Abstract
The DARC (Duffy antigen/receptor for chemokines) gene, also called Duffy or FY, encodes a membrane-bound chemokine receptor. Two malaria parasites, Plasmodium vivax and Plasmodium knowlesi, use DARC to trigger internalization into red blood cells. Although much has been reported on the evolution of DARC null alleles, little is known about the evolution of the coding portion of this gene or the role that protein sequence divergence in this receptor may play in disease susceptibility or zoonosis. Here, we show that the Plasmodium interaction domain of DARC is nearly invariant in the human population, suggesting that coding polymorphism there is unlikely to play a role in differential susceptibility to infection. However, an analysis of DARC orthologs from 35 simian primate species reveals high levels of sequence divergence in the Plasmodium interaction domain. Signatures of positive selection in this domain indicate that species-specific mutations in the protein sequence of DARC could serve as barriers to the transmission of Plasmodium between primate species.
Keywords: malaria, positive selection, arms race, species tropism, zoonosis
The DARC gene (Duffy antigen/receptor for chemokines), also called Duffy or FY, encodes a membrane-bound chemokine receptor. Different human alleles of this gene underlie the designation of different Duffy blood groups (Meny 2010), but DARC also plays a role in the biology of malaria. In humans, the most severe form of malaria is caused by Plasmodium falciparum, with less severe forms of the disease caused by the related organisms Plasmodium knowlesi, Plasmodium vivax, Plasmodium ovale, and Plasmodium malariae. Two of these, P. vivax and P. knowlesi, exploit DARC for internalization into red blood cells. Interaction with DARC is mediated by a Plasmodium surface ligand called Duffy-binding protein (DBP) (Haynes et al. 1988; Wertheimer and Barnwell 1989). A cis-regulatory polymorphism that silences DARC expression in erythrocytes has arisen independently in human populations from different parts of the world and is highly protective against P. vivax and P. knowlesi infection (Miller et al. 1975, 1976; Tournamille et al. 1995; Zimmerman et al. 1999). In a fascinating example of convergent evolution, polymorphisms in the cis-regulatory region of DARC in African baboons are also associated with resistance to a malaria-like parasite common in baboon populations, Hepatocystis kochi (Tung et al. 2009). Although much has been reported on the evolution of DARC regulatory elements, little is known about the evolution of the coding portion of this gene or the role that protein sequence divergence in this receptor may play in disease susceptibility or zoonosis.
Lack of Human Diversity in the Plasmodium Interaction Domain
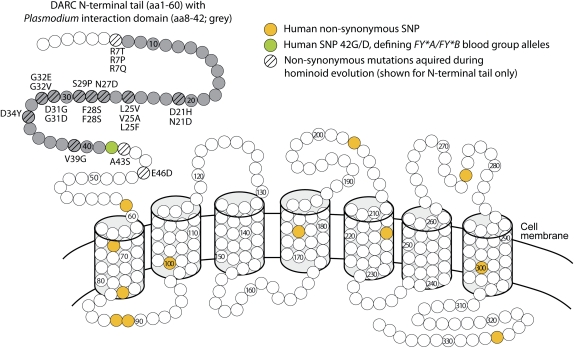
The N-terminal extracellular tail of DARC interacts with the DBP of P. vivax and P. knowlesi (fig. 1) and also contains some of the determinants for chemokine binding (Chitnis et al. 1996; Tournamille et al. 2003, 2005). In mutagenesis studies, several mutations in this domain have been shown to destroy the interaction with P. vivax while not interfering with chemokine binding (Tournamille et al. 2003, 2005). Given that P. vivax is the most widely distributed malaria parasite in the world (Mendis et al. 2001), it is intriguing that polymorphisms at these sites have not been reported in the human population. In fact, we find that no nonsynonymous single-nucleotide polymorphisms have ever been reported in the portion of DARC encoding the Plasmodium interaction domain (fig. 1). The only exception to this is the common polymorphism at position 42, which defines the FY*A and FY*B Duffy blood group alleles (Meny 2010). Thus, polymorphism in the coding sequence of DARC is unlikely to play a role in differential susceptibility of humans to P. knowlesi and P. vivax, since most human genomes will encode an identical DARC N-terminal tail. Destabilizing mutations elsewhere in the protein which convey reduced levels of DARC expression at the cell surface will still be important, and one such human polymorphism (R89C) is known (Meny 2010).
FIG. 1.
Divergence but not diversity shapes the Plasmodium interaction domain of DARC. A model of DARC embedded in the cell membrane is shown (Tournamille et al. 2003). In yellow and green are residue positions where a human nonsynonymous single-nucleotide polymorphism (SNP) has been reported (dbSNP and 1000 Genomes databases). The N-terminal tail contains the interaction domain with Plasmodium vivax and Plasmodium knowlesi (Chitnis et al. 1996). Labeled next to residues in the N-terminal tail are the nonsynonymous mutations predicted to have occurred during hominoid evolution by free-ratio analysis. F28S occurred twice, on separate branches.
Divergence of DARC Protein Sequence during Primate Evolution
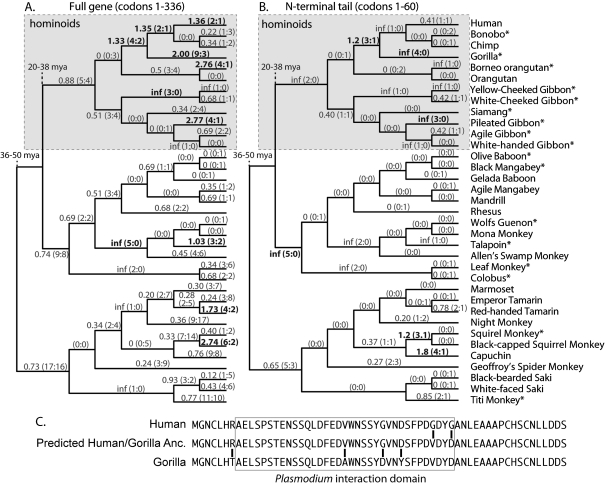
Plasmodium vivax and P. knowlesi, as well as P. falciparum, are all known to be zoonotic transmissions from nonhuman primates (Escalante et al. 2005; Mu et al. 2005; Liu et al. 2010; Lee et al. 2011). Furthermore, species-specific susceptibility of primate cells to P. vivax and P. knowlesi is well documented (Wertheimer and Barnwell 1989). There is evidence to suggest that this is at least partly attributable to species-specific differences in the DARC protein sequence. For example, the protein sequence of rhesus macaque DARC conveys a special glycosylation pattern that blocks binding of the P. vivax DBP (Chitnis et al. 1996). To better understand how the DARC protein varies between primates, we sequenced DARC from 17 simian primate species, including nine hominoid species (great apes and gibbons) closely related to humans. Primate cell lines were obtained and grown under standard conditions (supplementary table S1, Supplementary Material online), and polymerase chain reaction was performed on genomic DNA harvested from these cells (supplementary table S2, Supplementary Material online). The coding sequence of each ortholog was assembled from genomic sequence based on the structure of the human isoform “b” transcript, most of which (990 bases) is encoded by a single exon, with 21 bases coming from an upstream exon (Iwamoto et al. 1996). These novel sequences (starred in fig. 2; Genbank accessions JN544123–JN544139) were combined with 18 DARC sequences available in Genbank to create a final data set of 35 full-length simian primate DARC orthologs. The alignment of these sequences was straightforward with a single codon deletion being the only indel present, and their phylogenetic relationship was in agreement with accepted primate phylogeny (Perelman et al. 2011).
FIG. 2.
Divergence of DARC during simian primate speciation. dN/dS was calculated along each branch over either (A) the entire gene length or (B) the first 60 codons, which encode the N-terminal tail. Actual numbers of nonsynonymous and synonymous mutations predicted to have occurred along each branch are indicated in parentheses (N:S). dN/dS is indicated as “inf” (infinity) where dS = 0 and in bold where dN/dS > 1 (or, arbitrarily, where N:S ≥ 3:0 in cases where dS = 0). Asterisks indicate sequences acquired as part of this study. (C) An alignment of the N-terminal tail sequence of human and gorilla DARC, along with their predicted ancestral sequence. Vertical lines represent nonsynonymous mutations acquired since that ancestor.
We used the free-ratio model, implemented in PAML (Yang 1997), to calculate the dN/dS value on each branch of the DARC phylogeny. This ratio summarizes the accumulation of nonsynonymous and synonymous mutations, with dN representing the number of nonsynonymous mutations per nonsynonymous site and dS representing the number of synonymous mutations per synonymous site (Hurst 2002). It is well appreciated that, over evolutionary time, most protein coding genes accumulate nonsynonymous mutations at a rate far slower than synonymous mutations (dN/dS ≪ 1) (Meyerson and Sawyer 2011). However, on several branches DARC has accumulated nonsynonymous mutations at a rate as fast or faster than synonymous changes (dN/dS ≥ 1) (bold type; fig. 2A).
This pattern was also observed when the portion encoding the N-terminal tail of DARC was analyzed alone (fig. 2B), a region where high sequence divergence between primate orthologs has previously been noted (Tournamille et al. 2004). In these 60 codons, four nonsynonymous mutations have accumulated along the branch leading to gorilla from its last common ancestor with humans, whereas zero synonymous mutations became fixed during this time. An alignment of the human and gorilla N-terminal tail is shown along with the predicted sequence of DARC that existed at the time of their last common ancestor, illustrating these four nonsynonymous mutations (R7T, V25A, G31D, D34Y; fig. 2C). To illustrate the extent of mutation that has occurred in this N-terminal tail, all nonsynonymous mutations predicted to have occurred during the evolution of our closest ancestors, the hominoids, are diagrammed in figure 1. This diagram illustrates that there has been a constellation of divergent N-terminal tails encoded by primate genomes during their evolution. Divergence in this tail probably does not affect chemokine binding, as different primate orthologs of DARC are all able to bind a relevant human chemokine (Tournamille et al. 2004), but could potentially have affected Plasmodium susceptibility by modulating the interaction with DBP. If so, species-specific features of the DARC protein sequence could impede the movement of Plasmodium between primate species.
Residues under Positive Selection in the Plasmodium Interaction Domain
We tested the hypothesis that nonsynonymous mutations in DARC can confer an adaptive fitness advantage, regardless of what that might be. We did this by assessing whether DARC bears the signatures of positive natural selection for such mutations. Codon-based models in PAML were used to analyze the 35-species data set. Positive selection of DARC was supported (P ≤ 0.03) under all models of codon evolution (supplementary table S3, Supplementary Material online), which was verified with the program HyPhy (supplementary table S4, Supplementary Material online). We noticed that DARC evolution appeared to be least conservative in the hominoids (boxed in fig. 2). This could either be due to relaxed selection or to more acute positive selection in these species. However, positive selection of DARC was also supported in a codon-based analysis of only the hominoid species (P ≤ 0.003; supplementary table S5, Supplementary Material online). This result is more significant than it was with the full data set, even though fewer species and reduced tree length should reduce statistical power (Anisimova et al. 2001). A branch-sites analysis also supports hominoid-specific positive selection (P < 0.001; supplementary table S6, Supplementary Material online).
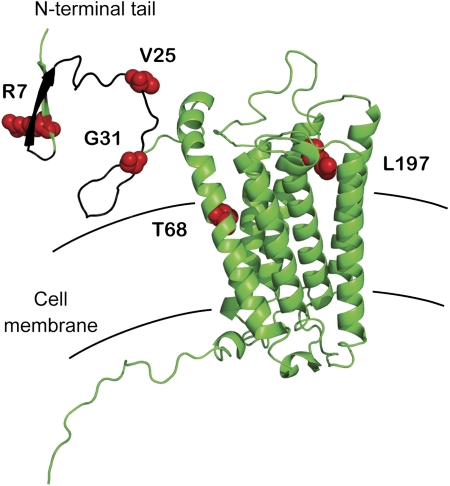
Five codons, R7, V25, G31, T68, and L197 (human coordinates), were identified in these analyses as belonging to the class of codons with dN/dS >1 (supplementary tables S3–S6, Supplementary Material online). Residues V25 and G31 fall within the Plasmodium interaction domain of DARC (comprised of residues 8–42), whereas residue R7 sits directly adjacent (fig. 3). Even though no other pathogen is currently known to interact with the N-terminal tail of DARC, it is possible that the rapid evolution of these codons may have been driven by a selective pressure other than Plasmodium. However, it has been demonstrated that mutating V25 to alanine disrupts the interaction between DARC and the P. vivax DBP without affecting chemokine binding (Tournamille et al. 2003, 2005), making Plasmodium a plausible selective force to explain the rapid evolution of at least some of these residues in DARC. Interestingly, nature has also made this very mutation (fig. 2C). Gorillas encode an alanine at this residue position, suggesting that P. vivax would not be capable of invading gorilla cells. Such a mutation could potentially serve as a species-specific barrier to infection.
FIG. 3.
Three residues under positive selection map to the N-terminal extracellular tail. A structural model of human DARC (de Brevern et al. 2005) is shown, with sites under positive selection indicated in red (human coordinates). The domain that interacts with the Plasmodium vivax and Plasmodium knowlesi ligand (residues 8–42) is colored black (Chitnis et al. 1996).
Although the zoonotic transmissions of P. vivax and P. knowlesi from Asian macaques are estimated to have occurred less than 250,000 years ago (Escalante et al. 2005; Mu et al. 2005; Lee et al. 2011), the signatures of positive selection that we observe are consistent with a far more ancient evolutionary “arms race” between Plasmodium and DARC, one that has lasted on the order of tens of millions of years (Meyerson and Sawyer 2011). In support of an arms race model, the DBP of P. vivax also shows evidence for positive selection (Baum et al. 2003). The stronger signal of positive selection of DARC observed in the hominoid clade may be evidence for a more intense evolutionary battle in these species, our closest relatives. Indeed, evidence exists that African apes are currently under intense pressure, as sampling projects in wild chimpanzees, bonobos, and gorillas have uncovered P. vivax variants and closely related Plasmodium species beyond those known to infect humans (Kaiser et al. 2010; Krief et al. 2010; Liu et al. 2010). Understanding the host genetics that define species tropism of these pathogens is extremely important, both for the development of primate model systems for studying malaria and for understanding how zoonotic processes give rise to new human diseases.
Supplementary Material
Supplementary tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We wish to thank Dianne Lou, Nick Meyerson, and Paul Rowley for helpful comments on the manuscript. This work was supported by grants (to S.L.S.) from the Norman Hackerman Advanced Research Program (003658-0250-2009) and the National Institutes of Health (R01-GM-093086). A.D. is supported by an American Cancer Society Postdoctoral Fellowship. S.L.S. holds a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and is an Alfred P. Sloan Research Fellow in Computational and Evolutionary Molecular Biology.
References
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Baum J, Thomas AW, Conway DJ. Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics. 2003;163:1327–1336. doi: 10.1093/genetics/163.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med. 1996;184:1531–1536. doi: 10.1084/jem.184.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brevern AG, Wong H, Tournamille C, Colin Y, Le Van Kim C, Etchebest C. A structural model of a seven-transmembrane helix receptor: the Duffy antigen/receptor for chemokine (DARC) Biochim Biophys Acta. 2005;1724:288–306. doi: 10.1016/j.bbagen.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA. A monkey's tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci U S A. 2005;102:1980–1985. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes JD, Dalton JP, Klotz FW, McGinniss MH, Hadley TJ, Hudson DE, Miller LH. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J Exp Med. 1988;167:1873–1881. doi: 10.1084/jem.167.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Iwamoto S, Li J, Omi T, Ikemoto S, Kajii E. Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood. 1996;87:378–385. [PubMed] [Google Scholar]
- Kaiser M, Löwa A, Ulrich M, et al. (15 co-authors) Wild chimpanzees infected with 5 Plasmodium species. Emerg Infect Dis. 2010;16:1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Escalante AA, Pacheco MA, et al. (18 co-authors) On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-S, Divis PCS, Zakaria SK, Matusop A, Julin RA, Conway DJ, Cox-Singh J, Singh B. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7:e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li Y, Learn GH, et al. (22 co-authors) Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- Meny GM. The Duffy blood group system: a review. Immunohematology. 2010;26:51–56. [PubMed] [Google Scholar]
- Meyerson NR, Sawyer SL. Two-stepping through time: mammals and viruses. Trends Microbiol. 2011;19:286–294. doi: 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- Mu J, Joy DA, Duan J, et al. (11 co-authors) Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, et al. (14 co-authors) A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournamille C, Blancher A, Le Van Kim C, Gane P, Apoil PA, Nakamoto W, Cartron J-P, Colin Y. Sequence, evolution and ligand binding properties of mammalian Duffy antigen/receptor for chemokines. Immunogenetics. 2004;55:682–694. doi: 10.1007/s00251-003-0633-2. [DOI] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron J-P, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- Tournamille C, Filipe A, Badaut C, Riottot M-M, Longacre S, Cartron J-P, Le Van Kim C, Colin Y. Fine mapping of the Duffy antigen binding site for the Plasmodium vivax Duffy-binding protein. Mol Biochem Parasitol. 2005;144:100–103. doi: 10.1016/j.molbiopara.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Tournamille C, Filipe A, Wasniowska K, Gane P, Lisowska E, Cartron J-P, Colin Y, Le Van Kim C. Structure-function analysis of the extracellular domains of the Duffy antigen/receptor for chemokines: characterization of antibody and chemokine binding sites. Br J Haematol. 2003;122:1014–1023. doi: 10.1046/j.1365-2141.2003.04533.x. [DOI] [PubMed] [Google Scholar]
- Tung J, Primus A, Bouley AJ, Severson TF, Alberts SC, Wray GA. Evolution of a malaria resistance gene in wild primates. Nature. 2009;460:388–391. doi: 10.1038/nature08149. [DOI] [PubMed] [Google Scholar]
- Wertheimer SP, Barnwell JW. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol. 1989;69:340–350. doi: 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zimmerman PA, Woolley I, Masinde GL, et al. (11 co-authors) Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc Natl Acad Sci U S A. 1999;96:13973–13977. doi: 10.1073/pnas.96.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.