Abstract
Salinity stress is one of the major factors negatively affecting growth and productivity in living organisms including plants and bacteria resulting in significant losses worldwide. Therefore, it would be fruitful to develop salinity stress tolerant useful species and also to understand the mechanism of stress tolerance. The pea DNA helicase 45 (PDH45) is a DNA and RNA helicase, homologous to eukaryotic translation initiation factor 4A (eIF-4A) and is involved in various processes including protein synthesis, maintaining the basic activities of the cell, upregulation of topoisomerase I activity and salinity stress tolerance in plant, but its role in salinity stress tolerance in bacteria has not heretofore been studied. This study provides an evidence for a novel function of the PDH45 gene in high salinity (NaCl) stress tolerance in bacteria (Eschericia coli, BL21 cells) also. Furthermore, it has been shown that the functionally active PDH45 gene is required to show the stress tolerance in bacteria because the single mutants (E183G or R363Q) and the double mutant (E183G + R363Q) of the gene could not confer the same function. The response was specific to Na+ ions as the bacteria could not grow in presence of LiCl. This study suggests that the cellular response to high salinity stress across prokaryotes and plant kingdom is conserved and also helps in our better understanding of mechanism of stress tolerance in bacteria and plants. It could also be very useful in developing high salinity stress tolerant useful bacteria of agronomic importance. Overall, this study provides an evidence for a novel function of the PDH45 gene in high salinity stress tolerance in bacteria.
Key words: bacteria, cellular stress response, PDH45, pea, plant DNA helicase, salinity stress
Introduction
The helicases are enzymes that catalyze the unwinding of energetically stable duplex DNA (DNA helicases) or duplex RNA secondary structures (RNA helicases). Most of the helicases contain nine conserved motifs (Q, I, Ia, Ib, II–VI) in their protein sequence.1–3 Helicases might be playing an important role in stabilizing growth in plants under stress by regulating stress-induced transcription and translation.4 Earlier it was shown that mRNA of pea DNA helicase 45 (PDH45) is induced in pea seedlings in response to high salinity and its overexpression in tobacco plants confers salinity tolerance, suggesting a new role of a plant helicase in stress tolerance.5 The PDH45 is homologous to eukaryotic translation initiation factor (eIF-4A) which is a prototypic member of the DEAD-box family.6,7 PDH45 is a unique member of this family, containing DESD and SRT motifs instead of DEAD and SAT in motif V and VI, respectively.6 The homolog of PDH45 in Medicago sativa was also reported to play important role in salt and drought tolerance in Arabidopsis.8 The overall properties of PDH45 suggest that it could be an important multifunctional protein involved in protein synthesis, maintaining the basic activities of the cell, upregulation of topoisomerase I activity and abiotic stress response.5,6 Though plants may have unique stress adaptive mechanisms against various stresses, but the basic cellular stress response (CSR) mechanisms are similar in prokaryotes, lower eukaryotes and plants.9 CSR may target a special set of cellular functions, including chromatin stabilization, DNA repair and removal of damaged proteins, cell cycle control, protein chaperoning and assembly, and few other aspects of cellular metabolism.9 As the cellular adaptive mechanisms are conserved, therefore, it is possible that overexpression of plant stress responsive genes in bacteria may also provide stress tolerance to them. Earlier, it has already been reported that overexpression of certain plant stress-induced genes confer increased stress tolerance in bacteria.10–14 To know whether PDH45 also provides salinity stress tolerance in bacteria, we have overexpressed the PDH45 gene in bacteria and checked the growth of transformed bacteria under salinity stress. Furthermore, we have also created single and double mutations in the conserved motifs II and VI of PDH45 gene and then overexpressed the mutant gene in bacteria and compared the growth of the bacteria transformed with mutant and non-mutant PDH45 genes.
Results
The schematic representation of all the nine conserved motifs along with conserved sequence of motif II (VLDESD) and motif VI (HRIGRSGR) of PDH45 are shown in Figure 1A. Based on the characteristics of conserved motifs, two highly conserved amino acids in motifs II and VI were selected for substitution. In motif number II (DESD box), glutamic acid (E) at position 184 was changed to glycine (G) (E183G) and in motif number VI the second arginine (R) at position 363 was changed to glutamine (Q) (R363Q) using PCR based site directed mutagenesis.
Figure 1.
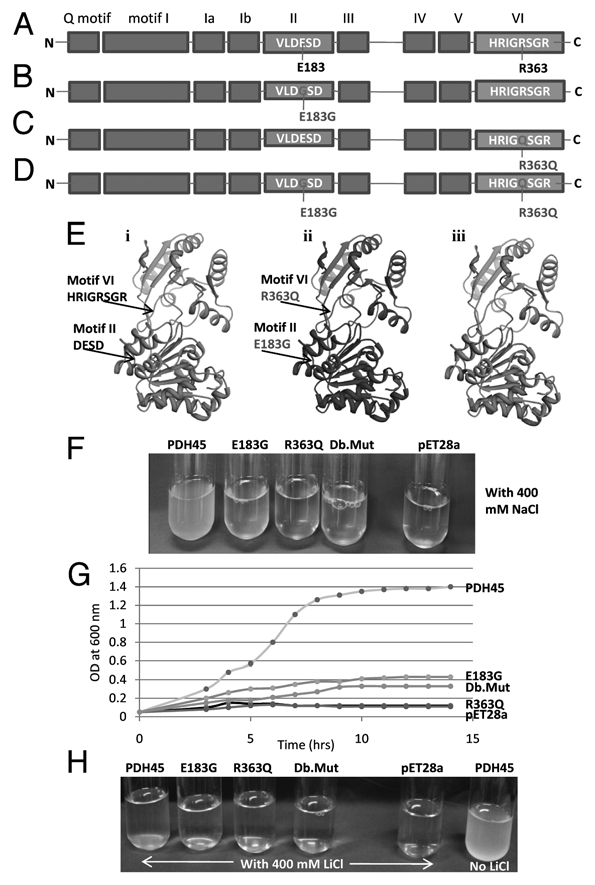
Site directed mutagenesis in two conserved domains of PDH45. (A) Schematic diagram shows nine conserved motifs of PDH45 and the sequence of two motifs (II and VI) on which mutations have been created is shown. (B) Schematic diagram of single mutant E185G. (C) Schematic diagram of single mutant R363Q. (D) Schematic diagram of double mutant E185G and R363Q. (E) Structure modeling and motifs selected for site directed mutagenesis of PDH45 protein. PDH45 sequence was submitted to Swissmodel server and the structure was obtained. The molecular graphic images were produced using the UCSF Chimera package from the resource for Biocomputing, Visualization and Informatics (www.cgl.ucsf.edu/chimera) at the University of California, San Francisco (supported by NIH P41 RR-01081). (i) Wild type PDH45; (ii) Double mutant PDH45; (iii) Superimposed image. Motif (II and VI) have been shown in wild type and mutant structures. (F and G) High Salinity (NaCl) stress response of PDH45 and its mutants in E. coli. A comparison of growth curves of E. coli (BL21) cells transformed with pET28a or pET28a-PDH45 or pET28a-mutant PDH45 genes in the presence of 400 mM NaCl reveals better survival ability of the cells containing pET28-PDH45. (F) The tubes picture shows the bacterial growth of all four constructs, single mutants (E183G and R263Q), Db.Mut, (double mutant, E183G + R363Q) and empty vector after 12 h. (G) The growth curves under salinity stress of all experimented samples reveals better survival ability of the E. coli (BL21) cells transformed with pET28a-PDH45. (H) High LiCl salt stress response of PDH45 and its mutants in E. coli. The E. coli (BL21) cells transformed with pET28a or pET28a-PDH45 or pET28a-mutant PDH45 genes in the presence of 400 mM LiCl reveals no survival of the bacterial cells in any case (first 5 tubes). The last tube shows the growth of the PDH45 gene transformed bacterial cells without LiCl.
Preparation of single and double mutants of PDH45 gene.
The single mutants (E183G or R363Q) and double mutant (E183G + R363Q) were prepared as described in materials and methods. All the PDH45 genes (mutated and non-mutated) were amplified with primers containing desired restriction sites and were first cloned in pGEM-T easy vector followed by cloning in pET28a vector. The sequencing result of the cloned single mutant (E183G) PDH45 gene confirmed the substitution of GAG (code for amino acid E) to GGA (code for amino acid G) in the PDH45 gene. Schematic representation of single mutant E185G is shown in Figure 1B. The sequencing result of the other cloned single mutant (R363Q) PDH45 gene confirmed the substitution of CGG (code for amino acid R) to CAA (code for amino acid Q) in the PDH45 gene. Schematic representation of single mutant R363Q is shown in Figure 1C. The sequencing result of double mutant (E183G + R363Q) also confirmed that clone PDH45 contains both the mutations. Schematic representation of double mutant E185G and R363Q is shown in Figure 1D.
Homology based modeling of mutated and nonmutated PDH45 genes.
For structural modeling the sequence of PDH45 was submitted to the Swissmodel homology-modeling server (swissmodel.expasy.org/).15 The structural model obtained was based on the crystal structure of a DEAD-box protein.16 Molecular graphic images were produced using the UCSF Chimera package (www.cgl.ucsf.edu/chimera) from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).17 The motif II (DESD) and motif VI (HRIGRSGR) which have been selected to create mutations in PDH45 are shown in Figure 1E-i. Figure 1E-ii shows the exact location of the selected amino acids in the double mutant PDH45. The merged picture of non-mutated and mutated PDH45 shows no significant difference (Fig. 1E-iii).
Salinity (NaCl) stress response of mutated and non-mutated PDH45 in E. coli bacteria.
The E. coli (BL21 cells) containing PDH45-pET28 and three different mutated constructs separately, were subjected to NaCl salt stress. The BL21 cells were grown to log phase OD600 = 0.5 and 1 mM IPTG was added to the culture to induce the expression of proteins. Along with IPTG, cells were grown with 400 mM NaCl concentration (Fig. 1F). The growth of the cells after 12 h was much better for wild type PDH45 (non-mutated) as compared with the single (E183G or R363Q) and double (E183G + R363Q) mutants (Fig. 1F). It is interesting to note that very little growth was also observed in case of E183G mutant.
A growth curve was made by taking OD600 of the bacterial culture after every 1 h interval till it reached the stationary phase (Fig. 1G). The comparison of the growth curves of the BL21 cells containing wild type PDH45-pET28a (non-mutated) with those containing mutated PDH45-pET28a and empty vector pET28a as a negative control showed that the wild type PDH45 was able to help E. coli tolerate salinity stress, while cells containing mutated proteins were not able to tolerate salinity stress condition. Under salinity stress condition, the mutated gene in vector and empty vector pET28a containing BL21 cells reached the stationary phase earlier (within 3–5 h) as compared with the wild type PDH45-pET28a containing BL21 cells, which grew linearly even till 8 h of stress (Fig. 1G).
Na+ ions-specific stress response of mutated and nonmutated PDH45 in E. coli bacteria.
In order to check if the stress response is specific to sodium ions (Na+), we have used another salt LiCl also. The E. coli (BL21 cells) containing PDH45-pET28 and three different mutated constructs separately, were subjected grow in presence of 400 mM LiCl salt concentration as described above. The results are shown in Figure 1H, which clearly show that the transformed BL21 bacterial cells with mutated (single, E183G or R363Q, and double E183G + R363Q mutants) or nonmutated PDH45 gene and vector (pET28a) alone could not grow in presence of 400 mM NaCl (first 5 tubes). The experiment was repeated with less concentration of LiCl (200 mM) and still the results were same, i.e., the bacterial cells could not grow (data not shown). However, PDH45 transformed cells could easily grow without the LiCl (Fig. 1H and last tube).
Discussion
In all living systems including bacteria the stress tolerance mechanism exists. Out of various stresses, the salinity stress is one of the major stresses that negatively affect the living organism including plants and bacteria.18,19 In response to the sub-lethal conditions, plants and other organisms activate the CSR mechanism which controls important cellular functions and thereby helps them to withstand otherwise non-permissive hostile growth conditions.9 Since the cellular adaptive mechanisms against the stress including the salinity stress are conserved across the prokaryotes and eukaryotes, therefore, it can be assumed that plant stress genes can be screened functionally by their overexpression in simple organisms.13,14 Through microarray studies in plants, it has been suggested that differences in stress tolerance are mainly due to a higher constitutive expression of several stress-related genes rather than de novo gene transcription.20,21 It is known that functionally analogous stress tolerant genes exist in both unicellular organisms and plants.22 In the present study, we have identified the novel function of PDH45 in salinity stress tolerance in bacteria with an unknown mechanism.
PDH45 has been included in a group designated helicase superfamily II (SFII) based on conservation of amino acid sequence in nine distinct motifs. These motifs represent sites of functional significance that have been evolutionarily conserved. Because of the presence of motif II (DEAD), they are also called as DEAD/H-box helicases family of proteins.23 Each of these conserved motifs has important characteristics. The motif II is also known as ‘Walker motif B’.24 Previous studies demonstrated that DEAD box motif has role in helicase and ATPase activity.25 The DE of DEAD sequence is highly conserved and is also present in many proteins, which play a role in DNA and RNA replication.26 The residue D of DE has been shown to interact with Mg2+, which is required for binding ATP.
The motif VI is suggested to be involved in interaction with RNA. This is also a part of the ATP-binding cleft and is also involved in coupling between helicase and NTPase activities of the protein.3 Mutation in motif VI leads to defects in the nucleic acid binding.27 Point mutation in motif VI of E. coli UvrD DNA has a negative impact on ATP hydrolysis and ligand induced conformational changes.3 Changing the basic residues H or R to the uncharged glutamine abolishes RNA binding and reduces ATP hydrolysis, which also results in reduced helicase activity.
In plants PDH45 has been shown to play important role in salinity stress tolerance,5 but to check whether the same gene can also provide the salinity stress tolerance in bacteria or not was the aim of this study. In this study we observed that overexpression of PDH45 also confers the salinity stress tolerance in the bacteria (E. coli; BL21 cells). In order to know whether the functionally active gene is required to show the stress tolerance we have created two single and a double mutation in the PDH45 gene and then checked their stress tolerance in bacteria. We observed that the mutant PDH45 gene does not provide the salinity stress tolerance in the same bacteria. These findings confirmed that the functionally active gene is required for the stress tolerance in bacteria. However, it remains to be tested whether these mutations affect the activities of the PDH45 protein or whether expression of these mutant genes will confer durable resistance to high salinity tolerance in plants. This stress response was specific to Na+ ions as the transformed bacterial cells could not grow if the NaCl was replaced with LiCl. Overall, this study indicates the first direct evidence for the role of PDH45 in promoting the salinity stress tolerance in bacteria which could be very useful in developing high salinity stress tolerant useful bacteria of agronomic importance. Therefore, this study may have a great application in agricultural biotechnology. Since the same gene (PDH45) can provide the salinity stress tolerance in both the plant5 and bacteria (this study), therefore, this study shall provide a significant contribution for our better understanding of mechanism of stress tolerance in bacteria and plants, which is not very well understood. These observations also give us new insights into its application in the plant biotechnology and microbiology fields and can provide a new tool for developing stress tolerant crop plants and bacteria.
Materials and Methods
Site-directed mutagenesis.
The detail of the PDH45 gene (Accession number Y17186) is described earlier in reference 6. Desired point mutations were created in PDH45 gene using specific designed forward and reverse primers (E183G-PDH45 and R363Q-PDH45, Table 1).
Table 1.
Primer sequences
| Name of the Primer | Sequence | Length (bases) |
| E183G-PDH45F | 5′-CAA GTT ACT AGT TCT GGA TGG ATC TGA TGA AAT GTT GAG CAG-3′ | 42 |
| E183G-PDH45R | 5′-CTG CTC AAC ATT TCA TCA GAT CCA TCC AGA ACT AGT AAC TTG-3′ | 42 |
| R363Q-PDH45F | 5′-GTA CAT TCA TCG GAT TGG TCA ATC TGG ACG TTT TGG ACG AAA G-3′ | 43 |
| R363Q-PDH45R | 5′-CTT TCG TCC AAA ACG TCC AGA TTG ACC AAT CCG ATG AAT GTA C-3′ | 43 |
| PDH45-F (NdeI) | 5′-AGA GGC ATA TGG CGA CAA CTT CTG TGG-3′ | 27 |
| PDH45-R (XhoI, EcoRI) | 5′-GAG GAA TTC CTC GAG TTA TAT AAG ATC ACC AAT ATT C-3′ | 37 |
E183G single mutation in DESD-box motif (motif II) of PDH45.
The QuikChange site-directed mutagenesis kit (Stratagene Cloning Systems, La Jolla, CA USA) was used to create the mutation. The 1.2 kb E183G mutated PDH45 DNA fragment was prepared by PCR using normal PDH45 gene (cloned in pGEMT easy vector) and primers (E183G-PDH45F or E183G-PDH45R, Table 1) with desired point mutations designed from PDH45 gene. Finally the E183G mutated 1.2 kb (1,224 bp) fragment was cloned in pGEM-T easy vector and sequenced to confirm the E183G mutation. The sequencing result of the cloned gene shows substitution of GAG (code for amino acid E) to GGA (code for amino acid G) in the PDH45 gene. Schematic representation of single mutant E185G is shown in Figure 1B.
R363Q single mutation in HRIGRSGR motif (motif VI) of PDH45.
The 1.2 kb R363Q mutated PDH45 DNA fragment was prepared by PCR using normal PDH45 gene (cloned in pGEMT easy vector) and primers (R363Q-PDH45F or R363QPDH45R, Table 1) with desired point mutation designed from PDH45 gene. Finally the R363Q mutated 1.2 kb (1,224 bp) fragment was cloned in pGEM-T easy vector and sequenced to confirm the R363Q mutation. The sequencing result of the cloned gene shows substitution of CGG (code for amino acid R to CAA) (code for amino acid Q) in the PDH45 gene. Schematic representation of single mutant R363Q is shown in Figure 1C.
E183G and R363Q double mutation in PDH45 gene.
Double mutants construct created by using E183G mutant gene in pGEM-T easy vector as a template and primers (R363Q-PDH45F or R363Q-PDH45R, Table 1) in two separate tubes for the first PCR and the second PCR with mixture of products from first PCR and gene specific primers (PDH45-F and PDH45-R, Table 1). Finally the E183G and R363Q mutated 1.2 kb (1,224 bp) fragment was cloned in pGEM-T easy vector and sequenced to confirm these mutations. The sequencing result confirmed that clone PDH45 gene contains both the mutations. Schematic representative of double mutant E185G and R363Q is shown in Figure 1D.
Homology based modeling of mutated and non-mutated PDH45 genes.
This modeling was done as described in results.
Cloning of mutated and non-mutated PDH45 genes into pET28a vector.
All PDH45 genes (mutated and non-mutated) which were amplified with primers containing desired restriction sites were cloned in pGEM-T easy vector, digested with NdeI and XhoI enzymes. The digested reaction mix was run on the agarose gel and the expected fall out of 1.2 was eluted from the gel. The eluted fall out band was ligated into the digested pET28a vector (NdeI and XhoI). Ligation reaction was transformed using DH5α competent cells and colonies were confirmed by PCR with the gene specific primers. The recombinant plasmids were confirmed by sequencing.
Growth of E. coli bacteria transformed with mutated and non-mutated PDH45 gene under salt (NaCl or LiCl) stress.
The E. coli (BL21 cells) were transformed with mutated and nonmutated PDH45-pET28 with the standard method. The transformed BL21 cells were first grown to log phase OD600 = 0.5. Equal amount of these cells were transferred to sterile culture tubes with 10 ml of LB medium containing 50 µg/ml kanamycin, 1 mM IPTG (final concentration) and 400 mM NaCl or LiCl (final concentration). The cells were allowed to grow at 37°C and the growth was monitored every hour taking the OD600.
Acknowledgments
Work on helicases and plant abiotic stress tolerance in N.T.'s laboratory is partially supported by Department of Biotechnology (DBT), Government of India.
References
- 1.Hodgman TC. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 2.Tuteja N, Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur J Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- 3.Tuteja N, Tuteja R. DNA helicases as molecular motors: an insight. Physica A: Statistical Mechanics and its Applications. 2006;372:70–83. doi: 10.1016/j.physa.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vashisht AA, Tuteja N. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol B. 2006;84:150–160. doi: 10.1016/j.jphotobiol.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA. 2005;102:509–514. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pham XH, Reddy MK, Ehtesham NZ, Matta B, Tuteja N. A DNA helicase from Pisum sativum is homologous to translation initiation factor and stimulates topoisomerase I activity. Plant J. 2000;24:219–229. doi: 10.1046/j.1365-313x.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 7.Tuteja N, Vashisht A, Tuteja R. Translation Initiation Factor 4A (eIF 4A): Prototype DEAD-Box RNA Helicase. Physiol Mol Biol Plants. 2008;14:101–107. doi: 10.1007/s12298-008-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y, Liu YB, Dong YX, Gao XQ, Zhang XS. Expression of a putative alfalfa helicase increases tolerance to abiotic stress in arabidopsis by enhancing the capacities for ros scavenging and osmotic adjustment. J Plant Physiol. 2009;166:385–394. doi: 10.1016/j.jplph.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Kultz D. Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J Exp Biol. 2003;206:3119–3124. doi: 10.1242/jeb.00549. [DOI] [PubMed] [Google Scholar]
- 10.Soto A, Allona I, Collada C, Guevara MA, Casado R, Rodriguez-Cerezo E, et al. Heterologous expression of a plant small heat-shock protein enhances Escherichia coli viability under heat and cold stress. Plant Physiol. 1999;120:521–528. doi: 10.1104/pp.120.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada A, Saitoh T, Mimura T, Ozeki Y. Expression of mangrove allene oxide cyclase enhances salt tolerance in Escherichia coli, yeast and tobacco cells. Plant Cell Physiol. 2002;43:903–910. doi: 10.1093/pcp/pcf108. [DOI] [PubMed] [Google Scholar]
- 12.Yamada A, Tsutsumi K, Tanimoto S, Ozeki Y. Plant RelA/SpoT homolog confers salt tolerance in Escherichia coli and Saccharomyces cerevisiae. Plant Cell Physiol. 2003;44:3–9. doi: 10.1093/pcp/pcg001. [DOI] [PubMed] [Google Scholar]
- 13.Joshi A, Dang HQ, Vaid N, Tuteja N. Isolation of high salinity stress tolerant genes from Pisum sativum by random overexpression in Escherichia coli and their functional validation. Plant Signal Behav. 2009;4:400–412. doi: 10.4161/psb.4.5.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi A, Vaid N, Dang HQ, Tuteja N. Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconj J. 2010;27:133–150. doi: 10.1007/s10719-009-9265-6. [DOI] [PubMed] [Google Scholar]
- 15.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a webbased environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 16.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 17.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.Tuteja N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007;428:419–438. doi: 10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- 19.Mahajan S, Tuteja N. Cold, salinity and drought stresses: An overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki S, Borchert C, Deyholos M, Wang H, Brazile S, Kawai K, et al. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, et al. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serrano R, Gaxiola R, Rios G, Forment J, Vicente O, Ros R. Salt stress proteins identified by a functional approach in yeast. Monatsh Chem. 2003;134:1445–1462. doi: 10.1007/s00706-002-0606-4. [DOI] [Google Scholar]
- 23.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, et al. Birth of the DEAD box. Nature. 1989;337:121. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesan M, Silver LL, Nossal NG. Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J Biol Chem. 1982;257:12426–12434. [PubMed] [Google Scholar]
- 25.Turner AM, Love CF, Alexander RW, Jones PG. Mutational analysis of the Escherichia coli DEAD box protein CsdA. J Bacteriol. 2007;189:2769–2776. doi: 10.1128/JB.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


