Abstract
Photobacterium damselae subsp. damselae (formerly Vibrio damsela) is a marine bacterium that causes infections and fatal disease in a wide range of marine animals and in humans. Highly hemolytic strains produce damselysin (Dly), a cytolysin encoded by the dly gene that is lethal for mice and has hemolytic activity. We found that Dly is encoded in the highly hemolytic strain RM-71 within a 153,429-bp conjugative plasmid that we dubbed pPHDD1. In addition to Dly, pPHDD1 also encodes a homologue of the pore-forming toxin HlyA. We found a direct correlation between presence of pPHDD1 and a strong hemolytic phenotype in a collection of P. damselae subsp. damselae isolates. Hemolysis was strongly reduced in a double dly hlyA mutant, demonstrating the role of the two pPHDD1-encoded genes in hemolysis. Interestingly, although single hlyA and dly mutants showed different levels of hemolysis reduction depending on the erythrocyte source, hemolysis was not abolished in any of the single mutants, suggesting that the hemolytic phenotype is the result of the additive effect of Dly and HlyA. We found that pPHDD1-encoded dly and hlyA genes are necessary for full virulence for mice and fish. Our results suggest that pPHDD1 can be considered as a driving force for the emergence of a highly hemolytic lineage of P. damselae subsp. damselae.
INTRODUCTION
Photobacterium damselae subsp. damselae (formerly Vibrio damsela) is a halophilic bacterium associated with marine environments that was initially isolated in 1981 as the causative agent of skin ulcers in damselfish (34). It is a primary pathogen causing ulcers and hemorrhagic septicemia in a variety of marine species as sharks, dolphins, and shrimps, as well as wild and cultivated fish (18, 20, 45). In addition, this pathogen can cause fatal infections in humans. Most of the reported infections in humans have their origin in wounds inflicted during the handling of fish, exposure to seawater and marine animals, and ingestion of raw seafood (1, 2, 26, 38, 53). In some of the human cases the infection progresses into an extreme variant of a highly severe necrotizing fasciitis that advances following a very aggressive course leading to a fatal outcome (7, 53).
P. damselae subsp. damselae shares species level status with P. damselae subsp. piscicida (formerly Pasteurella piscicida), the causative agent of fish pasteurellosis (21). Although subsp. damselae is pathogenic for marine animals and humans, subsp. piscicida is only pathogenic for fish, it does not grow at 37°C and lacks observable hemolytic activity on blood agar plates (36, 41, 42). Very little is known about the virulence factors that enable P. damselae subsp. damselae to cause septicemia in aquatic animals and humans. A correlation was initially observed between the ability of P. damselae subsp. damselae to cause disease in mice and to produce large amounts of a cytolytic toxin that was later named damselysin (28) (hereafter referred to as Dly). Partially purified Dly preparations from culture supernatants showed to be active against erythrocytes of 16 species of homeotherm animals, with rat and mouse being the more sensitive ones (28). Other studies reported that the extracellular products of this bacterium also had hemolytic activity on turbot erythrocytes (16). A deeper characterization of Dly showed that it has phospholipase D activity against sphingomyelin, phosphatidylcholine, and phosphatidylethanolamine (10, 29). The main molecular activity of Dly consists of the removal of the polar choline groups of choline-containing membrane lipids. Dly was found to enhance the hemolytic effect of staphylococcal delta-toxin by removing the polar choline phosphate head group of sphingomyelin (29), which constituted the first evidence that Dly can act synergistically with hemolysins produced by other cells. The Dly toxin gene, dly, was cloned (9), and its sequence was determined (GenBank accession no. L16584). However, the genomic context of dly gene remained elusive, and the initial observation that highly hemolytic strains yielded spontaneous mutants with markedly reduced hemolytic activity which had lost dly gene, along with extensive flanking sequences, suggested that this gene might be located on a mobile element (9).
Thin-layer isoelectric focusing assays showed one major and two minor components with hemolytic activity in P. damselae subsp. damselae supernatants (27). These observations suggested that other hemolysins in addition to Dly might be produced by this subspecies. The degree of hemolysis varies among P. damselae subsp. damselae isolates. Two main distinct hemolytic phenotypes could be observed on blood agar plates, with strains showing a large hemolysis halo (LH) and strains producing a small hemolysis halo (SH), although the type strain ATCC 33539 can be described as moderately hemolytic (MH) (7, 9, 16, 30, 42). Early studies demonstrated that the P. damselae subsp. damselae strains showing the highest values of hemolytic activity were also those more virulent for mice (28). In addition, it was later demonstrated that the strongly hemolytic strains against mouse erythrocytes were also those that hybridized to a dly DNA probe (9). Interestingly, both fish and human isolates were represented among the strongly hemolytic and highly virulent strains (28). Actually, previous studies reported that the human isolate CDC2227-81 and the fish isolate RM-71 showed almost identical 50% lethal doses for mice, whereas RM-71 was more virulent for fish than CDC2227-81 (17). Other studies reported that strains lacking dly gene still showed virulence for mice and fish, indicating that dly is not a prerequisite for virulence in this bacterium (16, 30, 42). In addition, the extracellular products of virulent strains regardless of presence of dly gene were lethal for fish and mice, as well as cytotoxic for homeotherm and poikilotherm cell lines (16, 30), suggesting that other virulence factors than Dly might play a role in the pathogenesis of this bacterium, but their nature remains unknown.
A significant diversity in plasmid content has been demonstrated in P. damselae subsp. damselae strains. Some studies reported that highly hemolytic strains harbor a plasmid of ca. 90 to 100 MDa (150 to 170 kb) that is absent in the weakly hemolytic strains (18, 47). Plasmids have been found associated to virulence in P. damselae subsp. piscicida (13), V. anguillarum (12), and V. nigripulchritudo (31), among others.
In the present study we sequenced and characterized pPHDD1, a novel 153-kb plasmid in P. damselae subsp. damselae strain RM-71 that carries dly gene. In addition to Dly, pPHDD1 encodes a pore-forming toxin hemolysin of the HlyA family. We provide evidence that Dly and HlyA contribute to the hemolysis and the virulence of P. damselae subsp. damselae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains used here and those derived from the present study are listed in Tables 1 and 2. The plasmids are listed in Table 1. P. damselae subsp. damselae strains were subjected to standard biochemical tests to corroborate their taxonomic position and PCR tested for subspecies-specific gene markers (43). P. damselae subsp. damselae cells were routinely grown at 25°C on tryptic soy agar supplemented with 1% NaCl (TSA-1) or trypic soy broth supplemented with 1% NaCl (TSB-1). Sheep blood agar plates (Oxoid) were used for conjugative matings and hemolysis assays. Human, rat, and turbot blood was aseptically collected and added to TSA-1 at a final concentration of 5% (vol/vol) to obtain human, rat, and turbot blood agar, respectively. For hemolysis assays on agar plates, a single colony of each strain grown on a TSA-1 plate was picked with the tip of a rounded wooden pick and seeded on the blood agar plate, and pictures were taken after 15 h of incubation at either 25 or 37°C. Escherichia coli strains were routinely grown at 37°C in Luria-Bertani (LB) broth and LB agar, supplemented with antibiotics when appropriate. Antibiotics were used at the following final concentrations: kanamycin at 50 μg ml−1, ampicillin sodium salt at 50 μg ml−1, tetracycline at 4 μg ml−1, gentamicin at 15 μg ml−1, and rifampin at 50 μg ml−1.
Table 1.
Strains and plasmids used and constructed in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| P. damselae subsp. damselae | ||
| RM-71 | Wild type, isolated from turbot (Psetta maxima), strongly hemolytic | 18 |
| AR57 | RM-71 derivate, spontaneous rifampin-resistant mutant; Rfr | This study |
| AR64 | AR57 with in-frame deletion of dly gene | This study |
| AR133 | AR57 with in-frame deletion of hlyA gene | This study |
| AR78 | AR57 with in-frame deletion of dly-hlyA genes | This study |
| AR61 | AR57 with a first cross-over of suicide vector for dly mutant construction; Kmr | This study |
| P. damselae subsp. piscicida | ||
| PC554.2 | Nonhemolytic; Tcr | 35 |
| AR83 | PC554.2 transconjugant that acquired pPHDD1 from AR61 and further selected for suicide plasmid loss | This study |
| E. coli | ||
| XL1-Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| DH5-α | Cloning strain, recA | Laboratory stock |
| S17-1 λpir | recA thi pro ΔhsdR hsdM+ RP4-2-Tc:Mu-Km:Tn7 λpir; Tpr Smr | Laboratory stock |
| Plasmids | ||
| pKD4 | Template for Kmr gene | 11 |
| pNidKan | Suicide vector, derived from pCVD442; Kmr | 39 |
| pHRP309 | lacZ reporter plasmid, mob; Gmr | 44 |
| pWKS30 | Low-copy-number cloning vector; Apr | 51 |
| pACYC184 | Low-copy-number cloning vector; Tcr | Stratagene |
| pAJR38 | pHRP309 with hlyA gene from RM-71 | This study |
| pAJR27 | pWKS30 with hlyA gene from RM-71 | This study |
| pAJR29 | pACYC184 with dly gene from RM-71 | This study |
Rfr, rifampin resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance; Apr, ampicillin resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance; Tpr, trimethoprim resistance.
Table 2.
PCR detection of pPHDD1 gene markers within the P. damselae subsp. damselae strain collectiona
| P. damselae subsp. damselae strain | Source | Presence (+) or absence (–) of various pPHDD1 markers |
Hemolytic halob | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| dly | hlyA | parA | orf2 | repA | vep07 | tolC | tadC | rcpA | |||
| RM-71 | Turbot, Spain | + | + | + | + | + | + | + | + | + | LH |
| RG-91 | Turbot, Spain | + | + | + | – | – | + | + | + | + | LH |
| RG-153 | Turbot, Spain | + | + | + | – | – | + | + | + | + | LH |
| RG-214 | Turbot, Spain | + | + | + | – | – | + | + | + | + | LH |
| CDC2227-81 | Human, United States | + | + | + | – | – | + | + | + | + | LH |
| ATCC 33539 | Damselfish, United States | + | + | + | – | – | + | + | + | + | MH |
| LD-07 | Seabream, Spain | – | – | – | – | – | – | – | – | – | SH |
| 340 | Seawater, Spain | – | – | – | – | – | – | – | – | – | SH |
| 309 | Mussel, Spain | – | – | – | – | – | – | – | – | – | SH |
| 158 | Eel, Belgium | – | – | – | – | – | – | – | – | – | SH |
| 162 | Eel, Belgium | – | – | – | – | – | – | – | – | – | SH |
| PG801 | Shrimp, Taiwan | – | – | – | – | – | – | – | – | – | SH |
| 192 | Dolphin, United States | – | – | – | – | – | – | – | – | – | SH |
| 238 | Dolphin, United States | – | – | – | – | – | – | – | – | – | SH |
| ATCC 35083 | Brown shark, United States | – | – | – | – | – | – | – | – | – | SH |
| J3G801 | Shrimp, Taiwan | – | – | – | – | – | – | – | – | – | NH |
| PC586.1 | Seabream, Spain | – | – | – | – | – | – | – | – | – | SH |
orf2 and repA are gene markers of the two RM-71-specific insertions A and B, respectively.
LH, large hemolytic halo; MH, medium hemolytic halo; SH, small hemolytic halo; NH, no hemolytic halo.
Cosmid library construction, DNA sequencing, and annotation.
Genomic DNA of P. damselae subsp. damselae RM-71 was purified using a genome DNA kit (Qbiogene). DNA was partially digested with Sau3AI and ligated into the compatible BamHI site of alkaline phosphatase-treated SuperCos 1 cosmid vector (Stratagene). The ligated products were packaged into bacteriophage lambda particles using an in vitro packaging kit (Gigapack III Gold packaging extract; Stratagene) and introduced into E. coli XL1-Blue MR cells. The nucleotide sequences of cosmid DNA were determined using a 454 GS-FLX platform (Roche) and assembled using Newbler software (Roche). Sequences were further analyzed with the online BLAST facility of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). Annotation of plasmid was carried out with the RAST server (3), and a plasmid graphical map was generated with the CGView Server (22). G+C content analyses were conducted using the Artemis comparison tool (6). Protein domains were searched using the Pfam database (http://pfam.sanger.ac.uk/). The nucleotide sequence of pPHDD1 determined in the present study was deposited in GenBank under accession number NC_014653.
Plasmid DNA.
Plasmid DNA was extracted from P. damselae subsp. damselae following a modification of a previously described method (54). Cells were harvested from a 1-ml overnight culture in tryptic soy broth. Pelleted cells were resuspended in 300 μl of TENS solution (0.09 N NaOH and 0.45% sodium dodecyl sulfate in Tris-EDTA buffer) plus 170 μl of 3 M sodium acetate (pH 5.2). This mixture was incubated on ice and centrifuged. Plasmid DNA in the supernatant was extracted twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with ethanol, and resuspended in ca. 20 to 40 μl of Tris-EDTA buffer with RNase (20 μg/ml). DNA samples mixed with loading buffer were electrophoresed through 0.7% agarose gels (type II; Sigma). Plasmid molecular sizes were estimated using the reference plasmids from E. coli 39R861 (plasmids of 154, 66.2, 37.6, and 7.4 kb) (48).
Mapping of pPHDD1 replication origin.
A pPHDD1 2,698-bp region (positions 94549 to 97247 in the annotated sequence) containing repB gene (open reading frame 105 [ORF105]) and the putative iteron sequences was PCR amplified with oligonucleotides containing BamHI sites and ligated to the BamHI-cut PCR-amplified kanamycin resistance gene from plasmid pKD4 (11). The ligation reaction was electroporated into E. coli DH5-α cells, and kanamycin-resistant colonies were selected. The plasmid DNA from a kanamycin-resistant colony was purified and subjected to DNA sequencing.
Conjugation.
Conjugations were performed by agar plate matings. Exponential growing cells of donor and recipient strains were mixed, a drop (100 μl) placed directly onto a sheep blood agar plate, followed by incubation at 25°C for 3 days. Cells were scraped off the plate and resuspended in TSB-1, and 100-μl aliquots of serial decimal dilutions were spread onto TSA-1 plates with the corresponding antibiotic combinations to select for donors and transconjugants. The frequency of conjugal transfer, when necessary, was calculated as the number of transconjugants per donor cell.
Mutant construction and gene complementation.
Single and double nonpolar deletions were constructed by using PCR amplification of the amino- and carboxy-terminal fragments of each gene which, when fused together, would result in an in-frame deletion. Amplification was carried out using Hi-Fidelity Kapa Taq (Kapa). Allelic exchange was performed as previously described using the Kmr suicide vector pNidKan, containing sacB gene conferring sucrose sensitivity and R6K ori requiring the pir gene product for replication (39). The plasmid constructions containing the deleted alleles were mated from E. coli S17-1 λpir into a rifampin-resistant derivative (AR57) of P. damselae subsp. damselae RM-71, selecting for kanamycin resistance for plasmid integration and subsequently for sucrose resistance (15% [wt/vol]) for a second recombination event. This led to the obtention of P. damselae subsp. damselae dly, hlyA, and double dly hlyA mutant strains (Table 1). The presence of the correct alleles was confirmed by PCR. For complementation, dly and hlyA ORFs, together with their respective promoter sequences, were PCR amplified with Hi-Fidelity Kapa Taq, cloned into pHRP309 vector and mobilized from E. coli S17-1 λpir into the respective P. damselae subsp. damselae dly and hlyA mutants, as well as into P. damselae subsp. damselae ATCC 33539.
Curation of pPHDD1.
Curation of pPHDD1 was attempted by using RM-71 derivative strains containing a first crossover of the suicide vector constructions used for generation of the hlyA and the dly mutants (Table 1). This approach takes advantage of the inability of the suicide vector pNidKan to replicate in P. damselae subsp. damselae and of its positive (Kmr) and negative (sucrose sensitivity) selectable properties. After several passages on LB without selection, the first crossover strains were grown on LB agar with 15% sucrose, and colonies were tested for kanamycin sensitivity. Colonies that had lost the suicide plasmid were further tested on sheep blood agar plates for hemolysis reduction, as well as for lack of pPHDD1 markers by PCR, until pPHDD1-negative clones were found. As explained below, curation of pPHDD1 was a very rare event that could only be achieved from first crossover strains that had undergone additional mutations or DNA sequence loss that altered the hemolytic phenotype and rendered the strain unable to grow at 37°C.
Mice and fish virulence assays.
Virulence assays were carried on with BALB/c mice (6 to 8 weeks old, 26 to 30 g), as well as with turbot (Psetta maxima) (average weight, 15 g), in groups of five animals. The inoculum was prepared by suspending several colonies from a 24-h TSA-1 culture into saline solution to achieve the turbidity of a no. 2 McFarland standard. Mice were inoculated at the tail vein with 50 μl of a 2.5 μM hemoglobin solution (8 μg hemoglobin per mouse) 2 h before inoculation with the bacterial suspension, as previously described (19). Mice were inoculated intravenously at the tail vein, and turbot were inoculated intraperitoneally, with 0.1 ml of 10-fold serial dilutions of the bacterial suspensions and the actual number of injected CFU was determined by plate count on TSA-1. The final doses assayed corresponded to 2.1 × 106 and 2.1 × 105 bacterial cells per mice and 2.1 × 104 and 2.1 × 103 bacterial cells per fish. The mortalities were recorded daily for 3 days (mice) and 4 days (turbot), and the degree of virulence was expressed as percent values.
RESULTS
Diversity of hemolytic phenotypes in P. damselae subsp. damselae isolates and correlation with dly gene presence.
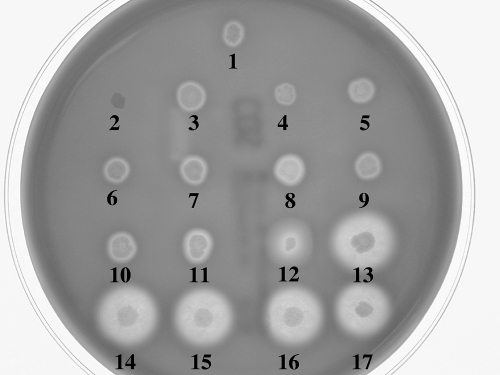
We conducted a hemolysis screening on sheep blood agar for a collection of 17 P. damselae subsp. damselae strains from different origins (Table 2), and four distinct phenotypes were found. Among the 17 strains, 6 showed a large hemolysis halo (LH), one was nonhemolytic (NH), one showed moderate hemolysis (MH), and the remaining 9 strains yielded a small hemolytic halo (SH) on sheep blood agar plates (Fig. 1 and Table 2). In order to assess the correlation between dly gene presence and hemolytic phenotype, we PCR tested the strain collection using specific primers for dly gene (42). We found that the strains positive for dly (strains 12 to 17) ((Fig. 1 and Table 2) showed a hemolytic halo whose radius was ca. 5 to 10 times larger than that produced by the dly-negative strains (Fig. 1). In light of these results, the presence of dly is linked to the ability of P. damselae subsp. damselae to cause a large hemolytic halo on blood agar plates (Fig. 1 and Table 2), whereas the small hemolytic halo is characteristic of strains that lack dly.
Fig. 1.
Hemolytic phenotypes of P. damselae subsp. damselae strains on sheep blood agar. Strains (spot number, strain): 1, PC586.1; 2, J3G801; 3, ATCC 35083; 4, 238; 5, 192; 6, PG801; 7, 162; 8, 158; 9, 309; 10, 340; 11, LD-07; 12, ATCC 33539; 13, CDC2227-81; 14, RM-71; 15, RG-214; 16, RG-153; 17, RG-91. Four distinct phenotypes are recognized: large hemolysis (LH) (strains at spots 13 to 17), moderate hemolysis (MH) (strain 12), small hemolysis (SH) (strains at spot 1 and spots 3 to 11) and no hemolysis (NH) (strain at spot 2). The strains at spots 12 to 17 are positive for pPHDD1 markers.
Damselysin is encoded on a large plasmid in P. damselae subsp. damselae RM-71.
In order to characterize the genetic context of dly, we constructed a cosmid library of the highly hemolytic strain RM-71. This strain was selected because of its strong hemolytic phenotype and because it was reported to be as virulent for mice as the clinical strain CDC2227-81 and more virulent for fish (17). A total of 316 clones were streaked on sheep blood agar and two beta-hemolytic cosmids that tested positive by PCR for dly gene were identified. We found five overlapping cosmids whose 5′ and 3′ end sequences tested negative by PCR in dly-negative strains and accounted for a circular structure. The five cosmids were subjected to 454 DNA sequencing, yielding a 153,429-bp circular molecule that constituted a novel P. damselae subsp. damselae plasmid which was dubbed pPHDD1 and that contains dly gene (Fig. 2). In order to have physical evidence of pPHDD1, plasmid DNA was isolated from three dly-positive strains (RM-71, ATCC 33539, and RG-91) and from one dly-negative isolate (PG801). An ∼150-kb plasmid band (100 MDa) was evident in agarose gels in the three dly-positive strains but not in the dly-negative strain (data not shown).
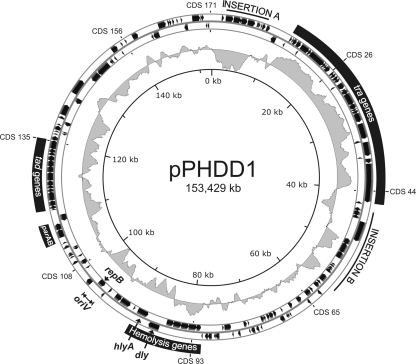
Fig. 2.
Circular representation of the P. damselae subsp. damselae pPHDD1 plasmid. The first circle (i.e., the outermost) represents pPHDD1 ORFs (in black, two rows of arrows corresponding to each of the two DNA strands, respectively), and specific genes, replication origin (oriV) as well as functional modules and insertions are highlighted. The next circle (moving inward) shows the G+C percent variation. The innermost circle shows the nucleotide positions in 20-kb intervals.
Genetic structure of pPHDD1.
The complete sequence of pPHDD1 consists of 153,429 bp and encodes 172 predicted ORFs (Fig. 2). The average G+C content was 37.9%, which is comparable to the G+C content of the P. damselae subsp. damselae reference strain genome (40%). The G+C content distribution is heterogeneous along the plasmid (Fig. 2), varying from 62.5 to 11.6% using a 120-bp window. Five modules can be highlighted in pPHDD1: a replication module, a partitioning module, a conjugation machinery module, a tad (tight adherence) module, and a hemolysin module (Fig. 2).
The nucleotide sequence upstream and downstream of the repB gene shows a high A+T value, and downstream of the repB there are three directed tandem repeats of the 9-mer TAAGATCTA that might correspond to iterons. These data suggest that the region surrounding repB might contain the pPHDD1 replication origin. In support of this, we found that a PCR-amplified 2.6-kb region that included repB, and the putative iteron sequences ligated to a kanamycin resistance gene was capable of independent replication into E. coli DH5-α (data not shown). The putative partitioning module (parA and parB genes) bears similarity to the par genes of Enterobacteriaceae plasmids. No evident addiction system genes, such as toxin/antitoxin genes, were found in pPHDD1 sequence by homology search.
pPHDD1 contains a set of tra genes encoding homologues of proteins of type IV secretion systems described in plasmids of other marine bacteria (Fig. 3A), suggesting that pPHDD1 might be conjugative. In this regard, we were able to detect conjugative transfer of a marked version of this plasmid to P. damselae subsp. piscicida although at a very low frequency (see below). pPHDD1 also contains a complete set of tad (tight adherence) genes that likely encode the biogenesis of the Flp subfamily of pili and show synteny with tad clusters of Vibrio species (Fig. 3B). These pili, initially described in Aggregatibacter actinomycetemcomitans as mediators of a tight-adherence phenotype, are involved in adhesion to abiotic surfaces and host cell attachment (49).
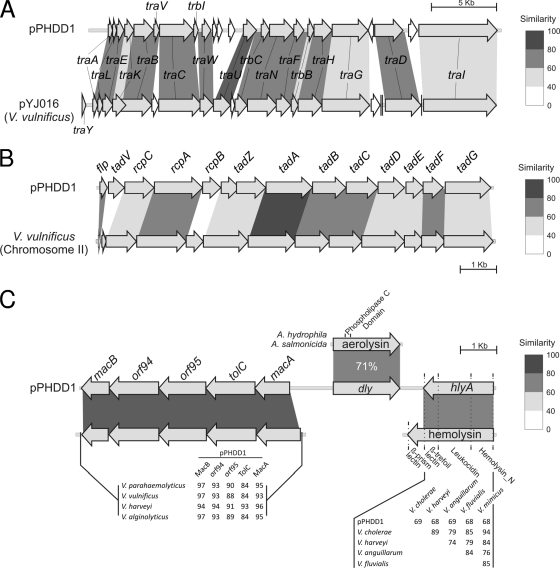
Fig. 3.
Physical maps of tra (A), tad (B), and hemolysin (C) clusters of P. damselae subsp. damselae pPHDD1 plasmid compared to related genes in other species of marine bacteria. pPHDD1 tra genes without counterpart in the species being compared are filled in white. The percent similarity refers to the amino acid sequences of the predicted proteins and is represented in similarity intervals in grayscale tones according to the scale depicted at the right of the figures. In panel C, similarity percent values between pairs of species are shown either in the shaded region (for Dly) or in data matrices.
One of the most notable aspects of pPHDD1 is the presence of a hemolysin cluster that contains five genes of a putative secretion system, as well as two hemolysin genes that encode the previously characterized Dly toxin and a hitherto uncharacterized pore-forming toxin HlyA, respectively (Fig. 3C). Although the activity of Dly was known for the past 2 decades, the complete sequence of dly gene remained unpublished until now. We found that the amino acid sequence of Dly bears little homology to known proteins and shows no conserved domains other than a putative phospholipase C domain. Only two homologues of Dly exist in databases: the phospholipase C aerolysins of Aeromonas hydrophila (37) and A. salmonicida (24), respectively (Fig. 3C). The gene for HlyA lies downstream of dly and is transcribed from the opposite strand. HlyA has similarity with HlyA pore-forming toxins with hemolytic activity described in Vibrio species, with similarity values ranging from 68 to 69% (Fig. 3C). HlyA hemolysins are predicted to form heptameric pore structures into the erythrocyte membrane, altering its permeability (23). A Pfam database search predicted in P. damselae subsp. damselae HlyA three conserved domains that are shared with HlyA of Vibrio cholerae (23, 40, 52) and other Vibrio species, albeit some Vibrio HlyAs contain an additional fourth domain at their C terminus that is absent in P. damselae (Fig. 3C).
The five genes upstream dly encode a putative secretion system that might be involved in the secretion of the two hemolysins. This system includes an inner membrane ATPase component homologous to macB (32), two ATP-binding/permease components, a membrane fusion protein, and TolC, respectively. In E. coli, TolC is specifically required for HlyA secretion (50). Interestingly, highly conserved homologues of these five proteins are found in several Vibrio genomes (Fig. 3C). It is noteworthy that these homologues have chromosomal locations in the Vibrio species and are not closely linked to Dly and HlyA homologues (data not shown).
Distribution and variability of pPHDD1 among P. damselae subsp. damselae isolates.
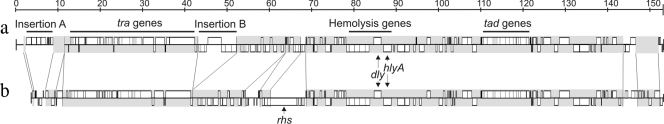
While the present study was under way, the genome sequence of the P. damselae subsp. damselae type strain ATCC 33539 was made available in public databases under several separate contigs (GenBank accession number ADBS00000000). One contig (contig 55) of 149,928 bp showed a high degree of synteny with the pPHDD1 sequence that we report here (strain RM-71). However, we found some differences between these two sequences, pPHDD1 being 3,501 bp larger (Fig. 4). Contig 55 harbors an rhs gene that is absent from pPHDD1, and two DNA regions of pPHDD1 are absent from the ATCC 33539 contig 55. These two DNA regions were dubbed insertion A (6.6 kb) and insertion B (11 kb), respectively (Fig. 2 and 4). Insertion A has a G+C content lower than the average of the plasmid and comprises 10 truncated ORFs that constitute pseudogenes. Insertion B comprises 11 predicted ORFs, two of them encoding distinct phage integrases with low homology to Shewanella integrases. Interestingly, we found that insertion B is unstable since a spontaneous deletion of a large part of it was observed in a transconjugant P. damselae subsp. piscicida clone that received pPHDD1 by conjugative transfer (see below). To get an insight into the molecular mechanism underlying this loss of DNA sequences, the region involved in the excision was PCR amplified and sequenced. We found that the excised region corresponded to a 9,202-bp sequence flanked by a perfect 12-bp DNA direct repeat (CGTGGGGTGTCA). It is tempting to speculate that this direct repeat might be the target of one of the integrases encoded by orf51 and orf52.
Fig. 4.
Conservation of synteny between P. damselae subsp. damselae RM-71 pPHDD1 plasmid (a) and contig 55 from P. damselae subsp. damselae ATCC 33539 (GenBank accession no. NZ_ADBS01000004) (b). Conserved blocks of synteny between the two sequences are indicated in gray. Vertical and diagonal lines connect regions of synteny that are separated by insertions specific to one of the two molecules. Two main pPHDD1 insertions (insertions A and B), functional modules (tra, tad, and hemolysis clusters), and gene names (rhs, dly, and hlyA) are highlighted.
In order to get an insight into the genetic diversity among pPHDD1-like plasmids, we conducted a PCR screening of seven interspersed pPHDD1 gene markers—dly, hlyA, parA, vep07, tolC, and the two Tad genes rcpA and tadC—in the collection of P. damselae subsp. damselae strains. Of the 17 strains tested, 6 gave a positive result for the seven markers, whereas the remaining 11 strains tested negative (Table 2). These results suggest the genetic linkage between the assayed genes and show the existence of differential distribution of pPHDD1-like plasmids among strains of this pathogen. Moreover, the three strains in which physical presence of pPHDD1 was demonstrated by gel electrophoresis (see above) tested positive for the markers, whereas PG801 strain tested negative, confirming the direct relationship between the presence of the ∼150-kb plasmid band and pPHDD1 markers. As expected, we found a correlation between presence of pPHDD1 markers and the production of a large or moderate hemolytic halo in the strains that had previously tested positive for the dly gene by PCR (strains 12 to 17) (Fig. 1, Table 2). When we PCR tested two pPHDD1 genes (repA and orf2) that are part of the two DNA insertions A and B, respectively, all of the strains except RM-71 yielded a negative result (Table 2), indicating that insertions A and B are unique to strain RM-71.
pPHDD1 confers hemolytic activity to P. damselae subsp. piscicida upon conjugative transfer.
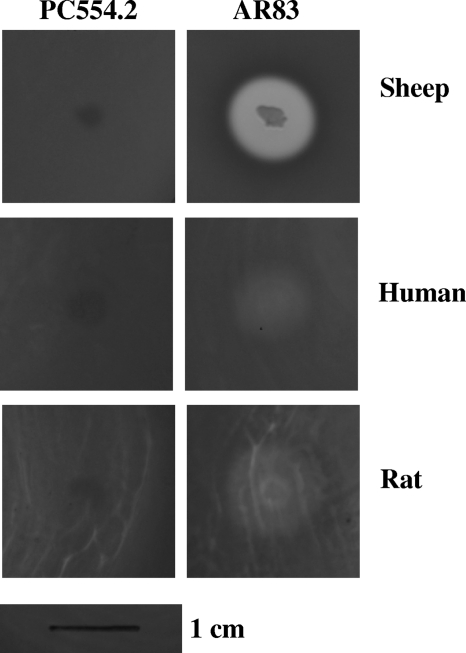
Further evidence of the pPHDD1 involvement in the hemolysis activity of P. damselae could be gained by attempting to transfer pPHDD1 by conjugation into a strain of the nonhemolytic subspecies P. damselae subsp. piscicida and analyzing the phenotypic changes in the recipient. As described above, pPHDD1 encodes a set of tra genes, suggesting that this plasmid is conjugative. We mated the kanamycin-labeled P. damselae subsp. damselae AR61 (Table 1) as a donor and the tetracycline-resistant P. damselae subsp. piscicida PC554.2 as a recipient. As a result, tetracycline/kanamycin-resistant P. damselae subsp. piscicida transconjugants were isolated at a low frequency of ∼10−8. These transconjugants showed a reduced hemolysis due to the fact that they contained the insertion of the suicide plasmid within the hlyA gene. We therefore selected for suicide plasmid loss on sucrose plates and searched for clones that had lost the suicide vector but that restored the wild-type gene. Doing this, we isolated the kanamycin-sensitive P. damselae subsp. piscicida AR83. This strain tested positive by PCR for eight pPHDD1 gene markers, as well as for several subsp. piscicida-specific gene markers (data not shown). This indicates that pPHDD1 is an independent replicon capable of undergoing conjugative transfer. We found that AR83 produced hemolysis on sheep blood agar plates (Fig. 5), although the halo was ca. 63% of that produced by subsp. damselae parental strain AR57 (rifampin-resistant derivative of RM-71). The hemolysis caused by AR83 on human and rat blood agar plates was very weak compared to AR57, suggesting that either additional factors non-pPHDD1 encoded are involved in the production of the strong hemolytic phenotype showed by AR57 or that the expression of the hemolytic determinants in subsp. piscicida cells does not achieve the optimal conditions.
Fig. 5.
Hemolytic activity in sheep, human, and rat blood agar plates of P. damselae subsp. piscicida PC554.2 and AR83 (the transconjugant that acquired pPHDD1 after conjugative transfer). Scale bar, 1 cm.
HlyA and Dly contribute to hemolysis in P. damselae subsp. damselae.
To date, the hemolytic activity of P. damselae subsp. damselae has been explained exclusively on the basis of dly gene, having been reported a direct relationship between highly hemolytic P. damselae subsp. damselae strains and the presence of the dly gene (9). However, our finding of an HlyA family hemolysin gene in pPHDD1 plasmid raises the question whether Dly is the only responsible of hemolysis or whether HlyA also plays a role. Based on previously described homologues in vibrios, the P. damselae subsp. damselae HlyA is predicted to be a protein with hemolytic activity. In order to unravel the contribution of Dly and HlyA, we analyzed single dly (AR64), single hlyA (AR133), and double dly hlyA (AR78) knockout mutants.
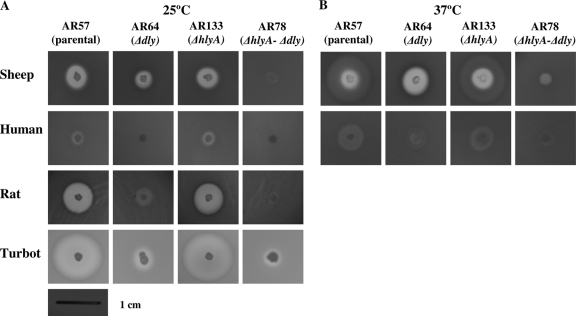
Since P. damselae subsp. damselae is a marine bacterium that causes disease in marine animals but is also known to cause opportunistic disease and death in humans, we evaluated the hemolytic phenotype on homeotherm (sheep, human, and rat) and poikilotherm (turbot) blood sources. Hemolysis was assayed at 25°C for the four blood types (Fig. 6A) and also at 37°C in two homeotherm blood types (Fig. 6B). We observed that each erythrocyte source had a different susceptibility to be hemolyzed by the parental strain. Turbot and rat erythrocytes, followed by sheep blood, showed the largest hemolysis halos at 25°C after 15 h, whereas human blood showed low susceptibility. Interestingly, when the assays were carried out at 37°C, a larger but less translucent halo was observed, and this halo disappeared in the dly mutants, which suggested that it is due to Dly (Fig. 6B).
Fig. 6.
Hemolytic activity in sheep, human, rat, and turbot blood agar plates of P. damselae subsp. damselae parental strain and mutants at 25°C (A) and 37°C (B). Strains: AR57 (parental strain), AR64 (Δdly), AR133 (ΔhlyA), AR78 (Δdly ΔhlyA double mutant). Pictures were taken after 15 h of growth. Scale bar, 1 cm.
Although different results were observed with the dly mutant depending on the erythrocyte source, we found that mutation of dly did not completely abolish hemolysis in any of the four erythrocyte sources. This indicates that Dly is not the only cause of the hemolytic phenotype in pPHDD1-harboring strains, although it clearly contributes to the production of a phenotype of large hemolytic halo (LH). We therefore wanted to test the contribution of hlyA to hemolysis. Interestingly, mutation of hlyA caused only a slight reduction in the radius of the hemolysis halo on the four blood sources assayed. Based on these results we can propose that hemolysis in P. damselae subsp. damselae is mainly due to the sum of the contributions of Dly and HlyA. In order to demonstrate this hypothesis, we assayed the effect of the deletion of the two hemolysis genes. As expected, the hlyA dly double mutant (AR78) showed a >80% reduction in the hemolytic halo on sheep, rat, human, and turbot blood, which decreased to levels similar to those observed for the pPHDD1-negative strains on sheep erythrocytes (Fig. 6; see also Fig. 1). These results suggest the existence of an additive effect between Dly and HlyA to produce hemolysis and also suggest that Dly is a major contributor to hemolysis on rat, human, and turbot erythrocytes.
In order to gain additional information on the contribution of Dly and HlyA to the hemolytic phenotype and to assess whether the two hemolysins show a synergistic as well as an additive effect, we assayed the hemolytic phenotype conferred by each individual gene and by the two genes together to E. coli DH5-α. Interestingly, E. coli cells harboring either hlyA or dly gene showed small hemolytic halos on sheep blood agar plates (Fig. 7A), the halo of the hlyA gene being more translucent than that conferred by dly. Interestingly, when the two genes were introduced into E. coli, the hemolytic halo produced was significantly larger than the mere addition of the two individual halos (Fig. 7A). This result suggests the existence of a synergistic effect between the two hemolysins.
Fig. 7.
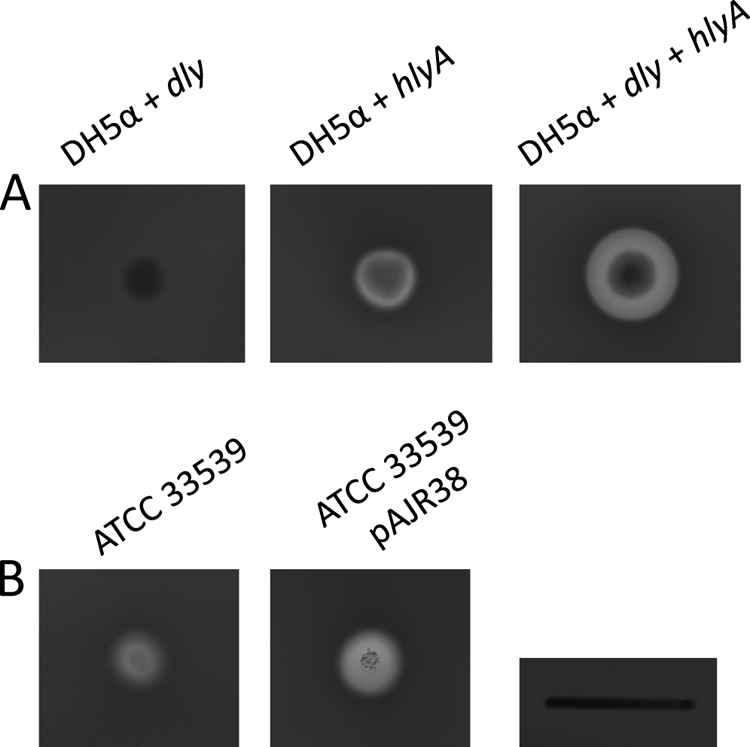
(A) Complementation of E. Coli DH5-α with dly and hlyA genes of strain RM-71, cloned in plasmids pAJR29 and pAJR27. (B) Complementation of P. damselae subsp. damselae ATCC 33539 with the hlyA gene of strain RM-71, cloned in plasmid pAJR38. Scale bar, 1 cm.
It is interesting that the type strain ATCC 33539 is positive for pPHDD1 markers, while it only shows moderate hemolysis (MH) on sheep blood agar, whereas all of the other pPHDD1-positive strains show a large hemolysis (LH) halo (Fig. 1). We compared the nucleotide sequences of hlyA genes between RM-71 and ATCC 33539 and found a number of amino acid substitutions (data not shown). Hence, we hypothesized that ATCC 33539 might produce a protein with reduced hemolytic activity. We therefore conjugally transferred into ATCC 33539 a plasmid (pAJR38) containing the cloned hlyA gene from RM-71 and found a restoration of the hemolytic phenotype at levels similar to those of RM-71 (Fig. 7B). This observation suggests that the moderate hemolytic halo of the type strain could be in part due to substitutions in the ATCC 33539 HlyA sequence with respect to RM-71 HlyA.
We tried to cure pPHDD1 from AR57 derivatives containing the first crossover insertions of the suicide plasmids used to obtain the dly and hlyA mutants (see Materials and Methods), but all of the attempts were unsuccessful. We finally obtained to some extent a first crossover insertion for the hlyA mutant construction that, when selecting for loss of the complete pPHDD1-suicide plasmid cointegrate, yielded cured colonies at a very high frequency (data not shown). However, we noted that this first crossover insertion was unable to grow at 37°C and showed a considerable reduction in the hemolytic activity that could not be restored by complementation with dly and hlyA (data not shown). Thus, curation of pPHDD1 could only be selected under circumstances that likely involved the loss of other genomic sequences or the occurrence of spontaneous mutations elsewhere on the genome.
Contribution of dly and hlyA to P. damselae subsp. damselae virulence for mice and fish.
Dly has been recognized as a cytolytic toxin with lethal activity for mice. We assayed the roles of dly and hlyA on the ability of P. damselae subsp. damselae to cause death in mice after tail vein inoculation, using two doses of 2.1 × 106 and 2.1 × 105 CFU per mouse. Mice died within 12 to 48 h postinfection, and P. damselae subsp. damselae was recovered from the spleen and liver as pure cultures. The parental strain caused death in 100% of mice inoculated with 2.1 × 106 cells and in 60% of mice inoculated with 2.1 × 105 cells (Fig. 8A). However, we found that all of the mutants showed some degree of reduction in their virulence. Interestingly, although the single ΔhlyA and dly mutants still maintained the ability to kill 60% of mice with the high dose, we found that the dly mutant was unable to cause death at the lower dose. Since the hemolytic activity results on blood agar suggested the existence of both an additive and synergistic effect between Dly and HlyA, we wanted to test the effect that mutation of the two hemolysin genes had on virulence. We found that the hlyA dly double mutant caused the death of only one animal at the high dose, suggesting that a synergistic effect between Dly and HlyA is necessary for maximal virulence in mice.
Fig. 8.
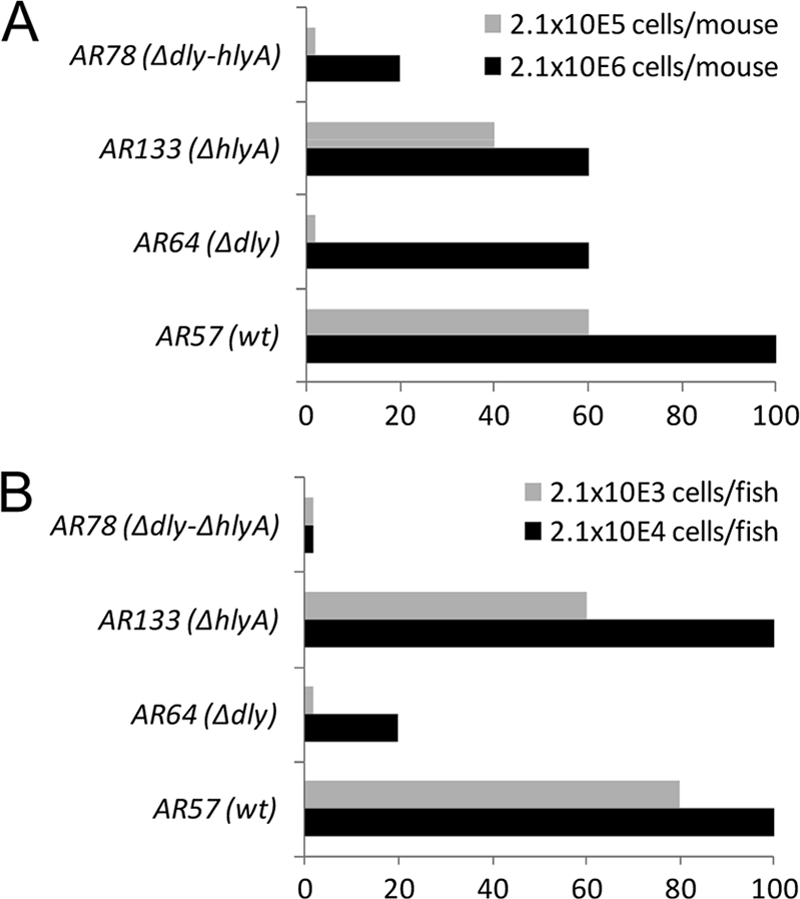
Mouse (A) and fish (B) virulence assays with P. damselae subsp. damselae strains, using two different doses. The results are expressed as the percent mortality.
Since P. damselae subsp. damselae RM-71 was isolated from diseased turbot, we also assayed the effect of dly and hlyA mutations on the virulence for fish by intraperitoneal inoculation, using two doses of 2.1 × 104 and 2.1 × 103 CFU per fish. The fish died within 24 to 72 h postinfection, and P. damselae subsp. damselae was recovered from the kidney as pure cultures. The parental strain caused death in 100% of fish inoculated with 2.1 × 104 cells and in 80% of fish inoculated with 2.1 × 103 cells (Fig. 8B). The ΔhlyA mutant only showed a slight reduction in virulence at the lower dose but maintained the same virulence values of the parental strain at the high dose. However, the Δdly mutant only killed 20% of fish at the high dose and no fish at the low dose. No fish deaths were recorded with the hlyA dly double mutant. These results suggest not only that Dly and HlyA play a synergistic effect in virulence for fish but also that Dly is the main contributor of the two hemolysins in P. damselae subsp. damselae virulence for fish. Based on the results obtained with the single and double mutants, it is clear that plasmid pPHDD1 is necessary for maximal virulence in mice and fish infected with P. damselae subsp. damselae.
DISCUSSION
Strains of P. damselae subsp. damselae have been isolated from aquatic environments and as causative agents of disease in a variety of aquatic animals and humans (41). Despite recent reports of fatal human cases due to this bacterium (2, 53), knowledge of the genetic basis of virulence of this bacterium remained quite limited to early studies on Dly toxin. The virulence of P. damselae subsp. damselae in mice has been previously correlated with the ability to produce Dly (28, 29). Similarly, the symptoms caused by this bacterium in fish have been related to the ability to produce extracellular products, which included phospholipase and hemolysin activities (16). The genomic location of Dly gene remained elusive until now. We found that dly is encoded on pPHDD1 in strain RM-71, a novel 153-kb plasmid of P. damselae subsp. damselae. In accordance with our findings, previous studies reported that all of the strongly hemolytic isolates contained a ca. 150- to 170-kb plasmid, but not the weakly hemolytic ones, with the exception of ATCC 33539 that contained a plasmid but was moderately hemolytic (18, 47). Like many large plasmids, pPHDD1 appears to have a mosaic-like structure due to modular evolution processes in which DNA sequence stretches are acquired by horizontal gene transfer and reorganized by general recombination, transposition, and site-specific recombination (14, 15). In this sense, it is noticeable that different pPHDD1 modules show similarity to plasmid-borne genes from different bacterial taxa. Although the tra and tad genes bear similarity to Vibrio plasmids, the par genes are highly similar to the sequences of Enterobacteriaceae plasmids.
Tad clusters are involved in the pathogenesis of several bacteria (8, 49). Interestingly, this is the first report of a plasmid-borne Tad cluster in a member of the Vibrionaceae, which suggests that conjugative transfer is one of the mechanisms for Tad cluster genes spread in the marine environment. We found that pPHDD1 can be mobilized to P. damselae subsp. piscicida but at a very low frequency. The reasons for this low transfer rate are unknown. Possible explanations include plasmid incompatibility or entry exclusion mechanisms due to residing plasmids or the necessity for SOS response induction or any environmental stress signal in order to trigger conjugative transfer (4). Although no evident addiction systems were found in pPHDD1 by protein homology searches, curation of this plasmid was only achieved as a rare event that likely involved the occurrence of additional mutations or DNA loss elsewhere in the genome. The reasons for this resistance to curation, as well as the mutations that likely allow pPHDD1 to be lost from cells, are currently unknown.
The existence of two main hemolytic categories among P. damselae subsp. damselae strains has been largely reported, with a clear distinction between strongly hemolytic and weakly hemolytic strains on blood agar plates (7, 30, 42). Southern blot analysis revealed that the dly gene was found only in highly hemolytic strains (9). In accordance with these previous observations, in the present study we found a direct correlation between pPHDD1 and the dly gene and the production of large hemolytic halos on sheep blood agar.
A variety of erythrocyte sources from homeotherms have been reported to be sensitive to P. damselae subsp. damselae cells and extracellular products (28). In addition, it was known that turbot erythrocytes were sensitive to the extracellular products of P. damselae subsp. damselae cells (16). To date, hemolysis caused by P. damselae subsp. damselae was explained exclusively in terms of Dly, although previous studies had suggested the possibility that other hemolysins might be produced (27). Our study has brought into play a hitherto-unknown factor, the pore-forming toxin HlyA. Our experiments with single and double mutants demonstrated that Dly is not only responsible for the strong hemolytic phenotype in P. damselae subsp. damselae. Rather, we found that both Dly and HlyA contribute to hemolysis. Interestingly, since it can be concluded from the results with the Δdly mutant, Dly makes a differential contribution to hemolysis according to the erythrocyte source, being the major contributor in rats, turbot, and humans. This selectivity might be explained by the different lipid compositions of erythrocyte membranes of each species (25). The lipid composition of mammal erythrocyte membranes is almost identical, with the exception of sphingomyelin being partially replaced equimolarly by phosphatidylcholine in various species and having a direct effect on membrane thermostability (25). That Dly toxin shows phospholipase D activity against sphingomyelin (29) might explain the different degrees of hemolysis observed in the ΔhlyA and Δdly mutants depending on the blood source tested. In addition, a possible connection between sphingomyelin content, Dly-mediated hemolysis, and temperature cannot be ruled out. Since sphingomyelin plays a role in membrane thermostability, the removal of sphingomyelin choline head groups by Dly might explain the wider turbid hemolytic halo observed at 37°C in parental and ΔhlyA strains (the two producing Dly) with respect to the observed halos at 25°C.
Although the hypothesis of an additive effect between Dly and HlyA might explain most of the hemolytic halos observed in the different mutant combinations and blood sources, there is also evidence that Dly and HlyA may act in a synergistic manner. The experiments carried out in E. coli suggest that the effect of the two hemolysins being produced at the same time in the same cell is stronger than the mere addition of their individual contributions. Synergistic effects between hemolysins have been well documented (5, 33, 46). An explanation of how Dly and HlyA interact synergistically can be drawn from the data available for V. cholerae cytolysin VCC. This hemolysin has specificity for cholesterol, which is essential for oligomerization and pore formation (56). It has been proposed that the choline head group of sphingomyelin has an inhibitory effect on VCC pore formation since it shields the cholesterol ring from VCC, constituting the so-called umbrella model (55, 56). This model might be applied to the synergistic effect between Dly and HlyA in P. damselae subsp. damselae, where Dly's removal of the choline head group from sphingomyelin would allow HlyA to enhance its hemolytic activity.
The finding that pPHDD1-negative strains produce weak hemolytic halos, together with the observation that the double mutant still produces small halos, clearly suggests the existence of additional hemolysins non-pPHDD1 encoded in P. damselae subsp. damselae strains. In this regard, previous studies had reported the existence of one major and two minor components with hemolytic activity in P. damselae subsp. damselae supernatants by thin-layer isoelectric focusing assays (27). An in silico analysis of the P. damselae subsp. damselae ATCC 33539 genome reveals the existence of two genes annotated as encoding putative hemolysin-like proteins (GenBank accession no. ADBS00000000; ORFs VDA_003208 and VDA_002420). These putative hemolysins might be responsible for the basal small hemolytic halos observed both in the double mutant and in the pPHDD1-negative strains, and further studies are necessary to ascertain the role of these proteins in hemolysis. Similarly, the observation that a transconjugant P. damselae subsp. piscicida that received pPHDD1 produced smaller hemolytic halos than did the subsp. damselae parental strain might also be partly explained by the existence of non-pPHDD1-encoded hemolysins that contribute to hemolysis.
Our results demonstrate that pPHDD1 is necessary for full virulence of P. damselae subsp. damselae for mice and fish. The double mutant, lacking pPHDD1-encoded hlyA and dly genes, caused death in only 20% mice at the high dose and 0% of fish at the two doses assayed, whereas the two single mutants proved to be more virulent than the double mutant. We found that mutation of hlyA had less effect on virulence than mutation of dly in the two animal models. This observation was consistent with the results obtained in the hemolysis assays, in which the hlyA mutant produced larger hemolysis halos than the dly mutant. The contribution to virulence of these two genes together was higher than the sum of their individual contributions, which suggests that a synergy between Dly and HlyA toxin is necessary for maximal virulence.
We have found that pPHDD1 is widespread in P. damselae subsp. damselae strains isolated from diseased marine fish. Thus, strains of this pathogen inhabiting the aquatic environments and infecting poikilotherm animals contain virulence factors that might be of potential concern for human health. Future comparative studies of the genomes of dly-positive and plasmidless P. damselae subsp. damselae strains isolated from mammals and poikilotherms will help to elucidate the key features that allow this marine bacterium to cause disease and explain how pPHDD1 constituted a driving force for the emergence of the highly hemolytic lineage of P. damselae subsp. damselae.
ACKNOWLEDGMENTS
This study was partially supported by grant INCITE08PXIB235028PR from Xunta de Galicia and by grants AGL2009-12266-C02-01 and CSD2007-00002 (Consolider Aquagenomics)—both cofunded by the FEDER Programme from the European Union—from the Ministry of Science and Innovation (MICINN) of Spain. A.J.R. is the recipient of an FPI fellowship from MICINN.
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Alvarez J. R., Lamba S., Dyer K. Y., Apuzzio J. J. 2006. An unusual case of urinary tract infection in a pregnant woman with Photobacterium damsela. Infect. Dis. Obstet. Gynecol. 80682:1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asato J., Kanaya F. 2004. Fatal infection of the hand due to Photobacterium damsela: a case report. Clin. Infect. Dis. 38:e100–e101 [DOI] [PubMed] [Google Scholar]
- 3. Aziz R. K., et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beaber J. W., Hochhut B., Waldor M. K. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74 [DOI] [PubMed] [Google Scholar]
- 5. Bernheimer A. W., Linder R., Avigad L. S. 1980. Stepwise degradation of membrane sphingomyelin by corynebacterial phospholipases. Infect. Immun. 29:123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carver T. J., et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 7. Clarridge J. E., Zighelboim-Daum S. 1985. Isolation and characterization of two hemolytic phenotypes of Vibrio damsela associated with a fatal wound infection. J. Clin. Microbiol. 21:302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clock S. A., Planet P. J., Perez B. A., Figurski D. H. 2008. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cutter D. L., Kreger A. S. 1990. Cloning and expression of the damselysin gene from Vibrio damsela. Infect. Immun. 58:266–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daniel L. W., King L., Kennedy M. 1988. Phospholipase activity of bacterial toxins. Methods Enzymol. 165:298–301 [DOI] [PubMed] [Google Scholar]
- 11. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Lorenzo M., et al. 2003. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J. Bacteriol. 185:5822–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. do Vale A., et al. 2005. AIP56, a novel plasmid-encoded virulence factor of Photobacterium damselae subsp. piscicida with apoptogenic activity against sea bass macrophages and neutrophils. Mol. Microbiol. 58:1025–1038 [DOI] [PubMed] [Google Scholar]
- 14. Erauso G., et al. 2011. Evidence for the role of horizontal transfer in generating pVT1, a large mosaic conjugative plasmid from the clam pathogen, Vibrio tapetis. PLoS One 6:e16759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernandez-Lopez R., et al. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942–966 [DOI] [PubMed] [Google Scholar]
- 16. Fouz B., Barja J. L., Amaro C., Rivas C., Toranzo A. E. 1993. Toxicity of the extracellular products of Vibrio damsela isolated from diseased fish. Curr. Microbiol. 27:341–347 [Google Scholar]
- 17. Fouz B., Conchas R. F., Magariños B., Toranzo A. E. 1992. Vibrio damsela strain virulence for fish and mammals. FHS/AFS Newsl. 20:3–4 [Google Scholar]
- 18. Fouz B., Larsen J. L., Nielsen B., Barja J. L., Toranzo A. E. 1992. Characterization of Vibrio damsela strains isolated from turbot Scophthalmus maximus in Spain. Dis. Aquat. Org. 12:155–156 [Google Scholar]
- 19. Fouz B., Toranzo A. E., Biosca E. G., Mazoy R., Amaro C. 1994. Role of iron in the pathogenicity of Vibrio damsela for fish and mammals. FEMS Microbiol. Lett. 121:181–188 [DOI] [PubMed] [Google Scholar]
- 20. Fujioka R. S., Greco S. B., Cates M. B., Schroeder J. P. 1988. Vibrio damsela from wounds in bottlenose dolphins Tursiops truncatus. Dis. Aquat. Org. 4:1–8 [Google Scholar]
- 21. Gauthier G., et al. 1995. Small-subunit rRNA sequences and whole DNA relatedness concur for the reassignment of Pasteurella piscicida (Snieszko et al.) Janssen and Surgalla to the genus Photobacterium as Photobacterium damsela subsp. piscicida comb. nov. Int. J. Syst. Bacteriol. 45:139–144 [DOI] [PubMed] [Google Scholar]
- 22. Grant J. R., Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36:181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He Y., Olson R. Three-dimensional structure of the detergent-solubilized Vibrio cholerae cytolysin (VCC) heptamer by electron cryomicroscopy. J. Struct. Biol. 169:6–13 [DOI] [PubMed] [Google Scholar]
- 24. Hirono I., Aoki T. 1993. Cloning and characterization of three hemolysin genes from Aeromonas salmonicida. Microb. Pathog. 15:269–282 [DOI] [PubMed] [Google Scholar]
- 25. Ivanov I. T. 2007. Allometric dependence of the life span of mammal erythrocytes on thermal stability and sphingomyelin content of plasma membranes. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 147:876–884 [DOI] [PubMed] [Google Scholar]
- 26. Kim H. R., Kim J. W., Lee M. K., Kim J. G. 2009. Septicemia progressing to fatal hepatic dysfunction in a cirrhotic patient after oral ingestion of Photobacterium damsela: a case report. Infection 37:555–556 [DOI] [PubMed] [Google Scholar]
- 27. Kothary M. H., Kreger A. S. 1985. Purification and characterization of an extracellular cytolysin produced by Vibrio damsela. Infect. Immun. 49:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kreger A. S. 1984. Cytolytic activity and virulence of Vibrio damsela. Infect. Immun. 44:326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kreger A. S., Bernheimer A. W., Etkin L. A., Daniel L. W. 1987. Phospholipase D activity of Vibrio damsela cytolysin and its interaction with sheep erythrocytes. Infect. Immun. 55:3209–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Labella A., et al. 2010. Toxicity of Photobacterium damselae subsp damselae strains isolated from new cultured marine fish. Dis. Aquat. Org. 92:31–40 [DOI] [PubMed] [Google Scholar]
- 31. Le Roux F., et al. 2011. Virulence of an emerging pathogenic lineage of Vibrio nigripulchritudo is dependent on two plasmids. Environ. Microbiol. 13:296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin H. T., et al. 2009. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J. Biol. Chem. 284:1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linder R. 1984. Alteration of mammalian membranes by the cooperative and antagonistic actions of bacterial proteins. Biochim. Biophys. Acta 779:423–435 [DOI] [PubMed] [Google Scholar]
- 34. Love M., et al. 1981. Vibrio damsela, a marine bacterium, causes skin ulcers on the damselfish Chromis punctipinnis. Science 214:1139–1140 [DOI] [PubMed] [Google Scholar]
- 35. Magariños B., Romalde J. L., Lopez-Romalde S., Morinigo M. A., Toranzo A. E. 2003. Pathobiological characterisation of Photobacterium damselae subsp. piscicida isolated from cultured sole (Solea senegalensis). Bull. Eur. Assoc. Fish Pathol. 23:183–190 [Google Scholar]
- 36. Magariños B., Toranzo A. E., Romalde J. L. 1996. Phenotypic and pathobiological characteristics of Pasteurella piscicida. Annu. Rev. Fish Dis. 6:41–64 [Google Scholar]
- 37. Merino S., et al. 1999. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67:4008–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris J. G., Jr., et al. 1982. Illness caused by Vibrio damsela and Vibrio hollisae. Lancet i:1294–1297 [DOI] [PubMed] [Google Scholar]
- 39. Mouriño S., Osorio C. R., Lemos M. L. 2004. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186:6159–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olson R., Gouaux E. 2003. Vibrio cholerae cytolysin is composed of an alpha-hemolysin-like core. Protein Sci. 12:379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osorio C. R., Lemos M. L. 2011. Photobacterium, p. 959–968. In Liu D. (ed.), Molecular detection of human bacterial pathogens. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 42. Osorio C. R., Romalde J. L., Barja J. L., Toranzo A. E. 2000. Presence of phospholipase D (dly) gene coding for damselysin production is not a pre-requisite for pathogenicity in Photobacterium damselae subsp. damselae. Microb. Pathog. 28:119–126 [DOI] [PubMed] [Google Scholar]
- 43. Osorio C. R., Toranzo A. E., Romalde J. L., Barja J. L. 2000. Multiplex PCR assay for ureC and 16S rRNA genes clearly discriminates between both subspecies of Photobacterium damselae. Dis. Aquat. Org. 40:177–183 [DOI] [PubMed] [Google Scholar]
- 44. Parales R. E., Harwood C. S. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram- bacteria. Gene 133:23–30 [DOI] [PubMed] [Google Scholar]
- 45. Pedersen K., Dalsgaard I., Larsen J. L. 1997. Vibrio damsela associated with diseased fish in Denmark. Appl. Environ. Microbiol. 63:3711–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ripio M. T., Geoffroy C., Dominguez G., Alouf J. E., Vazquez-Boland J. A. 1995. The sulfhydryl-activated cytolysin and a sphingomyelinase C are the major membrane-damaging factors involved in cooperative (CAMP-like) hemolysis of Listeria spp. Res. Microbiol. 146:303–313 [DOI] [PubMed] [Google Scholar]
- 47. Takahashi H., et al. 2008. Difference of genotypic and phenotypic characteristics and pathogenicity potential of Photobacterium damselae subsp. damselae between clinical and environmental isolates from Japan. Microb. Pathog. 45:150–158 [DOI] [PubMed] [Google Scholar]
- 48. Threlfall E. J., Rowe B., Ferguson J. L., Ward L. R. 1986. Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J. Hyg. (Lond.) 97:419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomich M., Planet P. J., Figurski D. H. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363–375 [DOI] [PubMed] [Google Scholar]
- 50. Wandersman C., Delepelaire P. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. U. S. A. 87:4776–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang R. F., Kushner S. R. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 52. Yamamoto K., Wright A. C., Kaper J. B., Morris J. G., Jr 1990. The cytolysin gene of Vibrio vulnificus: sequence and relationship to the Vibrio cholerae El Tor hemolysin gene. Infect. Immun. 58:2706–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamane K., et al. 2004. Two cases of fatal necrotizing fasciitis caused by Photobacterium damsela in Japan. J. Clin. Microbiol. 42:1370–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou C., Yang Y., Jong A. Y. 1990. Mini-prep in ten minutes. Biotechniques 8:172–173 [PubMed] [Google Scholar]
- 55. Zitzer A., et al. 2001. Coupling of cholesterol and cone-shaped lipids in bilayers augments membrane permeabilization by the cholesterol-specific toxins streptolysin O and Vibrio cholerae cytolysin. J. Biol. Chem. 276:14628–14633 [DOI] [PubMed] [Google Scholar]
- 56. Zitzer A., Zitzer O., Bhakdi S., Palmer M. 1999. Oligomerization of Vibrio cholerae cytolysin yields a pentameric pore and has a dual specificity for cholesterol and sphingolipids in the target membrane. J. Biol. Chem. 274:1375–1380 [DOI] [PubMed] [Google Scholar]