Abstract
Nitrogen-fixing root nodulation, confined to four plant orders, encompasses more than 14,000 Leguminosae species, and approximately 200 actinorhizal species forming symbioses with rhizobia (Rhizobium, Bradyrhizobium, etc.,) and Frankia bacterial species, respectively.
While several genetic components of the host-symbiont interaction have been identified in legumes, little is known about the genetic bases of actinorhizal symbiosis. However, we recently demonstrated the existence of common symbiotic signaling pathways in actinorhizals and legumes. Moreover, important data on the identification of flavonoids as plant signaling compounds and the role for auxins during Frankia infection process and nodule organogenesis have been acquired. All together these results lead us to propose a unified model for symbiotic exchange and genetic control of actinorhizal symbiosis.
Key words: actinorhizal symbiosis, Frankia, signaling, nodulation, transcriptomics, flavonoid, auxin
Introduction
Actinorhizal symbioses result from the interaction between actinobacteria of the genus Frankia and plants belonging to eight dicots plant families collectively called actinorhizal plants.1 While legume nodules have a stem-like morphology with their peripheral vascular system and infected cells in the central tissues, all actinorhizal nodules are structurally and developmentally related to lateral roots.2 Due to the lack of genetic tools on both bacteria and plant partners, signals exchange during the early stages of the actinorhizal symbiosis is still poorly understood.3 Recently, through comparative transcriptomics of two actinorhizal symbiotic plants, we identified several homolog genes of the symbiotic pathway known in legumes.4 This result, together with previous data showing activation of isoflavonoid pathway in actinorhizal symbioses5 and a role for auxin during the Frankia infection process6 leads us to propose a model of the early symbiotic steps of the Frankia-actinorhizal plants interaction.
Identification of Actinorhizal Genes that are Homologs of the Legumes Symbiotic Signaling Cascade
Around 30,000 expressed sequence tags (ESTs), corresponding to approximately 14,000 unigenes were recovered in roots and three-week old nodules of two actinorhizal species, Alnus glutinosa and Casuarina glauca.4,7 Unigenes classification into functional categories showed a similar distribution of genes in the two species suggesting that the actinorhizal C. glauca and A. glutinosa are closely related. Most of the genes of the common “SYM” pathway described for Arbuscular Mycorhizal (AM) and legumes-rhizobium symbioses were identified in C. glauca and A. glutinosa.4 This pathway contains a receptor-like kinase, nuclear pore proteins and potassium channels required for the induction of calcium oscillations.8,9 A putative calcium and calmodulin-dependent protein kinase (CCaMK) is also present and might thus recognize calcium “actinorhizal signatures”. Interestingly, our analysis also revealed the presence of genes linked to a “NOD”-specific pathway (not shared with AM symbiosis) as used by legumes in their symbiosis with rhizobia.10
Isoflavonoid Responding Genes are Activated during Actinorhiza Formation
Numerous studies have shown that flavonoids have evolved particular roles in legumes. They are not only required to signal symbiotic bacteria in the legume-bacterium symbiosis but also play important direct roles in root nodule organogenesis.11 Recent evidence has shown that flavonoids are also involved in actinorhizal nodulation.3 Eight structural genes involved in the flavonoid biosynthesis pathway were found in the C. glauca roots and nodules ESTs database.5 Quantitative real-time PCR was then used to monitor the relative transcript levels of these genes in roots of C. glauca from 12 h to 22 days after inoculation with Frankia. Transcripts of isoflavone synthase (IFR) accumulated specifically in roots very early after inoculation with Frankia (Signaling step). The fact that IFR transcripts accumulated as soon as 12 h after Frankia inoculation strongly suggests a role for isoflavonoids as signal molecules during actinorhizal symbiosis.5
Auxin is Involved in the Control of Actinorhizal Symbiosis
Recently it has been shown that the C. glauca auxin influx carrier gene (CgAux1) is expressed in Frankia infected cells during actinorhizal nodule formation.6,12 This result together with data showing auxins production by Frankia6,13 suggests a role for auxins in plant cell infection during actinorhizal symbiosis. Moreover, an accumulation of auxins within Frankia-infected cells in actinorhizal nodule of C. glauca was reported and it was established that this accumulation was driven by cell-specific expression of auxin transporters and by Frankia auxin biosynthesis in planta.6
Canonical nod Genes are Absent from Frankia Genomes
The recent determination of three Frankia genomes belonging to three of the four cross-infectivity groups permitted among other things to look for canonical nod homologs.14 If several nodC and nodB distant homologs were seen, these were spread all around the genomes. Furthermore, they did not form a SYM-island and finally no nodA gene could be detected.
Towards a Model for Early Frankia-Actinorhizal Plants Interaction
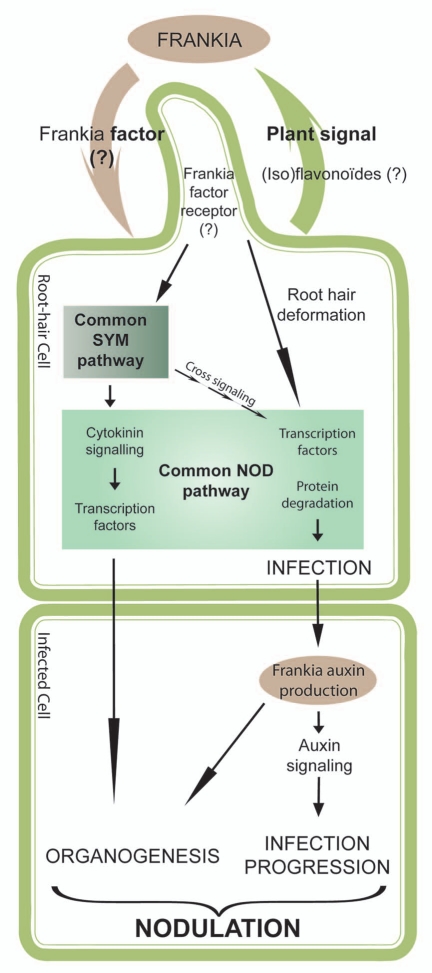
Taken together, all these recent available data on actinorhizal symbiosis allow us to propose a model of early Frankiaactinorhizal plants interaction (Fig. 1). Flavonoids are suggested as actinorhizal plant signal molecules that influence Frankia growth15 and Frankia symbiotic factor production, the nature of which remains unknown except that it has some biochemical similarities to the Rhizobium16 one, even though no canonical nod genes are present in the Frankia genomes published so far. Perception of Frankia factor by yet unknown plant root-hair cell receptor might induce signaling cascades of the “SYM” and “NOD” pathways. Following Frankia cell penetration, bacterial and/or plant auxin biosynthesis driven by cell-specific expression of auxin transporters would lead to auxin accumulation specifically in plant cell. Auxin might be necessary to control the infection process and nodule organogenesis. Our future work will focus on testing this model.
Figure 1.
A model for signal exchange in the actinorhizal symbiosis. Actinorhizal plant roots released flavonoids that induce production of the yet unknown Frankia symbiotic signal leading to the activation of the “SYM” and actinorhizal “NOD” pathways. Furthermore, upon Frankia penetration in root hair and cortical cells, auxin accumulates in infected cells driving infection and nodule organogenesis. Adapted from references 3 and 4.
References
- 1.Benson DR, Silvester WB. Biology of frankia strains, actinomycete symbionts of actinorhizal plants. Microbial Rev. 1993;57:293–319. doi: 10.1128/mr.57.2.293-319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pawlowski K, Bisseling T. Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell. 1996;8:1899–1913. doi: 10.1105/tpc.8.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrine-Walker F, Gherbi H, Imanishi L, Hocher V, Ghodhbane-Gtari F, Lavenus J, et al. Symbiotic signaling in actinorhizal symbioses. Curr Protein Pept Sci. 2011;12:156–164. doi: 10.2174/138920311795684896. [DOI] [PubMed] [Google Scholar]
- 4.Hocher V, Alloisio N, Auguy F, Fournier P, Doumas P, Pujic P, et al. Transcriptomics of actinorhizal symbioses reveals homologs of the whole common symbiotic signaling cascade. Plant Physiol. 2011;156:700–711. doi: 10.1104/pp.111.174151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auguy F, Abdel-Lateif K, Doumas P, Badin P, Guerin P, Bogusz D, et al. Isoflavonoids pathway activation in actinorhizal symbioses. Funct Plant Biol. 2011;38:690–696. doi: 10.1071/FP11014. [DOI] [PubMed] [Google Scholar]
- 6.Perrine-Walker F, Doumas P, Lucas M, Vaissayre V, Beauchemin NJ, Band LR. Auxin carriers localization drives auxin accumulation in plant cells infected by Frankia in Casuarina glauca actinorhizal nodules. Plant Physiol. 2010;154:1372–1380. doi: 10.1104/pp.110.163394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hocher V, Auguy F, Argout X, Laplaze L, Franche C, Bogusz D. Expressed sequence-tag analysis in Casuarina glauca actinorhizal nodule and root. New Phytol. 2006;169:681–688. doi: 10.1111/j.1469-8137.2006.01644.x. [DOI] [PubMed] [Google Scholar]
- 8.Capoen W, Den Herder J, Sunc J, Verplancke C, De Keyser A, De Rycke R, et al. Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell. 2009;21:1526–1540. doi: 10.1105/tpc.109.066233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markmann K, Parniske M. Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci. 2009;14:77–86. doi: 10.1016/j.tplants.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, et al. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:1–12. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian S, Stacey G, Yu O. Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 2007;12:282–285. doi: 10.1016/j.tplants.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Péret B, Swarup R, Jansen L, Devos G, Auguy F, Collin M, et al. Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol. 2007;144:1852–1862. doi: 10.1104/pp.107.101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammad Y, Nalin R, Marechal J, Fiasson K, Pepin R, Berry AM, et al. A possible role for phenyl acetic acid (PAA) on Alnus glutinosa nodulation by Frankia. Plant Soil. 2003;254:193–205. [Google Scholar]
- 14.Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, et al. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 2007;17:7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popovici J, Comte G, Bagnarol E, Alloisio N, Fournier P, Bellvert, et al. Differential effects of rare specific flavonoids on compatible and incompatible strains in the Myrica gale-Frankia actinorhizal symbiosis. Appl Environ Microbiol. 2010;76:2451–2460. doi: 10.1128/AEM.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceremonie H, Debelle F, Fernandez MP. Structural and functional comparison of Frankia root hair deforming factor and rhizobia Nod factor. Can J Bot. 1999;77:1293–1301. [Google Scholar]