Abstract
Some ABA-responsive genes are expressed only after ABA-dependent signaling, suggesting that their function may be required only transiently in the plant cell. The plant cell therefore needs an efficient regulatory mechanism to transcriptionally or post-translationally regulate the expression of such loci after stress. Although many plant stress-specific genes have been characterized to date, how plants readjust levels of a stress-induced protein when normal physiological conditions resume has not been addressed. By the same token, we know little about the molecular mechanism required for ABA homeostasis after signaling. We discuss recent findings showing that the ABA-induced TSPO protein in Arabidopsis is a hemoprotein and a potential porphyrin scavenger, and possible connection to ABA downregulation by the plant cell.
Key words: Arabidopsis thaliana, heme, TSPO, abiotic stress, abscisic acid, porphyrins trafficking, porphyrins metabolism
What goes up should come down and vice versa. Cellular homeostasis requires that every signaling process involving up or downregulation of a given pathway should only be transient and returning to steady state after a signaling process is as vital to living cells as being able to perceive and transduce changes of their environment. One of the best studied responses of plant cells subjected to water-related stress is the transient increase of the phytohormone abscisic acid (ABA). The increase in active ABA regulates the expression of ABA-responsive genes, some of which are strictly ABA-dependent in that their expression is almost undetectable in absence of elevated levels of cellular ABA. Since the function of these proteins may only be required transiently, a regulatory mechanism for transcriptionally and/or post-translationally regulate their expression should exist. In general during stress, molecular mechanisms aimed at shutting down the ABA-dependent signaling, as required at some point for the homeostasis of the plant cell, are poorly understood. The Arabidopsis TSPO (Translocator protein)-related protein is transiently induced by abiotic stresses and ABA treatment. Our recent work1 aiming at understanding the function and regulation of At-TSPO yielded exciting insights into the interplay among a stress-regulated protein, ABA responses, tetrapyrrole biosynthesis/scavenging and autophagy.2 We discuss these findings in relation to tetrapyrroles metabolism/trafficking and the regulation of ABA-dependent signaling by the plant cell.
ABA-Induced At-TSPO is Developmentally and Physiologically Regulated
At-TSPO is transcriptionally regulated by the master bZIP-type transcription factors AREB1, AREB2 and ABF3, which are involved in ABA-responsive elementdependent ABA signaling in Arabidopsis.3 At-TSPO transcripts are detectable in Arabidopsis developing seed as from the walking stick stage and culminate at the green cotyledons stage, and then decrease during after-ripening. At-TSPO expression ceases 24 hours after imbibition of dry seeds, probably as a direct consequence of ABA degradation. Expression of At-TSPO is almost undetectable after germination and seedling development. During pollen development, At-TSPO expression starts from the uninucleate microspore stage and culminates at the tricellular stage and then decrease thereafter, but is still present in mature pollen grain. The protein is abundant in desiccation-resistant plant structures, such as dry seeds and pollen grains, but can be induced transiently in vegetative tissues by abiotic stresses. After ABA treatment of Arabidopsis seedlings, At-TSPO expression is higher in the green tissues and lower in the root. We found that in the leaf, ABA-induced At-TSPO is relatively more abundant in guard cells than in pavement cells, and is almost undetectable in mesophyll cells. This was also true when At-TSPO was constitutively expressed in transgenic Arabidopsis plant.4,5 Overexpression of At-TSPO in cultured Arabidopsis cells rendered these cells more sensitive to salt stress. These observations suggest some level of developmental and cell-type-dependent post-transcriptional and/or post -translational regulation of At-TSPO.
At-TSPO Degradation Requires Heme-Binding and a Functional Autophagic Pathway
ABA-induced At-TSPO reached it maximum after about 24 hours of exogenous ABA treatment1,4 and declined thereafter to become undetectable as from 72 hours post induction. We showed previously that feeding δ-aminolevulinic acid, a precursor of tetrapyrroles, to plant tissues enhanced At-TSPO degradation.4 Given that TSPO proteins can bind porphyrins in vivo, this preliminary observation prompted us to investigate further a possible relationship between tetrapyrrole metabolism and At-TSPO stability in the plant cell.
We showed that At-TSPO can bind porphyrins including heme in vitro and in vivo. Heme binding required a conserved histidine residue (H91). Mutation of this histidine residue stabilized At-TSPO suggesting that heme binding capacity is required for the degradation of the protein. This degradation was mediated by autophagy, suggesting that the protein and its porphyrin ligand may be targeted to degradation. Indeed, overexpression of At-TSPO alleviated ALA-induced phototoxicity in plant cells indicating that plant TSPO may play a role in porphyrin binding and scavenging during stress in plants.
Despite the fact that tetrapyrroles including heme biosynthesis and their regulation have being extensively studied in eukaryotic cells, little is known about their intracellular trafficking with potentially very important consequences for the physiology of the cell. For example cytosolic unfettered heme can act as a signaling molecule in yeast, and in Chlamydomonas. It is not yet clear whether endogenous levels of unfettered porphyrins in the cytosol can mediate signaling for example between the prokaryotic organelles and the nuclear genome in plant cells.6,7 However, it stands to reason that amongst the cyclic porphyrins produced by the plastids in the plant cell, at least heme is transported to the cytoplasm and in other cellular compartment harboring apo-hemoproteins. In the cytosol, the lipophylic unfettered heme for instance can be a light-dependent trigger of lethal oxidative stress for the cell. We found that ABA can transiently upregulate the levels of free heme in the plant cell. It may be that the ABA-induced free heme is required by the parallel upregulation of reactive oxygen species scavengers (catalases, peroxidases) during stress. Since At-TSPO may also function as heme scavenger, its constitutive expression can be detrimental for ROS homeostasis and hemoproteins activities in the plant cell. Indeed, we found that an Arabidopsis transgenic cell line overexpressing At-TSPO accumulated more ROS than wild-type cells in presence or absence of ABA.
Heme Degradation by At-TSPO could Indirectly Regulate ABA 8′-hydroxylase Activity
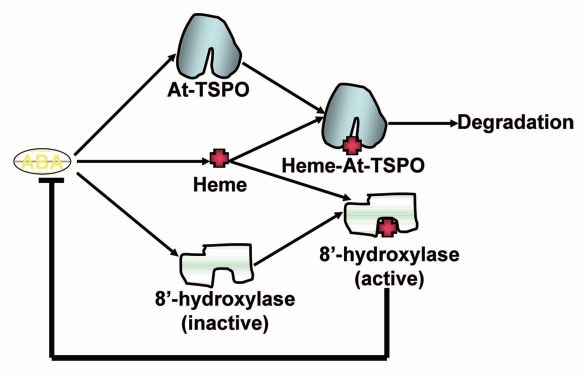
Endogenous ABA concentration in unstressed plant cell can be up to 0.7–1.5 µM (per guard cell for example), and can increase up to 30-fold during stress. This range of ABA concentration at steady state (unstressed plant) is sufficient to bind the core ABA receptors PYR/PYL/RCAR in vitro and activate the downstream signaling pathway. However, this amount of ABA appears not to induce the PYR/PYL/RCAR-dependent signaling in vivo, and it is likely that these ABA receptor complexes are antagonized in vivo in absence of stress.8 The maintenance of otherwise active amount of ABA in the plant cell also points to a potential physiological role of this phytohormone in absence of stress. ABA catabolism is required to restore ABA homeostasis after stress, and the main enzymes involved in irreversible inactivation of ABA in the plant cell are cytochrome P450 of the CYP707A subgroup. ABA-8′-hydroxylation is thought to play a predominant role in ABA catabolism. Plant ABA-8′-hydroxylases are endoplasmic reticulum-targeted membrane hemoproteins and are induced by ABA. The newly synthesized ABA-8′-hydroxylase needs heme as prosthetic group to be functional. We showed that exogenous ABA transiently upregulated heme biosynthesis in Arabidopsis cell. How the plastid-synthesized heme reaches the cytosol and how mechanistically ABA-8′-hydroxylases or any membrane-bound hemoproteins acquire it heme (co- orposttranslationally) is not yet clear. In any case, it may be that the ABA-dependent induction of ABA-8′-hydroxylases, concomitant to heme upregulation, is a prerequisite for ABA catabolism and homeostasis during stress. We showed that At-TSPO degradation required heme binding, suggesting that At-TSPO expression may partially regulate the level of unfettered heme in the cell during stress. Overexpression of At-TSPO might therefore sequester and divert some of the heme required by ABA-8′-hydroxylases (or other apo-hemoproteins), and hence affect ABA homeostasis in the plant cell (Fig. 1). Expression of At-TSPO could therefore interfere with ABA perception and signaling during stress. These potential connections may explain why the plant cell needs an effective and highly regulated pathway to induce and degrade At-TSPO during stress.
Figure 1.
Hypothetical model of ABA-dependent regulation of heme, At-TSPO and ABA signaling. ABA signaling induces At-TSPO and heme biosynthesis is upregulated transiently to accommodate the requirement for heme by ROS (reactive oxygen species) scavengers during stress; other hemoproteins including ABA degradation enzymes (8′-hydroxylase) also require heme for their activity; the ABA-induced hemoprotein At-TSPO is, at least in part, used as free porphyrins (heme) scavenger to prevent phototoxicity to the stressed cell; heme binding to the endoplasmic reticulum-localized 8′-hydroxylase allows efficient degradation and homeostasis of ABA in the cell.
References
- 1.Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23:785–805. doi: 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofmann NR. How a transient response becomes transient: Autophagy cleans up after abscisic acid. Plant Cell. 2011;23:429. [Google Scholar]
- 3.Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, et al. AREB1, AREB2 and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 4.Guillaumot D, Guillon S, Déplanque T, Vanhee C, Masquelier D, Morsomme P, et al. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 2009;60:242–256. doi: 10.1111/j.1365-313X.2009.03950.x. [DOI] [PubMed] [Google Scholar]
- 5.Guillaumot D, Guillon S, Morsomme P, Batoko H. ABA, porphyrins and plant TSPO-related protein. Plant Signal Behav. 2009;4:1087–1090. doi: 10.4161/psb.4.11.9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfannschmidt T. Plastidial retrograde signaling- a true “plastid factor” or just metabolite signature? Trends Plant Science. 2010;15:427–435. doi: 10.1016/j.tplants.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi Y, Imamura S, Hanoaka M, Tanaka K. A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat Cell Biol. 2011;13:483–487. doi: 10.1038/ncb2203. [DOI] [PubMed] [Google Scholar]
- 8.Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol. 2010;17:1102–1108. doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]