Abstract
Two component high affinity nitrate transport system, NAR2/NRT2, has been defined in several plant species. In Arabidopsis, AtNAR2.1 has a role in the targeting of AtNRT2.1 to the plasma membrane. The gene knock out mutant atnar2.1 lacks inducible high-affinity transport system (IHATS) activity, it also shows the same inhibition of lateral root (LR) initiation on the newly developed primary roots as the atnrt2.1 mutant in response to low nitrate supply. In rice, OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a to provide nitrate uptake over high and low concentration ranges. In rice roots OsNAR2.1 and its partner NRT2s show some expression differences in both tissue specificity and abundance. It can be predicted that NAR2 plays multiple roles in addition to being an IHATS component in plants.
Key words: NAR2, NRT2, nitrate transporter, root
Function of NAR2 as a Nitrate Accessory Protein
The plant NAR2-type genes were first named WR3 identified by expression that was induced by wounding1 and pathogens.2 The involvement of NAR2 in the high-affinity transport system (HATS) for nitrate was initially identified genetically using mutants in Chlamydomonas reinhardtii, in which NAR2 is next to NRT2.1 in the nitrate-related gene cluster.3,4 Functional analysis of the CrNAR2/CrNRT2.1 components for nitrate HATS was demonstrated in Xenopus oocytes.5,6 Numerous genes belonging to the NAR2 family have been identified during recent years in several plants species. At present, three very similar barley NAR2 genes (HvNAR2.1–HvNAR2.3),7 two Arabidopsis NAR2 genes (AtNAR2.1 and AtNAR2.2),8 and two rice NAR2s genes (OsNAR2.1 and OsNAR2.2),9 have been characterized. However, not all NAR2 family members demonstrated a function in two-component nitrate HATS. For example, among three barley NAR2 genes, only HvNAR2.3 formed a functional unit with HvNRT2.1.7 In rice OsNAR2.1, but not OsNAR2.2, functions in rice two-component nitrate HATS.9
Clarifying the function of NAR2 family members in nitrate transport requires the identification of partner pairs and the specificity of their interaction. In Arabidopsis, besides the well-characterized interaction with AtNRT2.1, AtNAR2.1 showed weak interaction with AtNRT2.3 in yeast, but the co-expression of AtNAR2.1 and AtNRT2.3 did not show nitrate uptake activity in oocytes.8 In rice, OsNAR2.1 can cooperate with both OsNRT2.1/2.2 and OsNRT2.3a in yeast,9 and the interaction provided NO3− uptake both in oocytes and rice over low to high concentration ranges.10 In tobacco NpNRT2.1, which is an ortholog of AtNRT2.1, can complement the atnrt2.1-1 mutant, but not the atnar2.1 mutants.11 The finding suggests that NpNRT2.1 can function in nitrate transport, but it is not known if NpNRT2.1 needs an accessory protein for its function.
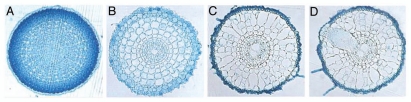
The NAR2s are a small proteins, possibly containing a single TM domain in CrNAR2 and two TM domains in HvNAR2.3.5,7 The NRT2s are typical carrier-type proteins, containing 12 putative TM domains.8 The NAR2 protein plays a role in establishing HATS activity by targeting NRT2 to the plasma membrane.12,13 In the rnc1 mutant of Arabidopsis, it was suggested that an aspartate residue in the central loop of NAR2 was important for HATS activity.14 Immunological results demonstrated an oligomer, proposed to contain a tetramer consisting of two subunits each of AtNRT2.1 and AtNAR2.1, was active in HATS.13 In barley, a direct protein-protein interaction was shown to occur between the HvNRT2.1 C-terminal, S463A, and the HvNAR2.3 central region.12 However, it is difficult to determine if this serine is a key residue because it is not conserved in AtNRT2.1 and HvNRT2.1 C-terminal.12 In rice roots we detected some differences in the tissue specific expression of OsNAR2.1 and its interacting NRT2 members.10 OsNAR2.1 expression was strongest in the epidermal cells and much lower in the cortical and stelar cells (Fig. 1), while OsNRT2.1, OsNRT2.2 and OsNRT2.3 were expressed abundantly in the stelar cells.10
Figure 1.
Root cross sections of plants transformed with OsNAR2.1 promoter-GUS genes. The expression specificity of OsNAR2.1 in the root of ten-day-old seedlings (Oryza sativa L. ssp. Japonica cv. Nipponbare) cultured with de-ionized water containing 25 mgL−1 hygromycin. Transverse sections are taken from approximately 0.5 mm (A), 1.5 mm (B), 1 cm (C) and 1.5 cm (D) from the root tip.
It was shown that the expression of AtNAR2.1, AtNRT2.1 and AtNRT2.2 in Arabidopsis was perfectly coordinated with nitrate HATS regulation: induction by low external nitrate concentration and sudden N deprivation, and suppression by high nitrate supply.8,15,16 In rice roots the expression of OsNAR2.1, OsNRT2.1, OsNRT2.2 and OsNRT2.3a genes was induced by both low and high nitrate concentrations.9 In the Arabidopsis atnar2.1 knockout mutant, the expression of AtNRT2.1 decreased when compared with WT grown at 0.2 mM nitrate, while it increased at 6 mM nitrate.8 The expression level of AtNRT1.1 did not change significantly between the atnar2.1 and wild type plants under a high nitrate concentration.8 In rice, RNAi-knockdown of OsNAR2.1 suppressed the expression of OsNRT2.1, OsNRT2.2 and OsNRT2.3a very largely, but not the expression of OsNRT1.1, OsNAR2.2, OsNRT2.3b and OsNRT2.4, at both 0.2 mM and 5 mM nitrate supply levels.9 It is interesting to know if there are the large different responses of NAR2/NRT2 members to the change of N supply among the genotypes of each rice or Arabidopsis species. If not, the expressional discrepancies of NAR2/NRT2 members between Arabidopsis and rice could link to the evolutionally adaptation of the plants to varied N form and concentrations in upland and flooded paddy soils.
Role of NAR2.1 in Root Development
Local stimulation of lateral root growth was shown to be due to a specific NO3− signaling pathway, involving the putative MADS-box transcription factor ANR1.17 NRT1.1 acts upstream of ANR1 in the NO3− signaling pathway governing lateral root growth.18 It has also been shown that AtNRT2.1 plays a crucial role in root branching independently of its NO3− uptake activity.19,20 The atnar2.1 and atnrt2.1-1 mutants display the same inhibition of lateral root (LR) initiation on the newly developed primary roots.8,11 The tobacco gene NpNRT2.1 cannot functionally replace the Arabidopsis gene AtNRT2.1 for the LR response while it can restore the HATS activity under nitrogen (N) limitation.8,11 Compared with the RolDNpNRT2.1 overexpresser (the plants overexpressing NpNRT2.1 gene) and wild type, RolDNpNRT2.1X atnar2.1 plants (the atnar2.1 mutant complemented with the NpNRT2.1 gene) and atnar2.1 mutant had significantly lower numbers of new LRs under a low nitrate concentration supply.11 The Arabidopsis atnar2.1 mutants showed decreased growth and lower numbers of new lateral roots (LR) only under a low nitrate supply condition.8,11
Does NAR2 Gene Regulate the Expression of Partner NRT2 s?
Members of the NRT2 family showed a variety of tissue expression patterns and regulation10 (Table 1). The expression patterns of some NAR2 and NRT2 family members of different species show common responses to different N forms, they are induced by nitrate and repressed by feedback regulation from N metabolites10 (Table 1). Furthermore, HATS activity and transcript levels of NAR2.1 and NRT2.1 in Arabidopsis and barley plants are repressed significantly by high nitrate concentrations.12,15 Interestingly, in rice the expression patterns of NAR2.1 and NRT2s in roots treated with high nitrate are similar to those treated with low concentrations.9 At present, no trans-acting factors (TFs) that are absolutely necessary to regulate nitrate-responsive transcription have been identified in higher plants. We found a nitrate regulated element (NRE) in the OsNAR2.1 promoter10 had a highly similar NRE to one located in the Arabidopsis NIR promoter, with only one base changed from A to C.10,21 However there was no such conserved sequence in the −1,000 bp promoter regions of OsNRT2 genes in rice, and AtNAR2.1/AtNAR2 genes in Arabidopsis.10 One possibility is that OsNAR2.1 can more directly sense nitrate than the OsNRT2s in rice plants. However, because the identity of the TFs or NRE(s) is still uncertain, another possibility is that OsNAR2.1 and OsNRT2s have their own independent and perhaps rice specific NREs.
Table 1.
Literature summary of the tissue expression and regulation of some NRT2s in higher plants
| Gene (Accession number) | Expression | Regulation | |
| Characterization | Technique | ||
| AtNRT2.1/2.2 (AF019748/AF01979) | Root cortical and epidermal cells | RT-PCR; GUS/GFP fusion | NO3−, sucrose and light induction; NH4+ and amino acids repression |
| AtNRT2.3 (AB015472) | Mainly in roots | RT-PCR | NO3− induction in shoots |
| AtNRT2.4 (AB015472) | Modest NO3− induction | ||
| AtNRT2.6 (AL353992) | - | ||
| AtNRT2.5 (AC012187) | NO3− repression; Glucose induction | ||
| AtNRT2.7 (AL163792) | Shoots; Embryo; seeds | RT-PCR; GUS/GFP fusion | NO3− insensity |
| ZmNRT2.1 (AY129953) | Root epidermis and cortex | RT-PCR; In situ hybridization | NO3− induction; Glucose down- and sucrose upregulation |
| ZmNRT2.2 (AY659965) | Root endodermis, centralcylinder and lateral primordia | ||
| NpNRT2 (Y08210) | Strong in epidermis and endodermis close to the root tip, week in the epidermis and lateral root primordia in the mature part of the root | RNA gel blot; In situ hybridization | NO3− induction |
| HvNRT2.1 | Root | RNA gel blot | NO3− and NO2− induction |
| OsNAR2.1 (AP008208) | Strongest in root epidermis and lower in cortex and stele | RT-PCR; GUS fusion | NO3−, light and sucrose upregulation; NH4+ repression |
| OsNRT2.1 (AP008519) | Whole roots; leaves; week in hulls and anthers | ||
| OsNRT2.2 (AP008519) | Whole roots; leaves; strong in hulls and anthers | ||
| OsNRT2.3a (AP003245) | Root stele; leaves; seed scutellum | ||
| OsNRT2.3b (AP003245) | light and sucrose upregulation | ||
| OsNRT2.4 (AP004614) | Adventitious root primordial; leaves; ends of the hull and in vascular tissue of the anther | NO3−, light and sucrose upregulation; NH4+ repression | |
The overexpression of NpNRT2.1 in tobacco plants failed to yield any increase in nitrate uptake.22 It may be that the overexpression of NRT2.1, without NAR2.1, is unlikely to increase nitrate uptake. AtNAR2.1 expression is similar in wild-type and atnrt2.1-1 plants growing under differing supplies of nitrate, while AtNRT2.1 expression is repressed in atnar2.1 mutants under low nitrate concentration.8 When both atnar2.1 and atnrt2.1 mutants were supplied with low nitrate concentrations (0.2 or 0.5 mM), the atnar2.1-1 mutant has a more severely stunted growth phenotype than the atnrt2.1-1 mutant.8 After 4 d supplied with low nitrate, the atnar2.1-1 mutant cannot sustain an increased growth rate while no limitation was reported for the atnrt2.1 mutant.8 An explanation is probably that atnar2.1-1 mutants suffer more N deficiency than atnrt2.1-1, indicating that AtNAR2.1 had a greater role in N uptake than AtNRT2.1.8 Some studies have suggested that AtNRT2.1 might act as a nitrate transceptor23 like AtNRT1.1.24 Because of the lack of confirmed nitrate sensing motifs, it is difficult to determine which gene in the NAR2/NRT2 partnership acts as the nitrate sensor/signal transducer.
Acknowledgments
Research in China was funded by the National Basic Research Program 973 (2011CB100300), the National Natural Science Foundation, Jiangsu Natural Science Foundation (BK2010227) and transgenic project (2008ZX08001-0052, 009ZX08009-126B).
References
- 1.Titarenko E, Rojo E, Leon J, Sanchez-Serrano J. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 1997;115:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marois E, Van den Ackerveken G, Bonas U. The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact. 2002;15:637–646. doi: 10.1094/MPMI.2002.15.7.637. [DOI] [PubMed] [Google Scholar]
- 3.Quesada A, Galvan A, Fernandez E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 1994;5:407–419. doi: 10.1111/j.1365-313X.1994.00407.x. [DOI] [PubMed] [Google Scholar]
- 4.Galván A, Quesada A, Fernandez E. Nitrate and nitrate are transported by different specific transport systems and by a bispecific transporter in Chlamydomonas reinhardtii. J Biol Chem. 1996;271:2088–2092. doi: 10.1074/jbc.271.4.2088. [DOI] [PubMed] [Google Scholar]
- 5.Zhou JJ, Fernandez E, Galvan A, Miller AJ. A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett. 2000;466:225–227. doi: 10.1016/S0014-5793(00)01085-1. [DOI] [PubMed] [Google Scholar]
- 6.Zhou JJ, Trueman LJ, Boorer KJ, Theodoulou FL, Forde BG, Miller AJ. A high affinity fungal nitrate carrier with two transport mechanisms. J Biol Chem. 2000;275:39894–39899. doi: 10.1074/jbc.M004610200. [DOI] [PubMed] [Google Scholar]
- 7.Tong Y, Zhou JJ, Li Z, Miller AJ. A two-component high-affinity nitrate uptake system in barley. Plant J. 2005;41:442–450. doi: 10.1111/j.1365-313X.2004.02310.x. [DOI] [PubMed] [Google Scholar]
- 8.Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, et al. Characterisation of a two component high affinity nitrate uptake system in Arabidopsis: physiology and protein-protein interaction. Plant Physiol. 2006;142:1304–1317. doi: 10.1104/pp.106.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan M, Fan XR, Feng HM, Miller AJ, Shen QR, Xu GH. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration range. Plant Cell Environ. 2011;34:1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x. [DOI] [PubMed] [Google Scholar]
- 10.Feng H, Yan M, Fan XR, Li BZ, Shen QR, Miller AJ, et al. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot. 2011;62:2319–2332. doi: 10.1093/jxb/erq403. [DOI] [PubMed] [Google Scholar]
- 11.Orsel M, Chopin F, Leleu O, Krapp SS, Daniel-Vedele F, Miller AJ. Nitrate Signaling and the Two Component High Affinity Uptake System in Arabidopsis. Plant Signal Behav. 2007;2:260–262. doi: 10.4161/psb.2.4.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa S, Ito Y, Sato Y, Fukaya Y, Takahashi M, Morikawa H, et al. Two-component high-affinity nitrate transport system in barley: membrane localization, protein expression in roots and a direct protein-protein interaction. Plant Biotechnol. 2009;26:197–205. doi: 10.5511/plantbiotechnology.26.197. [DOI] [Google Scholar]
- 13.Yong Z, Kotur Z, Glass ADM. Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 2010;63:739–748. doi: 10.1111/j.1365-313X.2010.04278.x. [DOI] [PubMed] [Google Scholar]
- 14.Kawachi T, Sunaga Y, Ebato M, Hatanaka T, Harada H. Repression of nitrate uptake by replacement of Asp105 by asparagine in AtNRT3.1 in Arabidopsis thaliana L. Plant Cell Physiol. 2006;47:1437–1441. doi: 10.1093/pcp/pcl010. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, et al. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-Like gene AtNRT3.1. Plant Physiol. 2006;140:1036–1046. doi: 10.1104/pp.105.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraisier V, Gojon A, Tillard P, Daniel-Vedele F. Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J. 2000;23:489–496. doi: 10.1046/j.1365-313x.2000.00813.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 18.Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, et al. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA. 2006;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006;140:909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konishi M, Yanagisawa S. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J. 2010;63:269–282. doi: 10.1111/j.1365-313X.2010.04239.x. [DOI] [PubMed] [Google Scholar]
- 22.Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, et al. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 2001;489:220–224. doi: 10.1016/S0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- 23.Gojon A, Krouk G, Perrine-Walker F, Laugier E. Nitrate transceptor(s) in plants. J Exp Bot. 2011;62:2299–2308. doi: 10.1093/jxb/erq419. [DOI] [PubMed] [Google Scholar]
- 24.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]