Abstract
The proteasome pathway regulates many aspects of biological processes in plants, such as plant hormone signaling, light responses, the circadian clock and regulation of cell division. Key cell cycle regulatory proteins including B-type cyclins, Cdc6, cyclin-dependent kinase inhibitors and E2Fc undergo proteasome-dependent degradation. We used the proteasome inhibitor MG132 to show that proteolysis of Arabidopsis RETINOBLASTOMA-RELATED 1 (AtRBR1) and three E2Fs is mediated by the proteasome pathway during sucrose starvation in Arabidopsis suspension MM2d cells. We found previously that estrogen-inducible RNAi-mediated downregulation of AtRBR1 leads to a higher frequency of arrest in G2 phase, instead of G1-phase arrest in the uninduced control, after sucrose starvation. Degradation of not only negative (AtRBR1 and E2Fc) but also positive (E2Fa and E2Fb) cell cycle regulators after sucrose starvation may be required for arrest in G1 phase, when cells integrate a variety of nutritional, hormonal and developmental signals to decide whether or not to commit to entry into the cell cycle.
Key words: arabidopsis, MM2d cells, RBR, E2F, sucrose starvation
Because of their sessile lifestyle, plants must be able to respond flexibly to changes in environmental conditions. Plants need to sustain a dynamic balance between growth rate and developmental pattern in response to intrinsic genetic cues and extrinsic environmental signals.1 One of the most important mediators of these signals is sucrose. Sucrose acts as an important signaling molecule that tightly regulates the patterns of gene expression in a spatiotemporal manner.2
In eukaryotes, cyclin-dependent kinases (CDKs) play pivotal roles in cell cycle control.3 Two major types of CDKs, CDKA and CDKB, participate principally in plant cell cycle control.4,5 Additionally, D-type cyclins (CYCDs) are thought to act as mediators linking extracellular and developmental signals to the cell cycle. GeneChip analysis revealed that expression of genes involved in S-phase entry decreases during sucrose starvation, a process which may be linked to G1-phase arrest.6
A number of cDNAs encoding E2Fs and RETINOBLASTOMA-RELATED (RBR) proteins have been identified in plants. In Arabidopsis, the E2F/DP family consists of six E2F and two DP proteins.7,8 E2Fa and E2Fb can bind with DPA to transcriptionally activate target genes,9,10 although E2Fc lacks transcriptional activation properties.11 It is assumed that RBR represses transcription by binding E2Fs, whereas CDKA-mediated phosphorylation of RBR during G1 phase releases a functional E2F-DP to activate target genes and allow cell cycle progression to S phase.
We reported previously that Arabidopsis suspension MM2d cells downregulating Arabidopsis RBR (AtRBR1) to evaluate the role of AtRBR1 in cell cycle arrest after sucrose starvation.12 Downregulation of AtRBR1 causes a higher frequency of arrest in G2 phase in response to stationary phase and sucrose starvation. Our data imply that AtRBR1 plays a key role in G1-phase arrest in sucrose starvation.
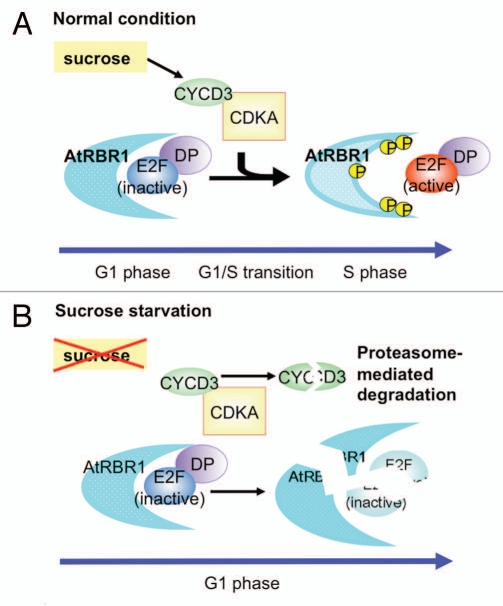
In our recent study, we found that AtRBR1 and three E2F proteins were degraded under limited sucrose conditions, while protein abundance increased in response to treatment with the proteasome inhibitor MG132.13 These results thus indicate that not only negative (AtRBR1 and E2Fc) but also positive (E2Fa and E2Fb) cell cycle regulators undergo proteasome-dependent degradation after sucrose starvation. The proteasome pathway regulates cell division by degrading key regulatory proteins including B-type cyclins,14 Cdc6,15 CDK inhibitors (CKIs),16–18 and E2Fc.19 The control of the transition from G1 to S phase is a critical step in cell cycle regulation, because before this G1/S transition cells have to integrate a variety of nutritional, hormonal and developmental signals to commit to entry into the cell cycle. CYCD3;1 is a highly unstable protein that undergoes proteasome-dependent degradation and disappears rapidly after sucrose starvation.20 In Arabidopsis MM2d cells, overexpressing CYCD3;1 partially overcomes the G1-phase arrest induced by sucrose removal. Given that AtRBR1 is most likely acting downstream of CYCD3;1 before the G1/S transition, overexpressing CYCD3;1 may cause inactivation of AtRBR1 by phosphorylation, leading to a higher frequency of arrest in G2 phase in response to sucrose availability. As observed in downregulation of AtRBR1, degradation of AtRBR1 after 12 h of sucrose-starved culturing may cause a higher frequency of arrest in G2 phase, thus suggesting a model in which degradation of E2F activators (E2Fa and E2Fb) may be required for maintaining G1-phase arrest after prolonged sucrose starvation (Fig. 1).
Figure 1.
Model of G1-phase arrest after sucrose starvation. (A) Under normal sucrose conditions, RBR represses transcription by binding E2Fs during early G1 phase. However, CDKA/CYCD3 phosphorylates RBR during late G1 phase, which results in releasing a functional E2F-DP to activate target genes involved in progression into S phase. (B) During sucrose starvation, CYCD3;1 undergoes proteasome-dependent degradation and disappears rapidly within several minutes. However, both AtRBR1 and E2Fs also undergo proteasome-dependent degradation, and this may be required for G1-phase arrest after prolonged sucrose starvation.
It has recently been shown that the plant hormone gibberellin promotes cell proliferation by stimulating the destruction of DELLA proteins, which restrain cell division by enhancing the accumulation of CKIs.21 It will be important to identify the mediator which links sucrose starvation and degradation of key cell cycle regulators.
References
- 1.Gutierrez C. Coupling cell proliferation and development in plants. Nat Cell Biol. 2005;7:535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- 2.Gupta AK, Kaur N. Sugar signaling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J Biosci. 2005;30:761–776. doi: 10.1007/BF02703574. [DOI] [PubMed] [Google Scholar]
- 3.Pines J. Four-dimensional control of the cell cycle. Nat Cell Biol. 1999;1:73–79. doi: 10.1038/11041. [DOI] [PubMed] [Google Scholar]
- 4.Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, Renaudi JP. CDK-related protein kinases in plants. Plant Mol Biol. 2000;43:607–620. doi: 10.1023/a:1006470301554. [DOI] [PubMed] [Google Scholar]
- 5.Inze D, De Veylder L. Cell cycle regulation in plant development. Ann Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 6.Contento AL, Kim SJ, Bassham DC. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 2004;135:2330–2347. doi: 10.1104/pp.104.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC. G(1) to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol. 2002;5:480–486. doi: 10.1016/s1369-5266(02)00301-1. [DOI] [PubMed] [Google Scholar]
- 8.Shen WH. The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 2002;7:505–511. doi: 10.1016/s1360-1385(02)02351-8. [DOI] [PubMed] [Google Scholar]
- 9.De Veylder L, Beeckman T, Beemster GT, de Almeida-Engler J, Ormenese S, Maes S, et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 2002;21:1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sozzani R, Maggio C, Varotto S, Canova S, Bergonioux C, Albani D, Cella R. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 2006;140:1355–1366. doi: 10.1104/pp.106.077990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosugi S, Ohashi Y. E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 2002;29:45–59. doi: 10.1046/j.1365-313x.2002.01196.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirano H, Harashima H, Shinmyo A, Sekine M. Arabidopsis G1 cell cycle proteins undergo proteasome-dependent degradation during sucrose starvation. Plant Mol Biol. 2008;66:259–275. [Google Scholar]
- 13.Hirano H, Shinmyo A, Sekine M. Arabidopsis RETINOBLASTOMA-RELATED PROTEIN1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Physiol Biochem. 2011;49:687–691. [Google Scholar]
- 14.Weingartner M, Pelayo HR, Binarova P, Zwerger K, Melikant B, de la Torre C, et al. A plant cyclin B2 is degraded early in mitosis and its ectopic expression shortens G2-phase and alleviates the DNA-damage checkpoint. J Cell Sci. 2003;116:487–498. doi: 10.1242/jcs.00250. [DOI] [PubMed] [Google Scholar]
- 15.Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell. 2001;13:2671–2686. doi: 10.1105/tpc.010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, et al. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase comlexes. Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakoby MJ, Weinl C, Pusch S, Kuijt SJ, Merkle T, Dissmeyer N, Schnittger A. Analysis of the subcellular localization, function and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 2006;141:1293–1305. doi: 10.1104/pp.106.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai J, Chen H, Teng K, Zhao Q, Zhang Z, Li Y, et al. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 2009;57:905–917. doi: 10.1111/j.1365-313X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
- 19.del Pozo JC, Boniotti MB, Gutierrez C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell. 2002;14:3057–3071. doi: 10.1105/tpc.006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planchais S, Samland AK, Murray JAH. Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 2004;38:616–625. doi: 10.1111/j.0960-7412.2004.02071.x. [DOI] [PubMed] [Google Scholar]
- 21.Achard P, Gustl A, Cheminant S, Alioua M, Dhondt S, Coppens F, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Cur Biol. 2009;19:1189–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]