 1 | 3 |
| MRL | 2 | 9 ± 3 | 8 |
| DBA/1 | 2 | 9 ± 3 | 6 |
| SJL/J | 2 | 9 ± 3 | 8 |
| LG/J | 2 | 9 ± 3 | 12 |
All strains were positive for the C5 allele.
DISCUSSION
A next-generation computational mapping program analyzed a murine genetic model of survival after hematogenous Candida albicans infection and indicated that genetic variation within early classical complement pathway components (C1q, C1r, and C1s) could affect survival. Abundant experimental data supported the genetic hypothesis that C1 alleles affect survival: C1 binding measurements provided a direct demonstration that C1 alleles affect C1 binding to the fungal pathogen, C1 (and C5) alleles could prospectively predict strain survival after infection, and C1 allelic substitutions in consomic strains had a major effect on the level of C1 binding to the fungal pathogen and on survival after infection. These data indicate that C1 alleles are at least some of the genetic factors that were postulated to affect survival but could not be identified using the available genetic discovery methods (2).
Although we have shown the combinatorial effect that C1 and C5 alleles have on survival in this model, their effect on the response to other infectious agents remains to be determined. It is also possible that other genetic factors, especially in different genetic backgrounds or with different phenotypes, could also impact the outcome after C. albicans infection. For example, a genetic analysis was performed using kidney fungal burden as an outcome after C. albicans infection, utilizing C5-deficient mouse strains, and this revealed that other genetic loci could affect the outcome (38). However, given the important role of the complement pathway in the response to multiple infectious agents, these findings should stimulate others to investigate whether this genetic mechanism impacts the response to other infectious agents. We do not yet know if a similar genetic mechanism involving the complement pathway will be found in humans. However, if an allelic effect is identified, genotyping at-risk individuals could identify those that would best benefit from increased monitoring or preventative therapy.
Beyond its potential applicability to other infectious diseases, this combinatorial genetic model could provide insight into the genetic architecture of susceptibility to autoimmune and neurodegenerative diseases. The relationship between complement alleles and autoimmune disease susceptibility in humans and mice has been puzzling. While kidney inflammation in human systemic lupus erythematosus and related murine models is driven by immune complex deposition and complement activation, paradoxically, C1q-deficient humans have the highest risk for lupus susceptibility (7, 31). Similarly, a C1q knockout can accelerate the development of renal disease in mice with certain genetic backgrounds, but its effect on autoimmune disease expression is highly strain dependent (6, 33). Investigators have partially explained these paradoxical observations by proposing that early complement proteins play multiple roles in autoimmune disease pathogenesis (reviewed in reference 31). Besides driving complement-dependent tissue inflammation, C1q is also a pattern recognition protein that facilitates the clearance of apoptotic cells (6, 15, 16, 33), and a deficiency in this activity could facilitate the development of autoimmunity.
The combinatorial allelic model described here provides a new mechanism for modulating complement pathway activity, which could explain the significant effect that genetic background has on the autoimmune phenotype in a C1q knockout mouse (6, 33). Also, the MRL and NZW strains provide prototypic models for human lupus, and both have C5 and C1 alleles that favor efficient classical complement pathway activation, which may partially explain why they spontaneously develop autoimmune disease (39, 39a). Despite intensive investigation, we do not fully understand the genetic basis for the renal disease that spontaneously develops in F1(NZB × NZW) mice but not in either parent (60). We previously demonstrated that the NZB Ifi202 allele promotes autoantibody production. However, congenic C57BL/6 mice expressing the NZB Ifi202 allele (B6.NZBIfi202) produce multiple autoantibodies but do not develop renal disease, while NZW mice expressing the NZB Ifi202 allele develop renal pathology at the same rate as F1(NZB × NZW) mice (41). Similarly, the lpr mutation promotes autoimmunity in MRL/lpr mice, and C57BL/6 mice expressing the lpr mutation develop high autoantibody titers but do not develop renal disease (53). This suggests that the complement alleles in NZW or MRL mice could be required for autoimmune disease expression. It was also demonstrated recently that early classical complement pathway components (C1q and C3) regulate synapse formation within the central nervous system and retina, that C1q binding “tags” selected synapses for elimination during development, and that C1q is an essential mediator of neurodegeneration in a murine glaucoma model mediated by retina-specific genetic factors (44). Polymorphisms in human C1q, C1r, and C1s are known to exist, and allelic associations with systemic lupus erythematosis and early Alzheimer's disease have been investigated (25, 32, 37, 40). Whether these polymorphisms influence the susceptibility of hospitalized patients to disseminated candidiasis or the outcome of this disease remains to be determined. Nevertheless, characterizing the combinatorial interaction between allelic variants in different complement proteins and with other genetic factors required for autoimmune disease expression (major histocompatibility complex [MHC], Ifi202, and Fas/lpr) in mice could increase our understanding of susceptibility to autoimmune and neurodegenerative diseases as well as disseminated candidiasis.
Supplementary Material
ACKNOWLEDGMENTS
G.P. was supported by funding from a transformative RO1 award (1R01DK090992-01) from the NIDDK, and S.G.F. was supported in part by NIH grants 5R21DE019414 and 5R01AI054928.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Ashman R. B., Bolitho E. M., Papadimitriou J. M. 1993. Patterns of resistance to Candida albicans in inbred mouse strains. Immunol. Cell Biol. 71:221–225 [DOI] [PubMed] [Google Scholar]
- 2. Ashman R. B., Fulurija A., Papadimitriou J. M. 1997. Evidence that two independent host genes influence the severity of tissue damage and susceptibility to acute pyelonephritis in murine systemic candidiasis. Microb. Pathog. 22:187–192 [DOI] [PubMed] [Google Scholar]
- 3. Ashman R. B., Fulurija A., Papadimitriou J. M. 1996. Strain-dependent differences in host response to Candida albicans infection in mice are related to organ susceptibility and infectious load. Infect. Immun. 64:1866–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashman R. B., et al. 2003. Role of complement C5 and T lymphocytes in pathogenesis of disseminated and mucosal candidiasis in susceptible DBA/2 mice. Microb. Pathog. 34:103–113 [DOI] [PubMed] [Google Scholar]
- 5. Bally I., et al. 2009. Identification of the C1q-binding sites of human C1r and C1s: a refined three-dimensional model of the C1 complex of complement. J. Biol. Chem. 284:19340–19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Botto M., et al. 1998. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 19:56–59 [DOI] [PubMed] [Google Scholar]
- 7. Botto M., et al. 2009. Complement in human diseases: lessons from complement deficiencies. Mol. Immunol. 46:2774–2783 [DOI] [PubMed] [Google Scholar]
- 8. Boxx G. M., Kozel T. R., Nishiya C. T., Zhang M. X. 2010. Influence of mannan and glucan on complement activation and C3 binding by Candida albicans. Infect. Immun. 78:1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boxx G. M., Nishiya C. T., Kozel T. R., Zhang M. X. 2009. Characteristics of Fc-independent human antimannan antibody-mediated alternative pathway initiation of C3 deposition to Candida albicans. Mol. Immunol. 46:473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown J. S., et al. 2002. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc. Natl. Acad. Sci. U. S. A. 99:16969–16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Butko P., Nicholson-Weller A., Wessels M. R. 1999. Role of complement component C1q in the IgG-independent opsonophagocytosis of group B streptococcus. J. Immunol. 163:2761–2768 [PubMed] [Google Scholar]
- 12. Chu L. F., et al. 2009. From mouse to man: the 5-HT3 receptor modulates physical dependence on opioid narcotics. Pharmacogenet. Genomics 19:193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ejzykowicz D. E., et al. 2010. Role of Aspergillus fumigatus DvrA in host cell interactions and virulence. Eukaryot. Cell 9:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferwerda B., et al. 2009. Human dectin-1 deficiency and mucocutaneous fungal infections. N. Engl. J. Med. 361:1760–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fraser D. A., Laust A. K., Nelson E. L., Tenner A. J. 2009. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J. Immunol. 183:6175–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fraser D. A., Pisalyaput K., Tenner A. J. 2010. C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J. Neurochem. 112:733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glocker E. O., et al. 2009. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med. 361:1727–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gregory L. A., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. 2003. X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement. J. Biol. Chem. 278:32157–32164 [DOI] [PubMed] [Google Scholar]
- 19. Guo Y. Y., et al. 2007. In vitro and in silico pharmacogenetic analysis in mice. Proc. Natl. Acad. Sci. U. S. A. 104:17735–17740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo Y. Y., et al. 2006. In silico pharmacogenetics: warfarin metabolism. Nat. Biotechnol. 24:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harrington D. P., Flemming T. R. 1982. A class of rank test procedures for censored survival data. Biometrika 69:553–566 [Google Scholar]
- 22. Hector R. F., Domer J. E., Carrow E. W. 1982. Immune responses to Candida albicans in genetically distinct mice. Infect. Immun. 38:1020–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu Y., et al. 2010. The role of IL-1 in wound biology. I. Murine in silico and in vitro experimental analysis. Anesth. Analg. 111:1525–1533 [DOI] [PubMed] [Google Scholar]
- 24. Hu Y., et al. 2010. The role of IL-1 in wound biology. II. In vivo and human translational studies. Anesth. Analg. 111:1534–1542 [DOI] [PubMed] [Google Scholar]
- 25. Kamboh M. I., Ferrell R. E. 1987. Genetic studies of low-abundance human plasma proteins. VII. Heterogeneity of the C1S subcomponent of the first complement component. J. Immunogenet. 14:231–238 [DOI] [PubMed] [Google Scholar]
- 26. LaCroix-Fralish M. L., et al. 2009. The β3 subunit of the Na+,K+-ATPase affects pain sensitivity. Pain 144:294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li X., et al. 2010. Expression genetics identifies spinal mechanisms supporting formalin late phase behaviors. Mol. Pain 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang D., et al. 2006. A genetic analysis of opioid-induced hyperalgesia in mice. Anesthesiology 104:1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang D. Y., Liao G., Lighthall G., Peltz G., Clark J. D. 2006. Genetic variants of the P-glycoprotein gene Abcb1b modulate opioid-induced hyperalgesia, tolerance and dependence. Pharmacogenet. Genomics 16:825–835 [DOI] [PubMed] [Google Scholar]
- 30. Liao G., et al. 2004. In silico genetics: identification of a functional element regulating H2-Ea gene expression. Science 306:690–695 [DOI] [PubMed] [Google Scholar]
- 31. Manderson A. P., Botto M., Walport M. J. 2004. The role of complement in the development of systemic lupus erythematosus. Annu. Rev. Immunol. 22:431–456 [DOI] [PubMed] [Google Scholar]
- 32. Martens H. A., et al. 2009. Analysis of C1q polymorphisms suggests association with systemic lupus erythematosus, serum C1q and CH50 levels and disease severity. Ann. Rheum. Dis. 68:715–720 [DOI] [PubMed] [Google Scholar]
- 33. Mitchell D. A., et al. 2002. C1q deficiency and autoimmunity: the effects of genetic background on disease expression. J. Immunol. 168:2538–2543 [DOI] [PubMed] [Google Scholar]
- 34. Mullick A., et al. 2004. Dysregulated inflammatory response to Candida albicans in a C5-deficient mouse strain. Infect. Immun. 72:5868–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nadeau J. H., Singer J. B., Matin A., Lander E. S. 2000. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 24:221–225 [DOI] [PubMed] [Google Scholar]
- 35a. National Research Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 36. Puel A., et al. 2011. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332:65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Racila E., et al. 2008. A polymorphism in the complement component C1qA correlates with prolonged response following rituximab therapy of follicular lymphoma. Clin. Cancer Res. 14:6697–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radovanovic I., Mullick A., Gros P. 2011. Genetic control of susceptibility to infection with Candida albicans in mice. PLoS One 6:e18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reilly C. M., Gilkeson G. S. 2002. Use of genetic knockouts to modulate disease expression in a murine model of lupus, MRL/lpr mice. Immunol. Res. 25:143–153 [DOI] [PubMed] [Google Scholar]
- 39a. Rigby R. J., Fernando M. M., Vyse T. J. 2006. Mice, humans and haplotypes—the hunt for disease genes in SLE. Rheumatology 45:1062–1067 [DOI] [PubMed] [Google Scholar]
- 40. Rosenmann H., et al. 2003. A polymorphism in the complement component C1r is not associated with sporadic Alzheimer's disease. Neurosci. Lett. 336:101–104 [DOI] [PubMed] [Google Scholar]
- 41. Rozzo S. J., et al. 2001. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity 15:435–443 [DOI] [PubMed] [Google Scholar]
- 42. Shepherd M. G., Poulter R. T., Sullivan P. A. 1985. Candida albicans: biology, genetics, and pathogenicity. Annu. Rev. Microbiol. 39:579–614 [DOI] [PubMed] [Google Scholar]
- 43. Smith S. B., et al. 2008. Quantitative trait locus and computational mapping identifies Kcnj9 (GIRK3) as a candidate gene affecting analgesia from multiple drug classes. Pharmacogenet. Genomics 18:231–241 [DOI] [PubMed] [Google Scholar]
- 44. Stevens B., et al. 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131:1164–1178 [DOI] [PubMed] [Google Scholar]
- 45. Thiel S. 2007. Complement activating soluble pattern recognition molecules with collagen-like regions, mannan-binding lectin, ficolins and associated proteins. Mol. Immunol. 44:3875–3888 [DOI] [PubMed] [Google Scholar]
- 46. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tregoning J. S., et al. 2010. Genetic susceptibility to the delayed sequelae of RSV infection is MHC-dependent, but modified by other genetic loci. J. Immunol. 185:5384–5391 [DOI] [PubMed] [Google Scholar]
- 48. van de Veerdonk F. L., et al. 2011. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N. Engl. J. Med. 365:54–61 [DOI] [PubMed] [Google Scholar]
- 49. Walport M. J. 2001. Complement. First of two parts. N. Engl. J. Med. 344:1058–1066 [DOI] [PubMed] [Google Scholar]
- 50. Walport M. J. 2001. Complement. Second of two parts. N. Engl. J. Med. 344:1140–1144 [DOI] [PubMed] [Google Scholar]
- 51. Wang J., Peltz G. Haplotype-based computational genetic analysis in mice, p. 51–70 Computational genetics and genomics: new tools for understanding disease. Humana Press Inc., Totowa, NJ [Google Scholar]
- 52. Waterston R. H., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562 [DOI] [PubMed] [Google Scholar]
- 53. Watson M. L., et al. 1992. Genetic analysis of MRL-lpr mice: relationship of the Fas apoptosis gene to disease manifestations and renal disease-modifying loci. J. Exp. Med. 176:1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wetsel R. A., Fleischer D. T., Haviland D. L. 1990. Deficiency of the murine fifth complement component (C5). A 2-base pair gene deletion in a 5′-exon. J. Biol. Chem. 265:2435–2440 [PubMed] [Google Scholar]
- 55. Winkelstein J. A. 1984. Complement and the host's defense against the pneumococcus. Crit. Rev. Microbiol. 11:187–208 [DOI] [PubMed] [Google Scholar]
- 56. Zaas A. K., et al. 2008. Plasminogen alleles influence susceptibility to invasive aspergillosis. PLoS Genet. 4:e1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang M. X., Lupan D. M., Kozel T. R. 1997. Mannan-specific immunoglobulin G antibodies in normal human serum mediate classical pathway initiation of C3 binding to Candida albicans. Infect. Immun. 65:3822–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X., et al. 2011. In silico and in vitro pharmacogenetics: aldehyde oxidase rapidly metabolizes a p38 kinase inhibitor. Pharmacogenomics J. 11:15–24 [DOI] [PubMed] [Google Scholar]
- 59. Zheng M., Shafer S. S., Liao G., Liu H.-H., Peltz G. 2009. Computational genetic mapping in mice: ‘the ship has sailed.’ Sci. Transl. Med. 1:3ps4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.