Abstract
Monocot plants produce numerous adventitious (crown) roots. The plant hormone auxin has positive effects on crown root formation, while cytokinin suppresses it. We have demonstrated that auxin-induced CROWN ROOTLESS5 (CRL5) regulates crown root initiation in rice through the induction of OsRR1, a negative regulator of cytokinin signaling. CRL5 overexpressing calli formed adventitious roots, although CRL5 overexpressing plants did not induce ectopic roots, suggesting that CRL5, which promotes de novo root initiation, might function only in de-differentiated cells. A radicle initiated normally in a crl5 mutant, in spite of the defect in crown root initiation, whereas crown roots, but not a radicle, were produced in a radicleless1 (ral1) mutant. A crl5 ral1 double mutant displayed an additive phenotype, showing that the formation of each root is regulated by different genetic mechanisms in rice.
Key words: crown (adventitious) root, radicle, initiation, AP2/ERF transcription factor, auxin signaling, cytokinin signaling, rice
Roots play essential roles in plant development such as absorbing water and nutrients and supporting the plant body. Recent progress in our understanding of the molecular mechanisms of root formation has revealed an elaborate system of regulation by plant hormones and their interactions.1,2 However, these analyses were mostly performed in Arabidopsis, a model dicot.
Monocot plants produce numerous adventitious (crown) roots from nodes (Fig. 1A) and form a fibrous root system (Fig. 1B). We reported a rice mutant,3 termed crl5, which produced fewer crown roots and displayed impaired initiation of crown root primordia. CRL5 encodes a member of the APETALA2 (AP2)/EHYLENE RESPONSIVE FACTOR (ERF) transcription factor family. We found that CRL5 functions downstream of the AUXIN (AUX)/INDOLE-3-ACETIC ACID (IAA) and AUXIN RESPONSE FACTOR (ARF)-mediated auxin signaling pathways involved in crown root initiation. Further analysis revealed that CRL5 upregulates a type-A response regulator (RR) of cytokinin signaling, OsRR1. These results indicated that auxin-induced CRL5 functions as a positive regulator of crown root initiation through repression of cytokinin signaling.
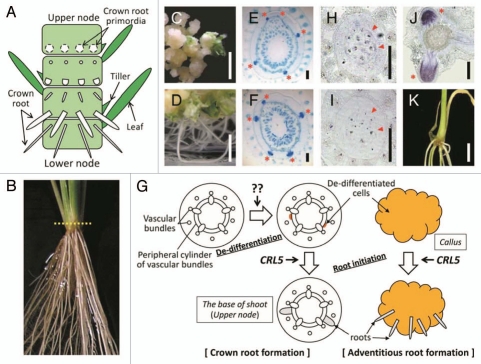
Figure 1.
The ability of CRL5 for de novo root initiation. (A) Schematic diagram of the base of shoot in rice. Nodes are not seen from outside because they are coverd by leaves. (B) One-month-old wild-type plant. Cross sections indicated in (E–J) were made associating the dotted line. (C and D) Wild-type calli containing empty vector (C) and CRL5 overexpressing calli (D) on regeneration media containing 1 µM NAA and 10 µM kinetin. Scale bars = 1 cm. (E and F) Cross sections through the nodes of 1-month-old wild-type containing empty vector (E) and CRL5 overexpressing plant (F). Asterisks indicate the crown root primordia. Scale bars = 200 µm. (G) Schematic diagram of the de novo root initiation. CRL5 may function in de-differentiated cells (highlighted) to promote root initiation, not in the de-differentiation process. The factor inducing the de-differentiation is unknown. (H–J) In situ hybridization with cross sections through the nodes of 7-d-old wild-type seedling. Localization of one of the OsPLT genes in the region where crown root initiation occurs using antisense (H) or sense probes (I) and in the region where crown root emergence occurs using antisense probe (J). Arrowheads and asterisks indicate the peripheral cylinder of vascular bundles and crown root primordia, respectively. Scale Bars = 200 µm. (K) Transgenic plant overexpressing one of the OsPLT genes. Scale bar = 1 cm.
The Ability of CRL5 for de novo Root Initiation
We showed that CRL5 shares significant sequence similarity with AINTEGUMENTA (ANT) containing 2 AP2 domains in Arabidopsis.4 Seven AINTEGUMENTA-like (AIL)/PLETHORA (PLT) proteins were reported to share high sequence similarity with ANT.5 Among them, PLT1, PLT2, AIL6/PLT3 and AtBABY BOOM (AtBBM) have been shown to act redundantly in root development and are thought to function as master switches for root initiation and development because of their ability, when overexpressed, to induce ectopic roots in cells that do not possess competence to produce roots (like shoot organs) in Arabidopsis.6,7 In spite of the similarity, CRL5 overexpressing plants did not induce ectopic roots, with the exception of the strong ability of CRL5 overexpressing calli to form adventitious roots (Fig. 1C–F). Therefore, it is possible that CRL5 is not involved in the de-differentiation process, but its involvement in root initiation might be restricted to de-differentiated cells (Fig. 1G). We used in situ hybridization to examine the expression of a gene similar to Arabidopsis PLTs in rice, OsPLT, at the base of shoots. This gene was expressed in the parenchyma cells adjacent to the peripheral cylinder of vascular bundles of the stem and the tip of crown root primordia (Fig. 1H–J). The expression of this gene in the root tip was similar to that of Arabidopsis PLTs (Fig. 1J), and overlapped the region where crown root initiation occurs (Fig. 1H). We also made transgenic plants that overexpressed this gene; however, ectopic root formation was not observed (Fig. 1K). We suggested that the function of PLT in rice may be different from that of Arabidopsis PLTs in root initiation and that other gene(s) may function as a master switch in rice root initiation.
Difference between Crown Root and Radicle Initiation in Rice
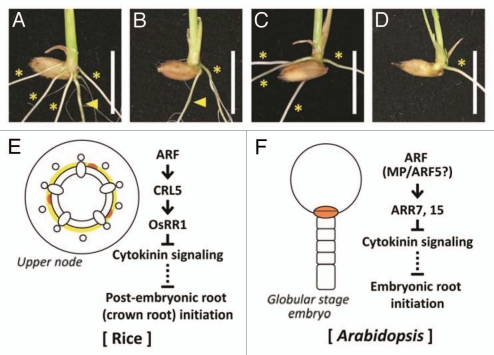
Roots are largely classified as embryonic roots and post-embryonic roots. Embryonic roots initiate during embryogenesis and post-embryonic roots initiate after embryogenesis. In rice, a radicle (a seminal root) is an embryonic root and crown roots are post-embryonic (Fig. 2A). Although the number of crown roots in crl5 was clearly lower than that in wild-type, crl5 normally produced a radicle (Fig. 2B).3 On the other hand, the crown root number was not decreased in radicleless1 (ral1), but ral1 cannot produce a radicle (seminal root) (Fig. 2C).8 In the crl5 ral1 double mutant, a seminal root was not produced and the crown root number was also clearly lower than in the wild type (Fig. 2D). However, the double mutant produced almost the same number of crown roots as crl5, showing that crown root initiation is not affected by ral1 mutation. The additive phenotype displayed by the crl5 ral1 double mutant indicated that formation of each root is regulated by different genetic mechanisms in rice.
Figure 2.
Difference between embryonic root and post-embryonic root initiation in rice. (A–D) Phenotype of wild-type (A), crl5 (B), ral1 (C) and crl5 ral1 double mutant (D). Arrowheads and asterisks indicate seminal roots (radicles, embryonic roots) and crown roots (post-embryonic roots), respectively. Scale bars = 1 cm. (E and F) Schematic diagram of post-embryonic root (crown root) initiation in rice (E) and embryonic root initiation in Arabidopsis (F). The regions where root initiation occurs are highlighted.
We demonstrated that interaction between OsARF and OsRR1 is mediated by CRL5 in rice crown root initiation (Fig. 2E).3 On the other hand, it has been reported that auxin signaling directly induces transcription of type-A ARABIDOPSIS RESPONSE REGULATOR7 (ARR7) and ARR15 genes, which negatively regulate cytokinin signaling through conserved TGTC elements and whose mutations prevent the establishment of a normal embryonic root.9 In addition, interaction between auxin signaling and these ARRs is essential for control of the shoot stem-cell niche.10 In the shoot apical meristem, MONOPTEROS (MP)/ARF5 directly relays auxin signals to ARR15 by binding TGTC elements in its promoter.10 Based on these reports, we hypothesized that MP/ARF5 is the most likely candidate to regulate ARR7 and ARR15 directly in Arabidopsis embryonic root initiation (Fig. 2F). We suspected that CRL5 might be a key factor to define the different genetic mechanisms between embryonic root (radicle) initiation and post-embryonic root (crown root) initiation in rice. Comprehensive understanding of root initiation in rice may progress by revealing the mechanism of embryonic and post-embryonic root initiation.
Acknowledgments
We thank H. Tazuke for kindly providing rice PLT cDNA clone. We thank the National Bioresource Project of Rice for providing the crl5 mutant line. This research was supported by Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan to Y.I. (numbers 21027015) and Grant-in-Aid for Japan Society for the Promotion of Science Fellows to Y.K. (numbers 21005221).
References
- 1.Fukaki H, Okushima Y, Tasaka M. Auxin-mediated lateral root formation in higher plants. Int Rev Cytol. 2007;256:111–137. doi: 10.1016/S0074-7696(07)56004-3. [DOI] [PubMed] [Google Scholar]
- 2.Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2010;2:1537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y. The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 2011;67:472–484. doi: 10.1111/j.1365-313X.2011.04610.x. [DOI] [PubMed] [Google Scholar]
- 4.Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nole-Wilson S, Tranby TL, Krizek BA. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol. 2005;57:613–628. doi: 10.1007/s11103-005-0955-6. [DOI] [PubMed] [Google Scholar]
- 6.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 8.Hong SK, Aoki T, Kitano H, Satoh H, Nagato Y. Phenotypic diversity of 188 rice embryo mutants. Dev Genet. 1995;16:298–310. doi: 10.1002/dvg.1020160403. [DOI] [Google Scholar]
- 9.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryo-genesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]