Abstract
Ehrlichia chaffeensis is an obligately intracellular bacterium that modulates host cell gene transcription in the mononuclear phagocyte, but the host gene targets and mechanisms involved in transcriptional modulation are not well-defined. In this study, we identified a novel tandem repeat DNA-binding domain in the E. chaffeensis 120-kDa tandem repeat protein (TRP120) that directly binds host cell DNA. TRP120 was observed by immunofluorescent microscopy in the nucleus of E. chaffeensis-infected host cells and was detected in nuclear extracts by Western immunoblotting with TRP120-specific antibody. The TRP120 binding sites and associated host cell target genes were identified using high-throughput deep sequencing (Illumina) of immunoprecipitated DNA (chromatin immunoprecipitation and high-throughput DNA sequencing). Multiple em motif elicitation (MEME) analysis of the most highly enriched TRP120-bound sequences revealed a G+C-rich DNA motif, and recombinant TRP120 specifically bound synthetic oligonucleotides containing the motif. TRP120 target gene binding sites were mapped most frequently to intersecting regions (intron/exon; 49%) but were also identified in upstream regulatory regions (25%) and downstream locations (26%). Genes targeted by TRP120 were most frequently associated with transcriptional regulation, signal transduction, and apoptosis. TRP120 targeted inflammatory chemokine genes, CCL2, CCL20, and CXCL11, which were strongly upregulated during E. chaffeensis infection and were also upregulated by direct transfection with recombinant TRP120. This study reveals that TRP120 is a novel DNA-binding protein that is involved in a host gene transcriptional regulation strategy.
INTRODUCTION
Ehrlichia chaffeensis is a Gram-negative obligately intracellular bacterium that resides in mononuclear phagocytes by subverting innate and adaptive host defense mechanisms (49). During E. chaffeensis infection, changes in transcription of host genes involved in early innate and cell-mediated immune responses, apoptosis regulation, membrane trafficking, signal transduction, and cell differentiation occur (67), but the molecular mechanisms involved have been largely undefined. Recently, we demonstrated that an E. chaffeensis nuclear translocated protein, Ank200, interacts with Alu elements associated with genes that regulate transcription, ATPase activity, and apoptosis, identifying a mechanism that involves Ank200 in a pathogen-host gene regulation strategy (69). Furthermore, we demonstrated that the E. chaffeensis 47-kDa tandem repeat (TR) protein (TRP47) interacts with the polycomb group ring finger protein-5 (PCGF5), a component of the polycomb repressive complex (PRC), suggesting that Ehrlichia may utilize a variety of mechanisms to modulate host cell gene transcription (60).
E. chaffeensis has two small groups of proteins associated with molecular host-pathogen interactions that contain tandem and ankyrin repeat domains (41, 61). Many of these proteins are major immunoreactive proteins, including E. chaffeensis TRP120, TRP47, TRP32, and Ank200. The TRPs contain long-period TR domains that are acidic, exhibit a high serine/threonine content, and have species-specific continuous antibody epitopes within the TRs (13, 35, 36). TRP120 and TRP47 are differentially expressed on dense-cored ehrlichiae, and both have been associated with defined molecular pathogen-host interactions (13, 47). TRP47 interacts with numerous host cell proteins and appears to be involved in modulation of host cell processes, including transcriptional regulation, signal transduction, and intracellular vesicle trafficking (60), and TRP120 is involved in binding and internalization, and its expression is regulated by second messenger cyclic di-GMP and protease HtrA (28, 47).
In human bacterial pathogens, TRs have been associated with pathogen-host interactions, including immune evasion (Mycoplasma, Streptococcus, and Neisseria) (9, 17, 58), actin nucleation (Chlamydia) (10), host adaptation (Helicobacter) (54), and binding and internalization (Ehrlichia and Anaplasma) (23, 28, 47). In Ehrlichia, tandem repeats are present in noncoding genomic regions as well as in regions coding for proteins and are associated with expansion and contraction of the genome (11). Moreover, a transcription activator-like effector family of TRPs has been identified in bacterial plant pathogens from the genera Xanthomonas and Ralstonia (26, 43). The type member of the TAL family, AvrBs3, is secreted by the type III secretion system and localizes to the plant cell nucleus, inducing expression of a subset of host genes that promote disease (26). AvrBs3 has a variable number of imperfect 34-amino-acid tandem repeats that bind to a UPA (upregulated by AvrBs3) DNA box that contains cytosine patches crucial for recognition of AvrBs3 (26). AvrBs3 binding of the UPA box activates host cell transcription by mimicking eukaryotic transcription factors (53).
In this study, we determined that E. chaffeensis TRP120 directly binds host cell DNA through the TR domain targeting a G+C-rich DNA motif. A large number of potential TRP120 binding sites were identified using a combination of chromatin immunoprecipitation and high-throughput DNA sequencing (ChIPSeq), and the majority of binding sites were mapped to intersecting regions of target genes primarily related to transcriptional regulation, signal transduction, and apoptosis. TRP120 was detected in the host cell nucleus and directly activated inflammatory chemokine genes. These findings demonstrate that TRP120 is involved in a molecular strategy to reprogram host cell gene transcription.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells. Uninfected and Ehrlichia-infected THP-1 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS; HyClone, Logan, UT), 1% HEPES, 4 mM l-glutamine, and 1% sodium pyruvate at 37°C in a 5% CO2 atmosphere. Ehrlichial growth was monitored for the presence of morulae by Diff-Quik staining.
Antibodies.
Anti-E. chaffeensis TRP120 was generated against synthetic keyhole limpet hemocyanin-conjugated peptide (SKVEQEETNPEVLIKDLQDVAS), which represents the major continuous epitope found in each respective TR unit, as previously described (35). Anti-E. chaffeensis Dsb antibody was produced in rabbits as previously described (40).
Expression and purification of recombinant Ehrlichia TRP120s.
The gene encoding TRP120 was amplified by PCR using Ehrlichia genomic DNA (gDNA) as a template. The amplified PCR products were cloned directly into the pBAD/Thio-TOPO expression vector (Invitrogen, Carlsbad, CA) as previously described (35). Recombinant TRP120 was expressed and purified at the University of Texas Medical Branch Protein Chemistry Core Laboratory. The recombinant TR domain (TRP120-TR), which represents the first two tandem repeats of E. chaffeensis TRP120, was expressed as a glutathione S-transferase (GST) fusion protein and purified as previously described (64, 65).
Nuclear extraction.
The cytoplasmic and nuclear fractions were extracted from E. chaffeensis-infected THP-1 cells using a nuclear extraction kit (Panomics, Fremont, CA) as previously described (69). Extractions were performed according to the manufacturer's protocol, and the samples of cells were collected at 3, 6, 12, 24, 48, and 72 h postinfection.
Immunofluorescent microscopy.
TRP120 nuclear translocation was determined as previously described (69), except that a rabbit anti-TRP120 (1:1,000) was used and the slides were viewed with an Olympus Fluoview 1000MPE system configured with an upright BX61 microscope. The fluorescent intensity of nuclei was determined with a random selection of 20 nuclei from infected and uninfected cells reacted with E. chaffeensis TRP120 or Dsb antibodies as previously described (69).
Western immunoblotting.
Approximately 10 μg of total cytoplasmic and nuclear extract was separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane using a semidry transfer apparatus, as previously described (69). Western immunoblotting was performed using rabbit anti-E. chaffeensis TRP120 (1:1,000) or anti-E. chaffeensis Dsb (1:100). Bound antibodies were detected by incubation with phosphatase-labeled goat anti-rabbit IgG (H+L; 1:1,000; Kirkegaard & Perry Laboratories, Gaithersburg, MD) and visualized after incubation with 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium (BCIP/NBT) substrate (Kirkegaard & Perry Laboratories).
ChIPSeq.
Chromatin immunoprecipitation (ChIP) was performed on highly infected (95%) THP-1 cells 4 days after infection using a ChIP-IT Express kit (Active Motif, Carlsbad, CA) with anti-E. chaffeensis TRP120 antibody or preimmune serum from the same rabbit according to the manufacturer's instructions. To prepare samples for ChIPSeq, the immunoprecipitated DNA was isolated and DNA fragments (200 ± 25 bp) were gel purified by Ambry Genetics (Aliso Viejo, CA). Approximately 10 ng of TRP120 and control ChIP DNA was sequenced by high-throughput DNA sequencing according to the library preparation protocol from Illumina using a Solexa/Illumina genome analyzer (Ambry Genetics).
ChIPSeq data analysis.
All sequence reads produced by the Illumina genome analyzer were analyzed by base calling and sequence quality filtering scripts using the Illumina Pipeline software (version 1.4.0; Illumina, Hayward, CA). Alignments were generated with efficient local alignment of nucleotide data (Eland) using the human genome (NCBI build 37), allowing a maximum of two mismatches to the reference genome. The sequence reads were further analyzed by the ArrayStar program (version 4.0; DNAStar, Madison, WI), and BED and wiggle (WIG) files were created and visualized using the University of California at Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/index.html).
TRP120 binding sites.
TRP120 binding sites were identified by comparing TRP120 and control ChIP samples and the identification of peaks, which were regions that were significantly more enriched in TRP120 samples. Peak detection was performed by running model-based analysis of ChIPSeq (MACS) (68) within ArrayStar (parameters were mfold [second structure] of 10, bandwidth of 300 bp, and a P-value threshold of 1 × 10−5).
TRP120 DNA motif analysis.
The DNA-binding motif was identified using the multiple em motif elicitation (MEME; version 4.3.0) software package (4). Sequences from a group of the highest peak ratios (top 10%) were used for motif analysis and identification. Potential TRP120 binding motifs were predicted on the basis of the highest number of occurrences with the lowest P value under the zero- or one-per-sequence option. Position-dependent letter-probability matrices (PWMs) generated by MEME were represented in the logo format by using the WebLogo application. The PWM was used to determine a score for any 6- to 20-bp sequence, and all motifs with a P value of <1 × 10−5 were examined (18).
Gene annotation and ontology.
All TRP120 binding sites were assigned to the nearest gene on the basis of the human NCBI build 37 genome assembly (GRCh37/hg19; February 2009) by ArrayStar (DNAStar). ArrayStar identifies upstream and downstream genes as those within 100 kb of the DNA fragment and intersecting genes as those that intersect the DNA fragment. Gene ontology (GO) analysis of TRP120 target genes was performed by using the Babelomics FatiGO+ tool (1, 2). This analysis was used to classify the target gene list into functionally related gene groups using biological process, molecular function, and cell structure terms.
Validation of TRP120 binding peaks by qPCR.
Quantitative real-time PCR (qPCR) was performed on TRP120 ChIP-enriched DNA, and the results were compared to those for ChIP DNA obtained with preimmune serum to normalize the enrichment levels. Primer pairs were designed to amplify regions (∼150 to 200 bp) within the binding site identified by ChIPSeq. Amplification of all genes was performed, using an initial denaturing step at 94°C for 2 min, followed by 40 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s, in triplicate with iQ SYBR green supermix (Bio-Rad Laboratories, Hercules, CA) in a Mastercycler ep realplex2 S apparatus (Eppendorf, Hamburg, Germany). A dissociation curve analysis was performed to verify amplicon identity. Relative enrichment for each gene was determined by the comparative threshold cycle (CT) method as previously described (33).
EMSA.
The genomic DNA was extracted and purified from THP-1 cells using a ChIP-IT Express kit (Active Motif). The purified DNA was digested with the restriction endonuclease CviJ I* (Chimerx) to create fragments of <1 kb. The digested DNA fragments were directly labeled with biotin using a Mirus Label IT μArray biotin labeling kit. Sequences of the oligonucleotide probes (P1, P2, and P3) used for electrophoretic mobility shift assays (EMSAs), corresponding to the conserved putative TRP120 motif, are underlined and in boldface in Table 1 and were biotinylated or unlabeled. P1 contained a consensus sequence [5′-CCT(G/C)CC-3′] in a repeat orientation, P2 contained a single consensus sequence motif (5′-CCTCCC-3′), and P3 did not contain the identified motif. All these probes were resuspended in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), and 10 μg each of sense and antisense probe was added to 100 μl 0.5× SSC (75 mM NaCl plus 7.5 mM sodium citrate, pH 7.0). The probes were denatured at 100°C for 10 min and annealed by slowly cooling them to room temperature. EMSA reactions were performed using a LightShift chemiluminescent EMSA kit (Pierce), following the manufacturer's instructions. Reactions were carried out in 1× LightShift binding buffer [10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 2.5% glycerol, 0.1% NP-40, 50 ng/μl of poly(dI-dC) or salmon sperm DNA] for 30 min on ice. Approximately 5 ng of biotinylated gDNA and 20 fmol of DNA probe were incubated with TRP120-TR and TRP120, respectively. Unlabeled gDNA and probes (P1 and P2) were used as competitors for competition experiments. The supershift assay was performed by incubating anti-TRP120 antibody for 30 min at 4°C. The probe-protein mixture was separated on a 6% DNA retardation gel at 100 V for 1.5 h at 4°C. The DNA was transferred to a precut modified nylon membrane (Biodyne B; Pierce, Rockford, IL) at 20 V for 1 h and cross-linked with UV light for 10 min. The membrane was then blocked, washed, and exposed to X-ray film following the manufacturer's protocol.
Table 1.
Probes used for TRP120 EMSA
| Probe | Sequence |
|---|---|
| P1 | GCGATTCTCCTGCCTCAGCCTCCCTAG |
| CTAGGGAGGCTGAGGCAGGAGAATCGC | |
| P2 | CTCCAGCCTCTGCCTCCCAGGTT |
| AACCTGGGAGGCAGAGGCTGGAG | |
| P3 | GTCACAAATTCTCATCTTTATATAAAGAT |
| ATCTTTATATAAAGATGAGAATTTGTGAC |
Direct protein transfection.
TRP120 and thioredoxin control recombinant proteins were transfected directly into THP-1 cells using the Chariot reagent (Active Motif). Protein (1 μg) was mixed with 6 μl of Chariot reagent in 200 μl water, and the mixture was incubated for 30 min. THP-1 cells were incubated at 37°C until the cells were 40 to 50% confluent. The cells were washed with phosphate-buffered saline and incubated with the Chariot-peptide complex in serum-free medium at 37°C for 4 and 72 h.
Host gene expression levels.
Total RNA was extracted from E. chaffeensis-infected THP-1 and uninfected cells, consisting of TRP120- and thioredoxin control recombinant protein-transfected THP-1 cells (3 × 106 cells/well), respectively, using an RNeasy total RNA purification kit according to the manufacturer's instructions (Qiagen). Purified total RNA was treated with DNase I (Applied Biosystems/Ambion) to remove contaminating genomic DNA, and cDNA was synthesized from 1 μg of total RNA using an iScript cDNA synthesis kit with oligo(dT) and random primers (Bio-Rad Laboratories). Transcript levels of selected host genes were quantitated by qPCR using iQ SYBR green supermix (Bio-Rad Laboratories) with gene-specific primers (Table 2) and a thermal cycling protocol consisting of an initial denaturation step of 95°C for 2 min and 40 cycles of 95°C for 10 s, 55°C for 30 s, and 65°C for 30 s. Gene expression values were calculated on the basis of the 2−ΔΔCT method and normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Table 2.
Primers for expression analysis of TRP120 target genes
| Gene product | Primer sequence (5′–3′) |
|---|---|
| TNFRSF14 | GCACAGTGTGTGAACCCTG |
| CACTTGCTTAGGCCATTGAGG | |
| Stat1 | ATGTCTCAGTGGTACGAACTTCA |
| TGTGCCAGGTACTGTCTGATT | |
| CD70 | CACTTGGGTGGGACGTAGC |
| CTGGTCCATGCAGGAAGGAG | |
| CCL22 | GCGTGGTGTTGCTAACCTTCA |
| AAGGCCACGGTCATCAGAGT | |
| CCL20 | GTCTGTGTGCGCAAATC |
| TGCAAGTGAAACCTCCAAC | |
| TNF-α | CTCTCTCTAATCAGCCCTCT |
| TATCTCTCAGCTCCACGC | |
| CXCL11 | GACATTATTACTGGAGTCAAGCCCTTA |
| GGAAAGAAGTGTGTATTTGCATGAA | |
| CIAS1 | GTCAGACAGAGAAGGCAG |
| CACAGTGGGATTCGAAAC | |
| CCL2 | From SABiosciences |
RESULTS
TRP120-TR binding to human genomic DNA.
To examine the TRP120-DNA interaction, we used recombinant TRP120-TR, which contains only two tandem repeats. The biotin-labeled host cell genomic DNA fragments were incubated with the recombinant TRP120-TR, and the formation of complexes was analyzed by an EMSA. A band indicating protein-DNA binding was observed with the TRP120-TR–DNA sample but not with the controls, including the recombinant GST (rGST)-DNA sample and labeled gDNA only (Fig. 1A). The specificity of the binding was examined using competition experiments with excess unlabeled gDNA, which resulted in a marked reduction in labeled DNA-protein complex formation (Fig. 1A). To examine whether this complex corresponds to TRP120-TR, we performed a supershift assay using rabbit anti-TRP120-TR antibody. A supershift band was observed using rabbit TRP120-specific antibody (Fig. 1B). The specificity of TRP120 antibody has been previously demonstrated (35).
Fig. 1.
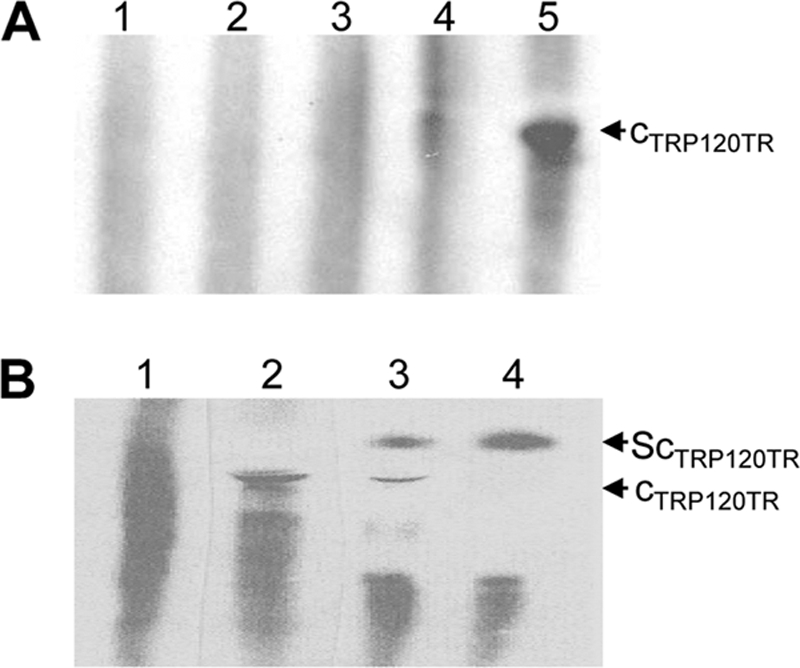
Analysis of recombinant TRP120-TR binding to human gDNA by EMSA. (A) Lane 1, free biotin-labeled gDNA; lane 2, GST protein with biotin-labeled gDNA; lane 3, recombinant TRP120-TR with biotin-labeled gDNA in the presence of a 200-fold molar excess of unlabeled competitor (gDNA); lane 4, recombinant TRP120-TR with biotin-labeled gDNA in the presence of a 100-fold molar excess of unlabeled competitor (gDNA); lane 5, recombinant TRP120-TR with biotin-labeled gDNA in the absence unlabeled competitor (gDNA); (B) lane 1, free biotin-labeled gDNA; lane 2, TRP120-TR with biotin-labeled gDNA; lane 3, TRP120-TR with biotin-labeled gDNA and antibody (1:40); lane 4, TRP120-TR with biotin-labeled gDNA and antibody (1:20). The putative TRP120-TR binding complex (CTRP120TR) and supershift binding complex (ScTRP120TR) are identified.
TRP120 ChIP and identification of DNA-binding site.
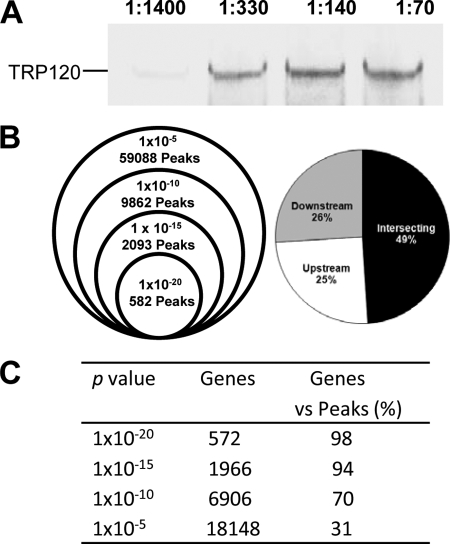
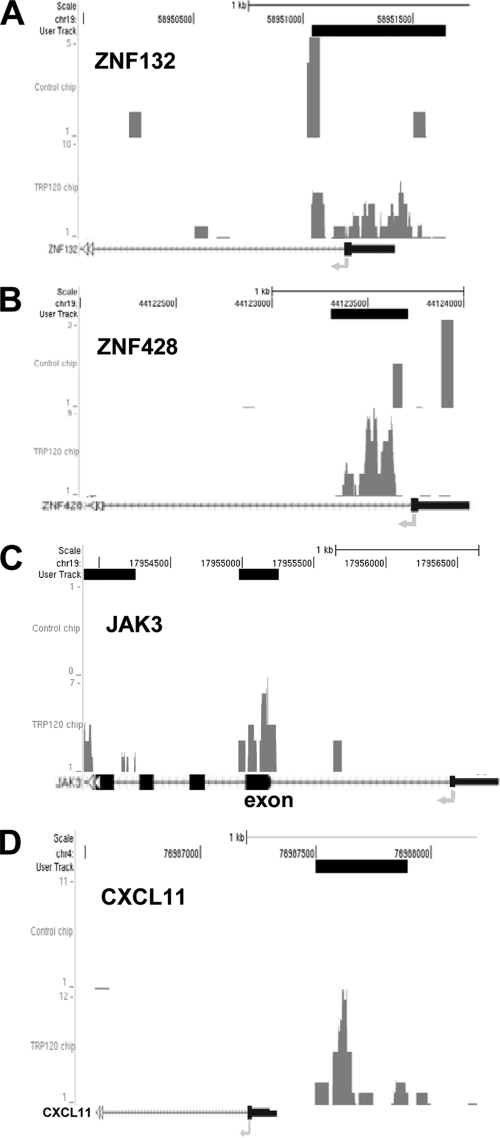
TRP120 was detected in ChIP protein-DNA complexes from E. chaffeensis-infected host cells by Western immunoblotting (Fig. 2A). ChIP and ChIPSeq were used to isolate E. chaffeensis TRP120 target sequences and map TRP120 binding positions. A total of 17 million ChIPSeq sequences were obtained from the Solexa/Illumina genome analyzer for both TRP120 and control antibody (preimmune serum). The high number of reads provided high sensitivity and signal-noise ratios, an even wider dynamic range, and a greater ability to detect rare DNA-protein interaction sites. To determine the peak which was bound by TRP120, we analyzed the sequence reads using the peak recognition algorithm of MACS (68). A range of different P values (10−20, 10−15, 10−10, and 10−5) was applied to identify TRP120 binding sites (Fig. 2B, left). With the highest P value (10−5), 59,088 interactions were identified, whereas 2,093 and 582 were identified with the lowest P values of 10−15 and 10−20, respectively (Fig. 2B, left). The detected peaks were further mapped independently to the human reference genome and normalized across the genome. With the highest P value (10−5), 18,148 TRP120 target genes were identified, whereas 572 were identified with the lowest P value (10−20; Fig. 2C; see Table S1 in the supplemental material). In order to visualize the peaks identified by MACS, the sequence reads of TRP120 ChIP and control ChIP were aligned as tracks onto the human genome using the UCSC Genome Browser (http://genome.ucsc.edu/index.html). Visual inspection of binding sites confirmed that the peaks identified by MACS correspond to the sequence coordinates. The lower-P-value peaks (∼2,000) were mapped to the human reference genome, and approximately ∼2,000 target genes that contain at least one binding site occupied by TRP120 with a P value of 10−15 were identified. A subset of these genes is shown in Table 3. The TRP120 binding regions were distributed in intersecting regions (49%), upstream (25%), and downstream (26%) (Fig. 2B, right). The same distribution was also observed by mapping selected subsets of genes representing other P values (10−5, 10−10, and 10−20). Several examples of TRP120 binding sites visualized as peaks using the UCSC Genome Browser are shown in Fig. 3.
Fig. 2.
ChIPSeq analysis of TRP120-DNA-binding sites. (A) Western immunoblot detection of TRP120 from chromatin immunoprecipitated samples prepared from E. chaffeensis-infected cells using TRP120 epitope-specific antibody. The four lanes contain increasing amounts of antibody, as indicated (1:1,400, 1:330, 1:140, 1:70). (B) Summary of TRP120 interactions displayed as a function of various P value thresholds applied to human whole-genome ChIPSeq and the locations of TRP120 binding peaks relative to the locations of identified genes. (C) Summary of TRP120 gene identification on the basis of peak analysis with various P value thresholds.
Table 3.
A subset of TRP120 binding sites present in the selected ChIPSeq-identified target genes
| Gene product | Peak | Chromosome | Gene identifier | Gene description |
|---|---|---|---|---|
| TNFRSF14 | 1 | 1 | NM_003820 | Tumor necrosis factor receptor superfamily, member 14 |
| TNFRSF9 | 1 | 1 | NM_001561 | Tumor necrosis factor receptor superfamily, member 9 |
| CD79 | 2 | 19 | NM_001784 | CD79 molecule |
| ZNF670 | 2 | 1 | NM_033213 | Zinc finger protein 670 |
| ZNF250 | 1 | 8 | NM_021061 | Zinc finger protein 250 |
| ZNF684 | 3 | 1 | NM_152373 | Zinc finger protein 684 |
| CARD9 | 2 | 9 | NM_052813 | Caspase recruitment domain family, member 9 |
| ADRBK1 | 2 | 11 | NM_001619 | Adrenergic, beta, receptor kinase 1 |
| LRP5 | 3 | 11 | NM_002335 | Low-density lipoprotein receptor-related protein 5 |
| IKBKB | 1 | 8 | NM_001556 | Inhibitor of kappa light polypeptide gene enhancer in B cells |
| PTK2 | 2 | 8 | NM_153831 | Protein tyrosine kinase 2 |
| Jak2 | 1 | 9 | NM_004972 | Janus kinase 2 |
| NOTCH1 | 9 | 9 | NM_017617 | Homo sapiens notch1 |
| TLR5 | 2 | 1 | NM_003268 | Toll-like receptor 5 |
| ZNF282 | 1 | 7 | NM_003575 | Zinc finger protein 282 |
Fig. 3.
TRP120 binding sites mapped near transcriptional start sites of cellular genes visualized by the UCSC Genome Browser. Given an mfold value of 10 and a sonication size (bandwidth) of 300 bp, MACS searched a window area across the genome to find genomic regions with tags more than mfold enriched relative to the expectation from a Poisson distribution. Several identified TRP120 target genes were identified, and the results are presented here. The bent arrows represent TSSs. The dark blue boxes represent the promoter regions. (A) ZNF132 (zinc finger protein 132); (B) ZNF428 (zinc finger protein 428); (C) Jak3 (Janus kinase 3); (D) CXCL11 (CXC motif ligand 11).
Motif analysis and validation of TRP120 binding sites.
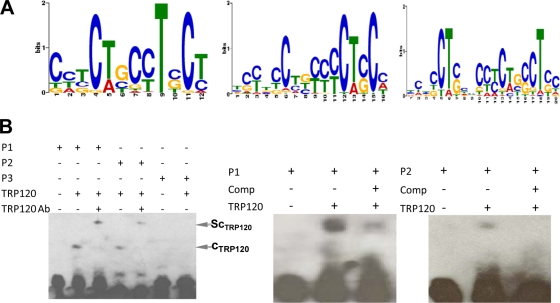
To determine TRP120 binding motifs, we used the sequence analysis tool MEME to identify common DNA sequence motifs among detected peak sequences representing genomic regions (4). This program can automatically compute optimal motif widths. The strongest peaks (top 10%) were used for searching enriched motifs, and the resulting TRP120 PWM was created on the basis of the different width settings (12, 16, and 20). A G+C-rich binding motif was shown in WebLogo format (Fig. 4A) using a more stringent cutoff (P < 10−5). By this stringent criterion, this motif was present in 75% of the 9,862 peaks identified in the TRP120 ChIP sample. This indicated that the TRP120 binding site was consistently present, on the basis of our genome-wide analysis. To determine if the TRP120 ChIPSeq site was bound directly by TRP120, we assayed the ability of purified TRP120 to bind candidate sequences in vitro using EMSA (Fig. 4B). An EMSA was performed using labeled (biotinylated) probes P1 and P2 [P1 contained consensus sequence 5′-CCT(G/C)CC-3′ in an repeat orientation, P2 contained consensus sequence 5′-CCTCCC-3′] representing the TRP120 binding site and one oligonucleotide probe (P3) which did not contain the putative binding site. Binding and a significant shift in migration of P1 and P2 were observed after incubation with TRP120, and binding to probe P3 was not observed (Fig. 4B, left). Formation of the protein complex was inhibited by competition with excess unlabeled P1 and P2 probes (Fig. 4B, center and right, respectively). This result confirmed that TRP120 directly binds the conserved G+C motif identified by MEME analysis.
Fig. 4.
E. chaffeensis TRP120 consensus DNA-binding motif and probe binding assay. (A) Consensus TRP120 binding motif that WebLogo identified in the TRP120 ChIPSeq peaks using the MEME program. Panels represent different motif width settings (12, 16, and 20 from left to right, respectively). (B) (Left) TRP120 binding to G+C-rich oligonucleotide probes representing MEME motif. The TRP120 complex (cTRP120) and supershift binding complex (ScTRP120) are indicated and were detected using EMSA for P1 and P2 probes. Ab, antibody. (Center) EMSA using biotinylated P1 and TRP120 in the presence of a 100-fold molar excess of unlabeled competitor (P1). (Right) EMSA using biotinylated P2 and TRP120 in the presence of a 100-fold molar excess of unlabeled competitor (P2).
The specificity of the binding motif was further examined by a supershift assay using TRP120-specific antibody. Two supershift bands were observed for P1 and P2, respectively (Fig. 4B, left). This result demonstrated that the migration of TRP120 complexes observed with the P1 and P2 probes was retarded by TRP120-specific antibody, indicating that TRP120 binds with probes that contain the defined G+C-rich motif.
Validation of TRP120 binding and ontology (GO) analysis.
The peak recognition by MACS is suggestive of binding regions based on the human genome, but in order to further assess the findings of ChIPSeq data analysis, validation of the targets identified by ChIPSeq experiments was performed. We selected 15 gene-associated peaks for manual site confirmation with specific primers and qPCR. The enrichment of these genes by immunoprecipitation relative to the serum amplicons was determined. Of the 15 genes selected, 12 were enriched (>1.5-fold) compared to negative serum controls, determined by qPCR (Table 4). The motif analysis by MEME revealed that all enriched genes contained the G+C-rich TRP120 DNA-binding motif.
Table 4.
ChIP enrichment of randomly selected ChIPSeq-identified TRP120 target genes by quantitative PCRa
| Gene product | GenBank accession no. | Chromosome | Fold change (vs serum) | Description |
|---|---|---|---|---|
| DNAJC16 | NM_015291 | 1 | 3.9 ± 0.3 | Homo sapiens DnaJ (Hsp40) homology, subfamily C, member 16 |
| TIE1 | NM_005424 | 1 | 5.5 ± 0.3 | Tyrosine kinase with immunoglobulin-like and EGFb-like domains 1 |
| IKBKB | NM_001556 | 8 | 3.6 ± 0.2 | Inhibitor of kappa light polypeptide gene enhancerin B-cells, kinase beta |
| LRP5 | NM_002335 | 11 | 5.5 ± 0.1 | Low-density lipoprotein receptor-related protein 5 |
| PTPRF | NM_130440 | 1 | 14.0 ± 0.5 | Protein tyrosine phosphatase, receptor type, F |
| ZNF292 | NM_015021 | 6 | 5.8 ± 0.3 | Zinc finger protein 292 |
| ZNF238 | NM_205768 | 1 | 12.8 ± 0.2 | Zinc finger protein 238 |
| ZNF276 | NM_152287 | 16 | 17.2 ± 0.2 | Zinc finger protein 276 |
| PTPRN2 | NM_002847 | 7 | 6.8 ± 0.2 | Protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| NOTCH1 | NM_017617 | 9 | 1.5 ± 0.1 | Homo sapiens notch1 |
| MAPK8IP3 | NM_001040439 | 16 | 1.5 ± 0.1 | Mitogen-activated protein kinase 8 interacting protein 3 |
| CTDP1 | NM_004715 | 18 | 3.4 ± 0.4 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
Mean fold gene enrichment ± standard error was calculated, and the TRP120 level was compared with serum control amplicon levels.
EGF, epidermal growth factor.
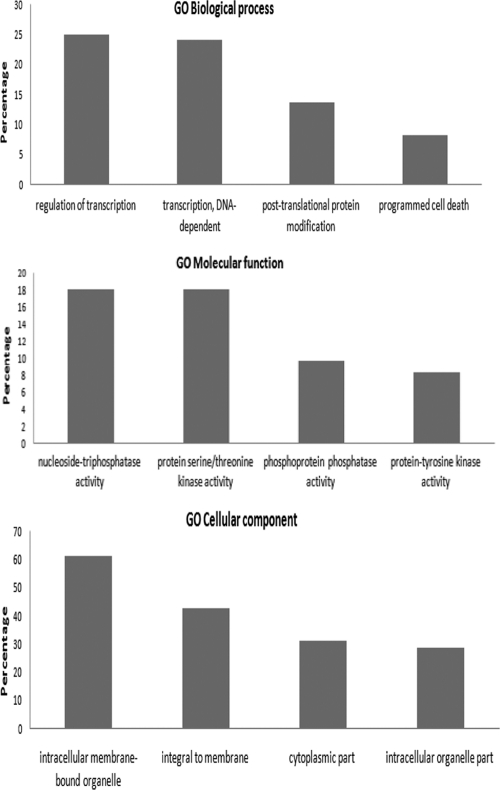
In order to reveal general biological processes that may be affected by TRP120, a gene functional analysis was performed by using the Babelomics FatiGO+ tool (1, 2). We used this powerful approach to extract gene ontology terms that are significantly represented in sets of genes within the context of our ChIPSeq data. The TRP120 putative target genes were classified on the basis of biological processes, molecular functions, and cellular components (Fig. 5). At the highest levels of the annotation hierarchy within the gene ontology databases, the most highly represented target genes were associated with biological functions, including transcription regulation, apoptosis, and phosphorylation (Table 5).
Fig. 5.
GO associations of TRP120 with host cell biological processes and molecular functions and cellular component genes identified by ChIP-Seq (level 7; P value, 10−20).
Table 5.
Top gene ontology categories of TRP120 target genes
| Term | Genes annotated to term |
|---|---|
| Regulation of transcription | GTF2H1, SYNGAP1, PPARA, ELL, ABCA2, BRF1, PRDM1, NFYC, ZNF644, FOXJ3, MYCL1, ATF5 ZNF358, NPAS2, TRIM33, HMGA1, CITED4, ZNF764, PRDM13, MLF1IP, TBX18, GJA10, TEF, ZNF707, AHRR, NFYA, KHDRBS1, MAF1, FOXA2, HIRA, FOXP4, ZNF623, SALL2, ELL2, CBFA2T3, ERN1, HIVEP3, NOTCH1, NFATC3, POLR2L, ZNF684, GTF2F1, RXRA, ZNF282, TNXB, ZBTB9, PKNOX2, LOC44179, ZNF395, TFB2 M, RLF, ZNF251, YBX1, CRABP2, RORC, PRDM15, KLF2, BACH2, HMG20A, LZTS1 |
| Apoptosis | PPT1, ERN1, GRM4, CLU, BARD1, NOTCH1, BAK1, PROC, PLEKHG5, PHLPP, PDCD6, TP53BP2, RTN4, MIF, ATF5, TNFRSF14, NLRP3, CARD9, HSPB1, NUAK2, AHRR |
| Protein phosphorylation | CSK, TNFRSF14, HIPK4, NTRK1, BCR, NUAK2, MAP3K2, IKBKB, TIE1, ERN1, PTK7, PMVK, PRKAR1B, RPS6KA2, PTK2, INSRR, ROR2, CAMK2B, OBSCN, ADRBK1 |
| Ras protein signal transduction | RASGRF1, ARHGEF4, PLEKHG5, TBC1D9B, OBSCN, TBC1D2, FGD2, BCR, TBC1D8, VAV1, TBC1D1, ITPKB |
| Chromatin modification | NPM2, HMG20A, SMYD3, HMGA1 |
| ATPase activity | DDX59, ABCA2, SEMA4A, CLPX, ATP2B2 |
| GTPase activity | GNAI2, EEF1A1, NUDT1, TUBG2, TUBB6 |
| Protein-tyrosine kinase activity | PTK2, OBSCN, PTK7, ROR2, CSK, IKBKB, TIE1 |
| Protein serine/threonine kinase activity | IKBKB, ADRBK1, TIE1, OBSCN, HIPK4, GTF2F1, BCR, RPS6KA2, GTF2H1, CAMK2B, ERN1, NUAK2, MAP3K2 |
Expression levels of TRP120 target genes.
Eight genes identified by ChIPSeq were selected for analysis by protein transfection of expression levels of TRP120 target genes relative to the levels for the thioredoxin control and of expression levels of TRP120 target genes for E. chaffeensis-infected cells relative to those for the uninfected cell control. Among these TRP120-targeted genes, four target genes (those for tumor necrosis factor alpha [TNF-α], CCL20, CXCL11, and CCL2) were strongly upregulated (∼5- to 30-fold) beginning at 4 h and increasing through 72 h (∼24- to 190-fold) after E. chaffeensis infection. Three genes (those for TNF-RSF14, signal transducer and activator of transcription 1 [Stat1], and CD70) were similarly upregulated at 4 and 72 h postinfection (∼2- to 5-fold). To determine the TRP120 role in the gene expression levels, purified TRP120 was introduced into THP-1 cells using the Chariot transfection reagent for 4 and 72 h. Interestingly, all of the genes examined except the gene for CCL20 (8-fold) exhibited a small increase (≤2-fold) in expression levels at 4 h postinfection; however, these genes were significantly upregulated (∼3- to 8.5-fold) at 72 h posttransfection with TRP120, except the gene for TNF-α (Fig. 6A and B). A gene (for CIAS1) which was not predicted to contain a TRP120 binding site was not regulated by E. chaffeensis infection or TRP120 posttransfection. This result demonstrated that TRP120 does directly activate the expression of these genes during E. chaffeensis infection, and the regulation of TNF-RSF14, Stat1, and CD70 by TRP120 was nearly identical to that with E. chaffeensis infection alone. The level of regulation of CCL20, CXCL11, and CCL2 by TRP120 was lower than that observed during E. chaffeensis infection.
Fig. 6.
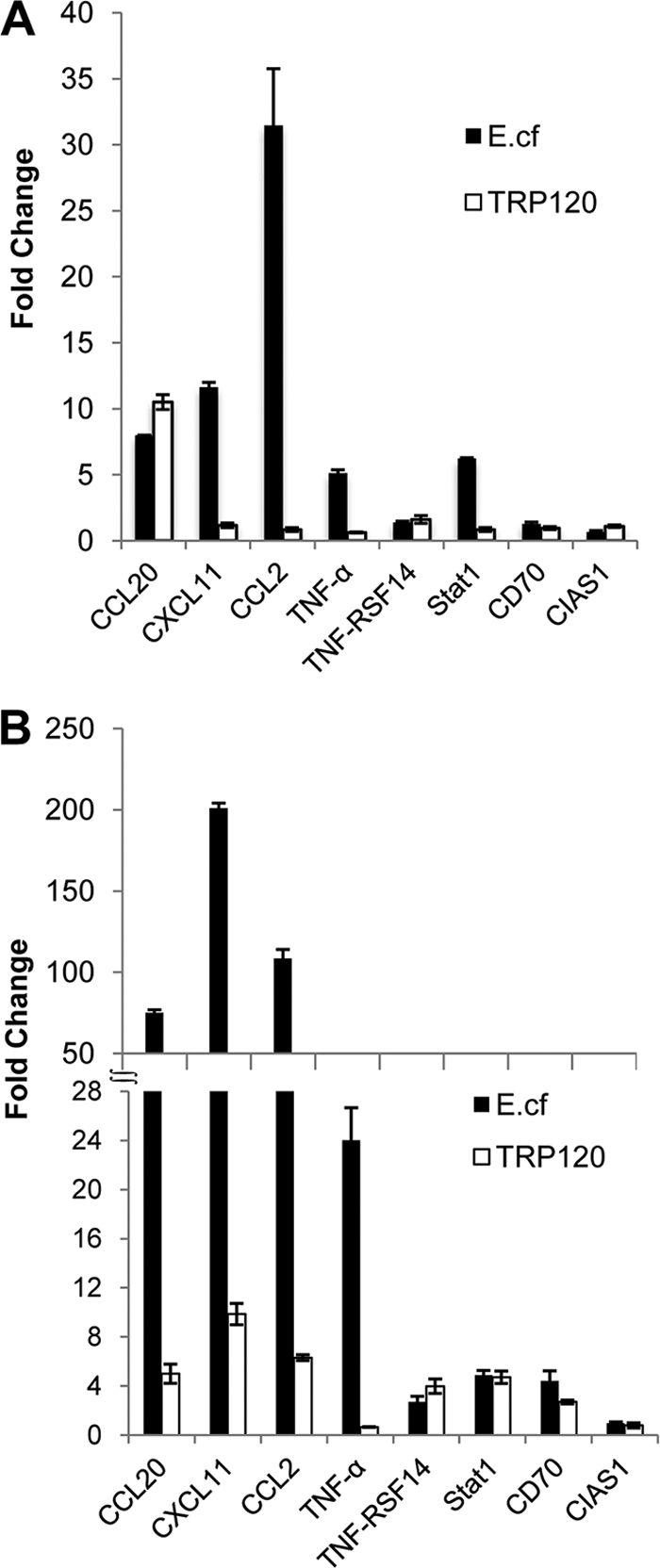
Expression levels of TRP120 target genes by protein transfection relative to those by the thioredoxin control and expression levels of TRP120 target genes for E. chaffeensis (E.cf)-infected cells relative to those for uninfected control cells. (A) Expression levels of TRP120 target genes in THP-1 cells 4 h after E. chaffeensis infection or recombinant TRP120 transfection; (B) expression levels of TRP120 target genes in THP-1 cells 72 h after direct transfection with recombinant TRP120 or infection with E. chaffeensis.
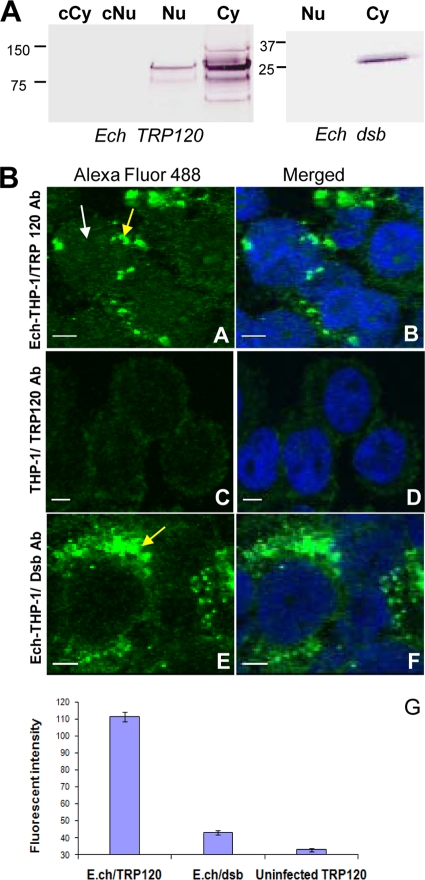
Ehrlichia TRP120 is translocated to the nuclei of infected monocytes.
TRP120 nuclear translocation was studied by Western immunoblotting using cytoplasmic and nuclear fractions which were extracted from E. chaffeensis-infected and uninfected THP-1 cells. TRP120 was detected in the fractions of the cytoplasm and nuclei of E. chaffeensis-infected cells but not in uninfected cells (Fig. 7A). The Ehrlichia Dsb (ehrlichial periplasmic protein) was detected only in the cytoplasmic fraction of infected cells (Fig. 7A), confirming that the nuclear extract preparation was not contaminated with ehrlichiae. Immunofluorescent microscopy with anti-TRP120 was performed to visualize the TRP120 localization in the host cell. TRP120 was detected in the nucleus of E. chaffeensis-infected host cells and cytoplasmic vacuoles (morula) containing ehrlichiae (Fig. 7B, sections A and B). TRP120 was not detected in the nucleus of uninfected THP-1 cells (Fig. 7B, sections C and D). To further demonstrate the specificity of the nuclear signal, an ehrlichial periplasmic protein (Dsb) that is not secreted, that is associated with the bacterial outer membrane, and that is not translocated to the nucleus was examined as a control. Dsb was not detected in the nuclei of E. chaffeensis-infected host cells (Fig. 7B, sections E and F). The quantity of nuclear translocated TRP120 was determined by the fluorescent intensity, represented from nuclei of infected cells (Fig. 7B, section G).
Fig. 7.
Detection of E. chaffeensis TRP120 in the host cell nucleus. (A) Western immunoblot detection of TRP120 and E. chaffeensis (Ech) periplasmic protein Dsb (not secreted) using nuclear extracts from E. chaffeensis-infected and uninfected THP-1 cells (3 days postinfection). cNu, control nuclear extract from uninfected cells; Nu, nuclear extract from infected monocytes; Cy, cytoplasmic fraction from infected cells; cCy, cytoplasmic fraction from uninfected cells. (B) E. chaffeensis-infected cells (sections A, B, E, and F) and uninfected THP-1 cells (sections C and D) were fixed with paraformaldehyde, permeabilized with Triton X-100, and incubated with anti-E. chaffeensis TRP120 and anti-Dsb. Host cell nuclei (blue) were counterstained with 4′,6-diamidino-2-phenylindole. The nuclei (white arrow in section A) of infected cells and E. chaffeensis morulae (yellow arrows in sections A and E) are identified. (Section G) Fluorescent intensity of protein nuclear translocation was quantified from infected and uninfected cells reacted with TRP120 or Dsb antibodies. Bar, 5 μm.
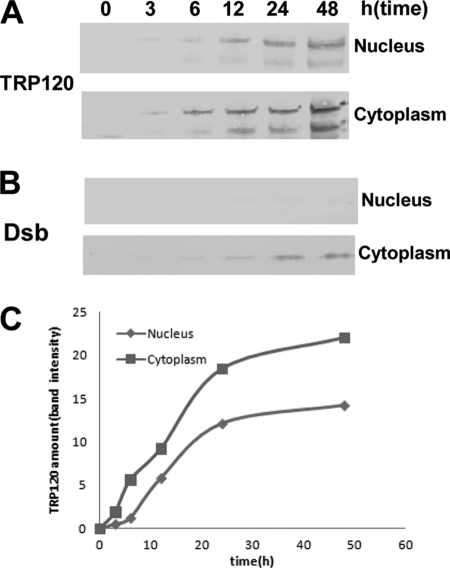
To study the kinetics of TRP120 accumulation in the nuclei of THP-1 cells, nuclear and cytoplasmic fractions of E. chaffeensis-infected cells were analyzed by immunoblotting using TRP120-specific antibody. TRP120 was detected in host cell nuclei as early as 3 h postinfection, and it accumulated in the nucleus over time (Fig. 8A). TRP120 was also detected in the host cell cytoplasm, and the amount of TRP120 detected in the nuclei of infected cells was less than that detected in the cytoplasm (Fig. 8C). Ehrlichial Dsb protein was detected in the cytoplasmic fraction but not in the nucleus, indicating that the nuclear extract preparation was not contaminated with ehrlichiae (Fig. 8B).
Fig. 8.
Accumulation of TRP120 in nuclei of host cells. (A and B) Western immunoblot detection of TRP120 accumulation and E. chaffeensis periplasmic protein Dsb (not secreted) using nuclear and cytoplasmic fractions of E. chaffeensis-infected THP-1 cells collected at different time points; (C) the amounts of TRP120 in nuclei and cytoplasm of infected THP-1 cells were calculated by densitometry of the TRP120 bands.
DISCUSSION
Immune subversion by chromatin manipulation is becoming recognized as an emerging and important new strategy in which the pathogen imposes its own transcriptional signature (3). Ehrlichia modulates host cell gene expression, and the recent finding that E. chaffeensis Ank200 and TRP120 interact with the host cell chromatin has revealed a molecular mechanism and associated effector proteins that are involved in host-DNA interactions and host cell gene transcriptional modulation. TRP120 is the second DNA-binding protein identified in Ehrlichia. Notably, TRP120 recognizes a DNA motif distinct from that of the previously described Ank200 and contains a novel TR DNA-binding domain that has not been previously described in any human pathogen. Other described TR DNA-binding proteins are limited to the recently described TAL effectors of the Gram-negative plant pathogens from the genera Xanthomonas and Ralstonia and the fibroin modulator binding protein-1 (FMBP-1) DNA-binding protein in the silkworm Bombyx mori (20, 22, 57).
The TRP120 DNA-binding motif and host target gene identification in this study were determined by ChIPSeq using high-throughput DNA sequencing to obtain TRP120-DNA interactions on a genome-wide scale. This approach differed from our previous and more limited approach using microarray technology to determine Ank200 binding sites in host gene promoter regions (69). Recent studies have concluded that ChIPSeq coupled with high-throughput sequencing provides high sensitivity and specificity and is rapidly becoming the method of choice for identification of protein-DNA interactions (25). Mapping of transcription factor-DNA interactions using ChIPSeq approaches has shown that transcriptional regulatory proteins bind thousands of active and inactive binding sites located in different target gene regions (32). Similarly, we determined that TRP120 has numerous binding sites that mapped to thousands of genes. According to our ChIPSeq analysis, the number of TRP120 binding sites ranged from 59,088 to as few as 582, depending on the statistical criteria applied. It is apparent that TRP120 binding affinity varies across the genome and some sites are bound more strongly, and such differences in transcription factor binding have generally been noted with other eukaryote transcription factors (32). In our study, a cutoff of 1 × 10−15 (P value) was considered to identify specific high-binding interactions with fewer false-positive results, because more than 90% of peaks were mapped to the human genome reference with a cutoff of 1× 10−15 (P value). Hence, approximately 2,000 genes were bound strongly by TRP120, and many of these genes had multiple TRP120 binding sites, suggesting that TRP120 exerts a more direct transcriptional regulatory effect on these host genes.
Cytoplasmic signaling pathways have been identified to be targets of ehrlichial host cell transcriptional modulation (60). However, directly targeting transcriptional machinery or posttranslational mechanisms also appears to be another important molecular host cell modulation strategy utilized by Ehrlichia (69). The effector proteins and mechanisms that E. chaffeensis utilizes to evade innate and adaptive immune responses are complex and involve many different effector proteins and host targets, most of which have not been defined. Previous studies have demonstrated altered transcription levels of host genes involved in a variety of functions (67). Notably, E. chaffeensis significantly manipulated genes related to the transcription of cytokines, apoptosis inhibitors, and membrane trafficking. Furthermore, the unique Ehrlichia-host interaction is illustrated by the fact that among intracellular bacteria, only a relatively few host genes were induced in common during E. chaffeensis and other intracellular bacterial infections, while no genes in common were repressed, suggesting that ehrlichial survival mechanisms have evolved distinctly from those of other intracellular pathogens (67). Although transcriptional modulation occurs, none of the ehrlichial effector proteins involved in this process had been identified, until we determined that the ankyrin repeat protein, Ank200, was translocated to the host cell nucleus in Ehrlichia-infected mononuclear phagocytes, where it interacts with an adenine-rich motif in promoter and intronic Alu elements (69). Host genes targeted by Ank200 were associated with transcription, the cell cycle, and apoptosis. Importantly, in this study, we have determined that other effector proteins are also involved in a direct transcriptional modulation strategy with distinct protein domains that recognize different DNA motifs. DNA-binding domains, such as the helix-turn-helix, zinc finger, and leucine zipper domains, are well-known and serve to structurally classify DNA-binding domains (56). However, this study and the excellent descriptions by others demonstrate that TR DNA-binding domains occur in prokaryotes and eukaryotes and represent a new class of DNA-binding proteins (9, 46, 52).
Bacterial TRPs have been linked to important host-pathogen interactions, and we have identified numerous and distinct interactions between E. chaffeensis TRP47 and host cell targets (60). It appears that TRP120 is a multifunctional protein, a finding illustrated by a recent study demonstrating a role for TRP120 in ehrlichial internalization and our recent findings that it interacts with numerous host cell proteins (28, 34). Interestingly, our new findings establish a functional relationship between ehrlichial TRPs, the TAL proteins of plant pathogens of the genera Xanthomonas and Ralstonia, and FMBP-1 of the silkworm Bombyx mori (20, 22, 57). The TR domain mediates protein-DNA binding, and in Xanthomonas TAL effectors, each repeat unit is associated with the recognition of two nucleotides on the DNA strand (39). Although E. chaffeensis TRP120 has four tandem repeat units, we determined that a two-repeat unit of TRP120 could directly bind host DNA, demonstrating that the TR region contains a DNA-binding domain. The TRP120 TR region of different E. chaffeensis strains contains variable numbers of TR units (2 to 5), and TR number variation has also been described in Xanthomonas TAL proteins (26, 66). The TR unit of TRP120 is 80 amino acids (36), while the repeat units for TAL (AvrBs3) and FMBP-1 are 34 and 23 amino acids, respectively (26, 57). Thus, there is variation in the length of TRs capable of binding DNA and in the DNA motif recognized by each. Further studies are needed to determine molecular interactions between specific TRP120 amino acids and DNA nucleotides, but in Xanthomonas, each imperfect 34-amino-acid repeat unit interacts with two nucleotides and governs DNA recognition by TAL effectors (26).
Interestingly, most of the TRP120 binding sites were located in intersecting regions of genes, including introns and exons. Studies have suggested that these binding regions may be associated with an alternative promoter (24) and influence transcriptional regulation (38). The finding that TRP120 has binding sites within intersecting gene regions is consistent with our previous findings in which we detected Ank200 binding sites in promoter and intron locations (69). Notably, the current findings are also consistent with binding sites reported for other transcription factors and nuclear receptors, including farnesoid X receptor (FXR) (7), p63 (47), Stat1 (7, 27, 50), peroxisome proliferator-activated receptor (PPAR) (31, 45), estrogen receptor (6), androgen receptor (5), and nuclear vitamin D receptor (48). Thus, TRP120 intersecting binding sites may play a role in potential transcriptional activation and influence transcriptional regulation of the associated genes.
The G+C motif was consistently represented in TRP120 peaks, and we found that the motif is highly represented throughout the human genome, particularly in the enhancer and promoter regions. The TRP120 binding motif is similar to the binding motifs of other well-characterized eukaryotic transcription factors, such as Sp1, which has a binding motif known as the GC box (5′-CCGCCC-3′) (29). Sp1 specifically binds to GC box promoter sequences of various cellular and viral genes and regulates gene expression (29). The GC box motif of Sp1 occurs randomly every 2 kbp in the human genome, but only a few Sp1 motifs are transcriptionally active. Moreover, G+C-rich motifs have also been identified in human liver prothrombin gene and hepatitis B virus enhancers (8), the α-1-antitrypsin promoter (21), and the fibrinogen β-chain promoter (12), and G+C-rich promoter/enhancer sequences have been associated with tissue-specific transcription (8). Notably, the TRP120 motif is also similar to the cytosine-rich (CCN2-3CC) domain in the UPA box of the well-characterized AvrBs3 DNA-binding protein motif. Binding to this motif specifically activates expression of numerous plant genes, including upa20, a plant helix-turn-helix transcription factor (52). Therefore, the TRP120 G+C-rich DNA motif is not unique and appears to be similar to other G+C motifs associated with other eukaryotic transcription factors.
The gene ontological analysis performed in this study indicates that the TRP120 influence is directed at host genes related to specific biological and molecular processes and cellular components. Transcriptional regulation was the most highly represented biological process linked to the TRP120 target genes, a finding that was consistent with the biological processes targeted by Ank200 (69). There is evidence that a large number of genes associated with transcription are modulated during E. chaffeensis infection (67). In addition, a number of other important biological processes were identified as having a high TRP120 gene target representation, including transcriptional regulation, apoptosis, protein modification, and signaling. This finding is also consistent with the biological processes identified to be targets of Ank200, and Ehrlichia is known to alter host cell gene transcription and modulate apoptosis, protein modification, and signaling pathways (30, 62). Identification of the biological and molecular processes that TRP120 targets provides some insight into the role of TRP120 in modulating biological events critical for ehrlichial survival (67). These results suggest that TRP120 plays an important role in regulating a diverse array of cellular processes that are known to be modulated by E. chaffeensis and suggest that Ank200 and TRP120 converge on similar host cell processes by binding to unique and overlapping biological process genes.
There were many host genes of interest that were identified to be TRP120 targets, but of particular interest was the identification of genes that encode inflammatory cytokines/chemokines and members of the Jak/Stat signaling pathway, a finding which suggests that TRP120 plays a role in transcriptional modulation of immune subversion mechanisms and immunopathology. In our previous report, inflammatory cytokines genes such as TNF-α and members of the Jak/Stat signaling pathway were also identified to be Ank200 targets (69). We previously demonstrated that TNF-α production is strongly upregulated in monocytes/macrophages, and we suggested that induction of the gene for TNF-α may be modulated, in part, by Ank200 interaction with the Alu element (69). Direct transfection with TRP120 did not induce TNF-α expression, suggesting that TRP120 does not independently activate TNF-α during E. chaffeensis infection. However, Ehrlichia strongly activates inflammatory chemokine production. Chemokines not only attract inflammatory cells to sites of infection or inflammation, but they are also responsible for selective migration (homing) of leukocyte subtypes, such as lymphocytes and dendritic cells, to lymphoid organs, allowing their differentiation and maturation (37, 55). Chemokines constitute a large family of chemotactic cytokines which have been subdivided into two predominant groups, the CXC and CC families (51). It has been reported that chemokines CCL2, CCL5, CCL20, CXCL1, CXCL2, and CXCL11 and cytokines TNF-α and the TNF-α receptor superfamily (TNFRSF9) were highly upregulated during E. chaffeensis infection (42). In this study, we demonstrated that THP-1 cells directly transfected with TRP120 showed significantly increased expression of CCL2 and CCL20, which are known to be highly upregulated during E. chaffeensis infection (42), suggesting that TRP120 is an effector sufficient to induce the transcriptional changes observed in THP-1 cells during infection and may play a role in the immunopathology associated with Ehrlichia infection.
Interestingly, TRP120 binding sites in CCL2, CCL20, and CXCL11 were detected in upstream regions which were distantly located upstream (≥20 kb) from transcription start sites (TSSs). The distant upstream binding site may be functional, which is supported by the recent observation that distal enhancers which are located more than 10 to 20 kb from the TSS can be important drivers of gene expression (44, 63). Both conserved and nonconserved sites also play important roles in determining transcription levels (38). This new model of transcriptional regulation addresses the effect of distant binding sites on gene expression and suggests that such TRP120 binding sites are functionally relevant.
It is known that the subversion of the gamma interferon (IFN-γ) response by Ehrlichia appears to be a major mechanism of immune escape. E. chaffeensis blocks phosphorylation of Stat1 and Jak2 during infection (30), and this study suggests that during E. chaffeensis infection, Jak/Stat genes are controlled by a direct transcriptional regulation mechanism that involves TRP120. Another strategy that Ehrlichia utilizes to modulate the IFN-γ response is direct transcriptional regulation. The TRP120 binding site in Stat1 was detected in an intronic region which is close to the TSS, and the regulation of Stat1 by TRP120 is nearly identical to that with E. chaffeensis infection, suggesting that TRP120 may modulate Stat1 expression directly during E. chaffeensis infection.
Active nuclear transport of macromolecules occurs via the nuclear pore complex. Proteins undergoing nuclear importing usually contain a mono- or bipartite basic-type nuclear localization sequence (NLS), which binds to the specific NLS receptor, α-importin (59). We did not identify a conventional NLS sequence in TRP120, suggesting that it gains access to the monocyte nucleus by a nonclassical mechanism. In contrast, NLSs have been identified on Xanthomonas AvrBs3, which also dimerizes prior to nuclear import (19). Other nuclear translocation mechanisms exist, including a vesicle-mediated secretory pathway (16), and a glycan-dependent nuclear import pathway has been described and characterized (14, 15). Further study for a deeper understanding of TRP120 nuclear translocation will be necessary to conclusively determine its transport mechanism.
E. chaffeensis TRP120 and TRP47 are differentially expressed on dense-cored ehrlichiae (the infectious form), secreted extracellularly within the ehrlichial endocytic vacuole, and strongly recognized by the host immune response (13, 47). As the functional role of TRPs is beginning to emerge, TRP47 interactions with important host cell proteins involved in signaling, modulation of gene expression and intracellular vesicle trafficking (60), and direct binding to specific gene targets of E. chaffeensis TRP120 provide new insight into the effector role of TRPs in manipulating host cell processes such as transcription. Further studies are needed to fully understand the complexity of pathogen-host interactions that involve TRPs and how these proteins act synergistically to modulate host cell processes critical for ehrlichial survival.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Allergy and Infectious Diseases grant AIAI069270, and additional support was provided by the Clayton Foundation for Research.
We thank David H. Walker and Xue-jie Yu for reviewing the manuscript and providing helpful suggestions.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 22 August 2011.
REFERENCES
- 1. Al-Shahrour F., et al. 2007. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 35:W91–W96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Shahrour F., et al. 2006. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 34:W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arbibe L. 2008. Immune subversion by chromatin manipulation: a ‘new face’ of host-bacterial pathogen interaction. Cell. Microbiol. 10:1582–1590 [DOI] [PubMed] [Google Scholar]
- 4. Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 5. Bolton E. C., et al. 2007. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 21:2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carroll J. S., et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- 7. Chong H. K., et al. 2010. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 38:6007–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chow B. K., Ting V., Tufaro F., MacGillivray R. T. 1991. Characterization of a novel liver-specific enhancer in the human prothrombin gene. J. Biol. Chem. 266:18927–18933 [PubMed] [Google Scholar]
- 9. Citti C., Kim M. F., Wise K. S. 1997. Elongated versions of Vlp surface lipoproteins protect Mycoplasma hyorhinis escape variants from growth-inhibiting host antibodies. Infect. Immun. 65:1773–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clifton D. R., et al. 2005. Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect. Immun. 73:3860–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins N. E., et al. 2005. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. U. S. A. 102:838–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Courtois G., Morgan J. G., Campbell L. A., Fourel G., Crabtree G. R. 1987. Interaction of a liver-specific nuclear factor with the fibrinogen and alpha 1-antitrypsin promoters. Science 238:688–692 [DOI] [PubMed] [Google Scholar]
- 13. Doyle C. K., Nethery K. A., Popov V. L., McBride J. W. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duverger E., Carpentier V., Roche A. C., Monsigny M. 1993. Sugar-dependent nuclear import of glycoconjugates from the cytosol. Exp. Cell Res. 207:197–201 [DOI] [PubMed] [Google Scholar]
- 15. Duverger E., Pellerin-Mendes C., Mayer R., Roche A. C., Monsigny M. 1995. Nuclear import of glycoconjugates is distinct from the classical NLS pathway. J. Cell Sci. 108(Pt 4):1325–1332 [DOI] [PubMed] [Google Scholar]
- 16. Gardella S., et al. 2002. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 3:995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gravekamp C., Horensky D. S., Michel J. L., Madoff L. C. 1996. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infect. Immun. 64:3576–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S. 2007. Quantifying similarity between motifs. Genome Biol. 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gurlebeck D., Szurek B., Bonas U. 2005. Dimerization of the bacterial effector protein AvrBs3 in the plant cell cytoplasm prior to nuclear import. Plant J. 42:175–187 [DOI] [PubMed] [Google Scholar]
- 20. Gurlebeck D., Thieme F., Bonas U. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163:233–255 [DOI] [PubMed] [Google Scholar]
- 21. Hardon E. M., Frain M., Paonessa G., Cortese R. 1988. Two distinct factors interact with the promoter regions of several liver-specific genes. EMBO J. 7:1711–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heuer H., Yin Y. N., Xue Q. Y., Smalla K., Guo J. H. 2007. Repeat domain diversity of avrBs3-like genes in Ralstonia solanacearum strains and association with host preferences in the field. Appl. Environ. Microbiol. 73:4379–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang B., et al. 2010. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect. Immun. 78:1864–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Impey S., et al. 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119:1041–1054 [DOI] [PubMed] [Google Scholar]
- 25. Johnson D. S., Mortazavi A., Myers R. M., Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502 [DOI] [PubMed] [Google Scholar]
- 26. Kay S., Hahn S., Marois E., Wieduwild R., Bonas U. 2009. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 59:859–871 [DOI] [PubMed] [Google Scholar]
- 27. Kouwenhoven E. N., et al. 2010. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 6:e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumagai Y., Matsuo J., Hayakawa Y., Rikihisa Y. 2010. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J. Bacteriol. 192:4122–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuwahara J., Yonezawa A., Futamura M., Sugiura Y. 1993. Binding of transcription factor Sp1 to GC box DNA revealed by footprinting analysis: different contact of three zinc fingers and sequence recognition mode. Biochemistry 32:5994–6001 [DOI] [PubMed] [Google Scholar]
- 30. Lee E. H., Rikihisa Y. 1998. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 66:2514–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lefterova M. I., et al. 2008. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22:2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li X. Y., et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 34. Luo T., Kuriakose J. A., Zhu B., Wakeel A., McBride J. W. 2011. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect. Immun. 79:4382–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo T., Zhang X., McBride J. W. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin. Vaccine Immunol. 16:982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo T., Zhang X., Wakeel A., Popov V. L., McBride J. W. 2008. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 76:1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luster A. D. 1998. Chemokines—chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338:436–445 [DOI] [PubMed] [Google Scholar]
- 38. MacIsaac K. D., et al. 2010. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput. Biol. 6:e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matthew J., et al. 2009. Distribution and seasonality of rhinovirus and other respiratory viruses in a cross-section of asthmatic children in Trinidad, West Indies. Ital. J. Pediatr. 35:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McBride J. W., Ndip L. M., Popov V. L., Walker D. H. 2002. Identification and functional analysis of an immunoreactive DsbA-like thio-disulfide oxidoreductase of Ehrlichia spp. Infect. Immun. 70:2700–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McBride J. W., Walker D. H. 2011. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev. Mol. Med. 13:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miura K., Rikihisa Y. 2009. Liver transcriptome profiles associated with strain-specific Ehrlichia chaffeensis-induced hepatitis in SCID mice. Infect. Immun. 77:245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mukaihara T., Tamura N., Iwabuchi M. 2010. Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23:251–262 [DOI] [PubMed] [Google Scholar]
- 44. Nerenz R. D., Martowicz M. L., Pike J. W. 2008. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol. Endocrinol. 22:1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nielsen R., et al. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22:2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nonaka Y., et al. 2010. STPR, a 23-amino acid tandem repeat domain, found in the human function-unknown protein ZNF821. Biochemistry 49:8367–8375 [DOI] [PubMed] [Google Scholar]
- 47. Popov V. L., Yu X., Walker D. H. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28:71–80 [DOI] [PubMed] [Google Scholar]
- 48. Ramagopalan S. V., et al. 2010. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 20:1352–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rikihisa Y. 2006. Ehrlichia subversion of host innate responses. Curr. Opin. Microbiol. 9:95–101 [DOI] [PubMed] [Google Scholar]
- 50. Robertson G., et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4:651–657 [DOI] [PubMed] [Google Scholar]
- 51. Rollins B. J. 1997. Chemokines. Blood 90:909–928 [PubMed] [Google Scholar]
- 52. Romer P., et al. 2007. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318:645–648 [DOI] [PubMed] [Google Scholar]
- 53. Romer P., et al. 2009. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150:1697–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shak J. R., Dick J. J., Meinersmann R. J., Perez-Perez G. I., Blaser M. J. 2009. Repeat-associated plasticity in the Helicobacter pylori RD gene family. J. Bacteriol. 191:6900–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sozzani S., Mantovani A., Allavena P. 1999. Control of dendritic cell migration by chemokines. Forum (Genoa, Italy) 9:325–338 [PubMed] [Google Scholar]
- 56. Struhl K. 1989. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem. Sci. 14:137–140 [DOI] [PubMed] [Google Scholar]
- 57. Takiya S., et al. 2009. DNA-binding property of the novel DNA-binding domain STPR in FMBP-1 of the silkworm Bombyx mori. J. Biochem. 146:103–111 [DOI] [PubMed] [Google Scholar]
- 58. Tonjum T., Caugant D. A., Dunham S. A., Koomey M. 1998. Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol. Microbiol. 29:111–124 [DOI] [PubMed] [Google Scholar]
- 59. Ushijima R., et al. 2005. Extracellular signal-dependent nuclear import of STAT3 is mediated by various importin alphas. Biochem. Biophys. Res. Commun. 330:880–886 [DOI] [PubMed] [Google Scholar]
- 60. Wakeel A., Kuriakose J. A., McBride J. W. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77:1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wakeel A., Zhu B., Yu X. J., McBride J. W. 2010. New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect. 12:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xiong Q., Bao W., Ge Y., Rikihisa Y. 2008. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J. Infect. Dis. 197:1110–1118 [DOI] [PubMed] [Google Scholar]
- 63. Yeamans C., et al. 2007. C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood 110:3136–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu X. J., Crocquet-Valdes P., Cullman L. C., Walker D. H. 1996. The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J. Clin. Microbiol. 34:2853–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu X. J., McBride J. W., Diaz C. M., Walker D. H. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yu X. J., McBride J. W., Walker D. H. 2007. Restriction and expansion of Ehrlichia strain diversity. Vet. Parasitol. 143:337–346 [DOI] [PubMed] [Google Scholar]
- 67. Zhang J. Z., Sinha M., Luxon B. A., Yu X. J. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72:498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Y., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhu B., et al. 2009. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect. Immun. 77:4243–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.