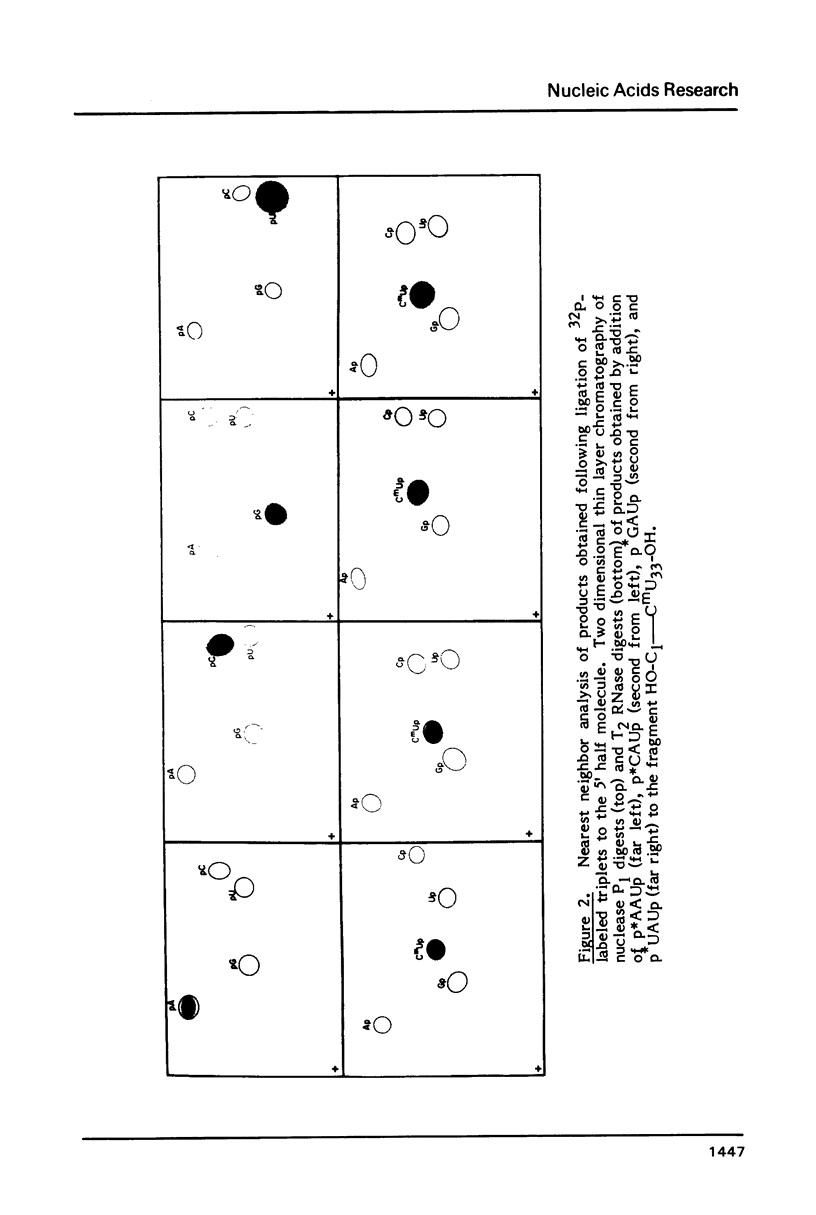
Abstract
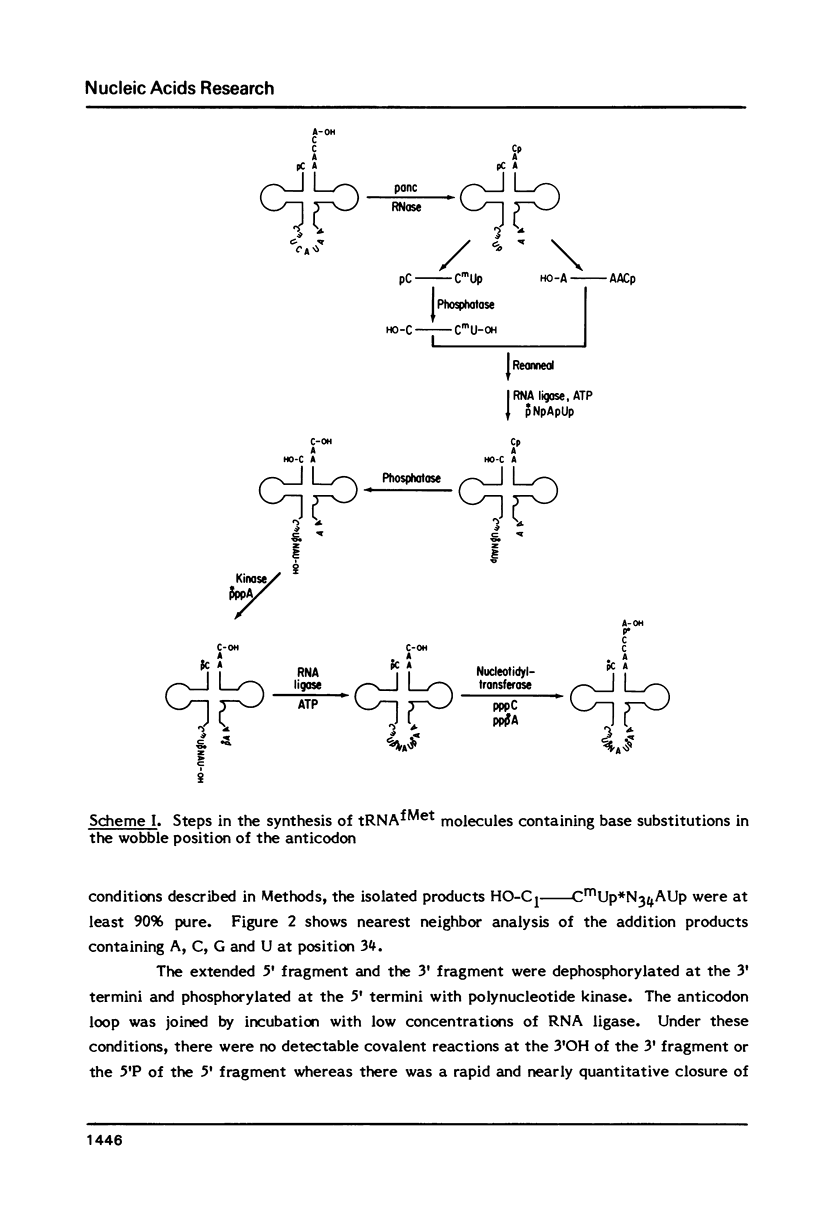
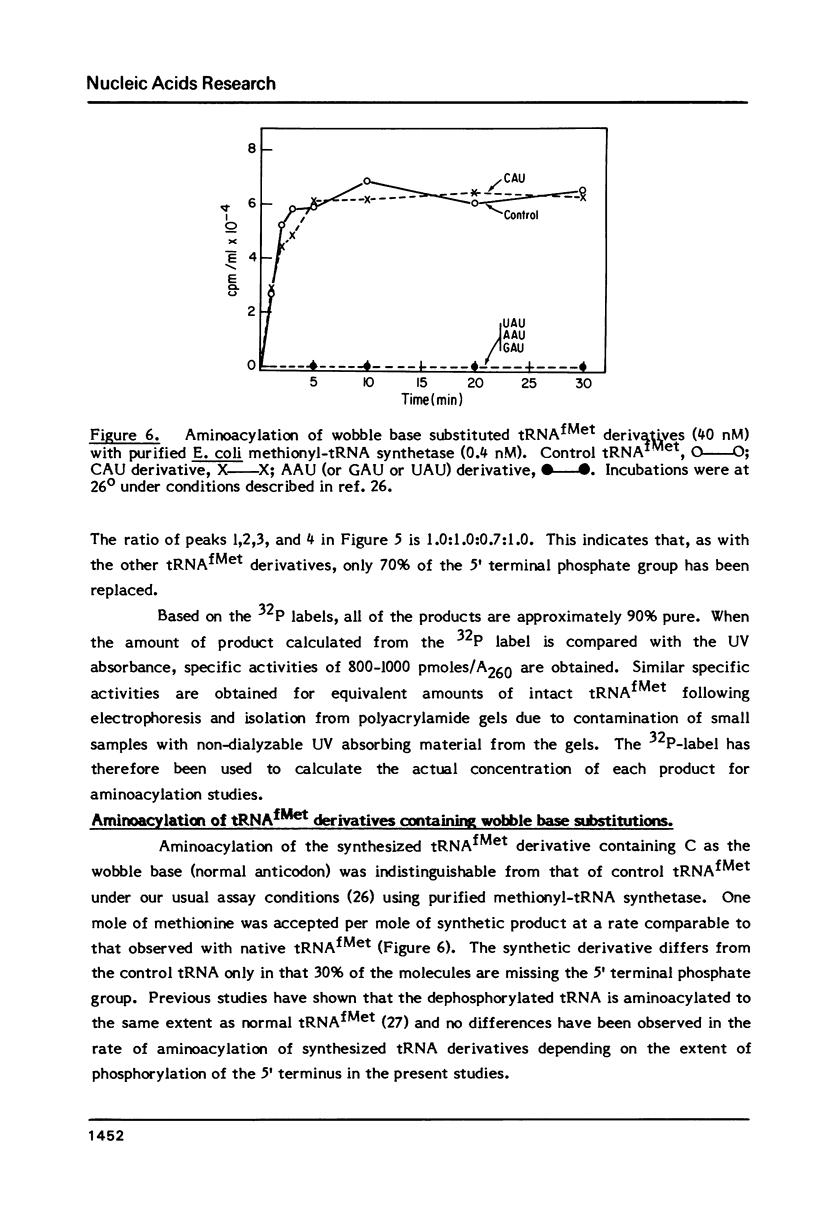
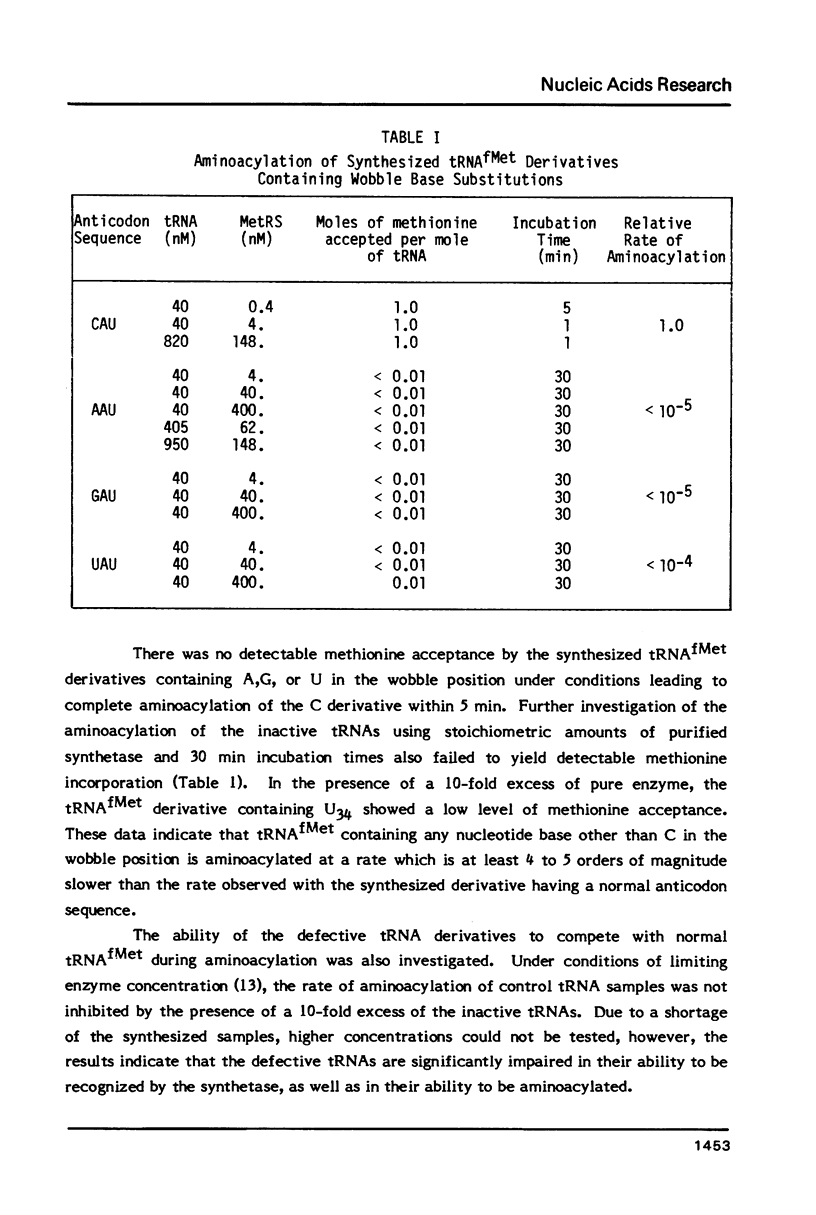
Derivatives of E. coli tRNAfMet containing single base substitutions at the wobble position of the anticodon have been enzymatically synthesized in vitro. The procedure involves excision of the normal anticodon, CAU, by limited digestion of intact tRNAfMet with RNase A. RNA ligase is then used to join each of four trinucleotides, NAU, to the 5' half molecule and to subsequently link the 3' and modified 5' fragments to regenerate the anticodon loop. Synthesis of intact tRNAfMet containing the anticodon CAU by this procedure yields a product which is indistinguishable from native tRNAfMet with respect to its ability to be aminoacylated by E. coli methionyl-tRNA synthetase. Substitution of any other nucleotide at the wobble position of tRNAfMet drastically impairs the ability of the synthetase to recognize the tRNA. Measurement of methionine acceptance in the presence of high concentrations of pure enzyme has established that the rate of aminoacylation of the AAU, GAU and UAU anticodon derivatives of tRNAfMet is four to five orders of magnitude slower than that of the native or synthesized tRNA containing C as the wobble base. In addition, the inactive tRNA derivatives fail to inhibit aminoacylation of normal tRNAfMet, indicating that they bind poorly to the enzyme. These results support a model involving direct interaction between Met-tRNA synthetase and the C in the wobble position during aminoacylation of tRNAfMet.
Full text
PDF










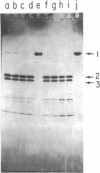
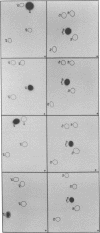
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruce A. G., Uhlenbeck O. C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982 Mar 2;21(5):855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Specific interaction of anticodon loop residues with yeast phenylalanyl-tRNA synthetase. Biochemistry. 1982 Aug 17;21(17):3921–3926. doi: 10.1021/bi00260a003. [DOI] [PubMed] [Google Scholar]
- Cameron V., Soltis D., Uhlenbeck O. C. Polynucleotide kinase from a T4 mutant which lacks the 3' phosphatase activity. Nucleic Acids Res. 1978 Mar;5(3):825–833. doi: 10.1093/nar/5.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Haumont E., De Henau S., Keith G., Grosjean H. Enzymatic replacement in vitro of the first anticodon base of yeast tRNAAsp: application to the study of tRNA maturation in vivo, after microinjection into frog oocytes. Nucleic Acids Res. 1982 Jun 25;10(12):3715–3732. doi: 10.1093/nar/10.12.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R. W., Aoyagi S., Furukawa Y., Zawadzka H., Bhanot O. S. Inactivation of valine acceptor ativity by a C-U missense change in the anticodon of yeast valine transfer ribonucleic acid. J Biol Chem. 1973 Aug 10;248(15):5549–5551. [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Goddard J. P., Schulman L. H. Conversion of exposed cytidine residues to uridine residues in Escherichia coli formylmethionine transfer ribonucleic acid. J Biol Chem. 1972 Jun 25;247(12):3864–3867. [PubMed] [Google Scholar]
- Hassur S. M., Whitlock H. W., Jr UV shadowing--a new and convenient method for the location of ultraviolet-absorbing species in polyacrylamide gels. Anal Biochem. 1974 May;59(1):162–164. doi: 10.1016/0003-2697(74)90020-7. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Geballe A. P., Snopek T. J., Sugino A., Cozzarelli N. R. Bacteriophage T4 RNA ligase: preparation of a physically homogeneous, nuclease-free enzyme from hyperproducing infected cells. Nucleic Acids Res. 1977 Sep;4(9):3175–3186. doi: 10.1093/nar/4.9.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M., Uhlenbeck O. C. Reversal of T4 RNA ligase. Biochemistry. 1982 Apr 13;21(8):1858–1864. doi: 10.1021/bi00537a024. [DOI] [PubMed] [Google Scholar]
- Krug M., de Haseth P. L., Uhlenbeck O. C. Enzymatic synthesis of a 21-nucleotide coat protein binding fragment of R17 ribonucleic acid. Biochemistry. 1982 Sep 14;21(19):4713–4720. doi: 10.1021/bi00262a030. [DOI] [PubMed] [Google Scholar]
- McCoy M. I., Lubben T. H., Gumport R. I. The purification of nuclease-free T4-RNA ligase. Biochim Biophys Acta. 1979 Mar 28;562(1):149–161. doi: 10.1016/0005-2787(79)90134-5. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Hecht S. M. A structurally modified yeast tRNAPhe with six nucleotides in the anticodon loop lacks significant phenylalanine acceptance. J Biol Chem. 1982 Sep 25;257(18):10536–10539. [PubMed] [Google Scholar]
- Ohtsuka E., Tanaka S., Tanaka T., Miyake T., Markham A. F., Nakagawa E., Wakabayashi T., Taniyama Y., Nishikawa S., Fukumoto R. Total synthesis of a RNA molecule with sequence identical to that of Escherichia coli formylmethionine tRNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5493–5497. doi: 10.1073/pnas.78.9.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Goddard J. P. Loss of methionine acceptor activity resulting from a base change in the anticodon of Escherichia coli formylmethionine transfer ribonucleic acid. J Biol Chem. 1973 Feb 25;248(4):1341–1345. [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Alteration of the kinetic parameters for aminoacylation of Escherichia coli formylmethionine transfer RNA by modification of an anticodon base. J Biol Chem. 1977 Feb 10;252(3):814–819. [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Structural requirements for aminoacylation of Escherichia coli formylmethionine transfer RNA. Biochemistry. 1977 Sep 20;16(19):4256–4265. doi: 10.1021/bi00638a020. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H., Sundari R. M. Structural requirements for recognition of Escherichia coli initiator and non-initiator transfer ribonucleic acids by bacterial T factor. J Biol Chem. 1974 Nov 25;249(22):7102–7110. [PubMed] [Google Scholar]
- Schulman L. H. Structure and function of E. coli formylmethionyl tRNA. I. Effect of modification of pyrimidine residues on aminoacyl synthetase recognition. Proc Natl Acad Sci U S A. 1970 Jun;66(2):507–514. doi: 10.1073/pnas.66.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber R., Malathi V. G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Squires C., Carbon J. Normal and mutant glycine transfer RNAs. Nat New Biol. 1971 Oct 27;233(43):274–277. doi: 10.1038/newbio233274a0. [DOI] [PubMed] [Google Scholar]
- Stern L., Schulman L. H. Role of anticodon bases in aminoacylation of Escherichia coli methionine transfer RNAs. J Biol Chem. 1977 Sep 25;252(18):6403–6408. [PubMed] [Google Scholar]
- THACH R. E., DOTY P. SYNTHESIS OF BLOCK OLIGONUCLEOTIDES. Science. 1965 Mar 12;147(3663):1310–1311. doi: 10.1126/science.147.3663.1310. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Lowary P. T., Wittenberg W. L. Role of the constant uridine in binding of yeast tRNAPhe anticodon arm to 30S ribosomes. Nucleic Acids Res. 1982 Jun 11;10(11):3341–3352. doi: 10.1093/nar/10.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel M., Koh C. K., Cohn W. E. Rapid ion-exchange chromatographic microanalysis of ultraviolet-absorbing materials and its application to nucleosides. Anal Biochem. 1968 Oct 24;25(1):77–98. doi: 10.1016/0003-2697(68)90083-3. [DOI] [PubMed] [Google Scholar]
- Wang G. H., Zhu L. Q., Yuan J. G., Liu F., Zhang L. F. Joining of yeast alanine transfer ribonucleic acid half molecules to form a whole molecule by T4 RNA ligase. Biochim Biophys Acta. 1981 Jan 29;652(1):82–89. doi: 10.1016/0005-2787(81)90211-2. [DOI] [PubMed] [Google Scholar]