Abstract
Autophagy is a conserved intracellular recycling system that traffics cellular organelles and cytosolic proteins within lysosomes for reuse or breakdown in eukaryotes. Increased evidence indicates that autophagy is involved in programmed cell death and disease resistance in plants. We recently showed that atg2, atg5, atg7 and atg10 displayed early senescence and cell death in later growth stage under nutrient-rich conditions in Arabidopsis thaliana. These mutants also exhibited powdery mildew resistance and mildew-induced cell death. Salicylic acid (SA) signaling is required for atg2-mediated powdery mildew resistance, however, inactivation of SA signaling is not sufficient to fully suppress powdery mildew-induced cell death in atg2 mutant.1 Here, we show that atg18a-2 is also resistant to the powdery mildew pathogen, Golovinomyces cichoracearum, and it shows mildew-induced cell death similar to the atg2 mutant. Taken together, our study reveals that autophagy plays important roles in suppression of cell death and defense response to the biotrophic pathogen, the powdery mildew fungus. Future work on autophagy in plants will shine light on how autophagy is involved in cell death and defense response in plants.
Key words: autophagy, cell death, defense response, ATG2, ATG18a, powdery mildew
Autophagy is a highly conserved biological process in eukaryotes from yeast to human. During autophagy, autophagosome (a double membrane structure) facilitates traffic of cellular organelles and cytosolic macromolecules into vacuole/lysosome for recycling or degradation.2–5 A number of genes required for autophagosome formation were isolated by genetic studies in yeast.5–7 Most of the core autophagy-related (ATG) genes are well conserved in higher eukaryotes, and to date, more than 30 Arabidopsis ATG genes have been identified.8–10
In plants, several atg mutants have been shown to display early senescence and/or spontaneous cell death in nutrient deficient or even in nutrient-rich conditions.8,9,11–18 Recently, we showed that atg2, atg5, atg7 and atg10 exhibited early senescence and spontaneous cell death under nutrient rich conditions.1 In addition, we showed that these mutants displayed enhanced disease resistance to the powdery mildew pathogen, G. cichoracearum. However, the atg9 mutant was similar to wild type in the absence or presence of the powdery mildew pathogen, indicating that different ATG genes may have different or redundant function.
One interesting finding in our previous report was that the atg2-mediated resistance is dependent on SA signaling, however, inactivation of SA signaling can not fully suppress atg2-mediated powdery mildew-induced cell death, indicating that cell death could be uncoupled from resistance, and that cell death is not sufficient for powdery mildew resistance.1
Several studies show that autophagy contributes to pathogen-induced hypersensitive response (HR).12,17,19,20 For instance, the restriction of HR cell death at the infection site was affected in ATG6 silenced tobacco plants and ATG6 antisense Arabidopsis plants.12,19 To further study the role of autophagy in disease resistance, we examined the atg18a-2 mutant for powdery mildew resistance. Previously, it has been shown mutation in ATG18a leads to early senescence phenotype.14 To assess atg18a-2 powdery mildew resistance, the atg18a-2 mutant was grown under standard growth conditions for 4 weeks and the plant was infected with G. cichoracearum. As shown in Figure 1A and B, atg18a-2 exhibited more pronounced cell death in the infected leaves than the wild type plant. To further investigate atg18a-2 resistance phenotype, we monitored the fungal growth by counting the conidiophores in the infected leaves. The atg18a-2 mutant supported significantly fewer conidiophores than wild type plants (Fig. 1C). These observations indicate that ATG18a also negatively affects both cell death and powdery mildew resistance, similar to ATG2, which was studied as well in Figure 1, as a positive control.
Figure 1.
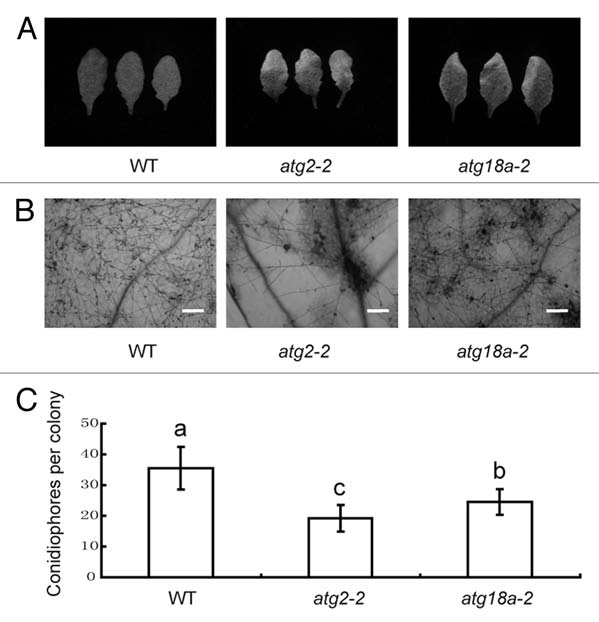
The atg18a-2 mutant displays enhanced powdery mildew resistance and mildew-induced cell death. (A) Four-week-old wild type, atg2, atg18a-2 plants were inoculated with G. cichoracearum. Leaves were removed from plants and photographed at 7 days post inoculation (dpi). (B) Trypan blue staining of leaves in (A). Bar = 100 µm. (C) Quantification of disease resistance by calculating the number of conidiophore per colony at 7 dpi in each genotype. Results represent mean ± SD in one experiment (n > 25). One-way AN OVA was performed for statistical analyses. Statistically significant differences between different genotype were indicated with different letters. Similar results were obtained from three independent experiments.
The interaction between autophage, cell death and disease resistance has emerged as an important topic in plant science. However, how autoghage affects cell death and resistance is not well understood. Hofius et al. reported that autophagy positively regulated disease resistance against virulent Pto DC3000, the Emwa isolate of H. arabidopsidis and avirulent bacteria,20 while Lenz et al. showed that loss-of-function of ATG5, ATG7, ATG10 and ATG18a conferred enhanced disease resistance to virulent bacteria Pto DC3000.21,22 In another report, Yoshimoto et al. did not observe obvious difference in response to avirulent Pto DC3000 avrRpm1.17 Similarly, we did not find significant differences between atg2 and wild type upon infection with virulent or avirulent Pto DC3000. However, atg2, atg5, atg7, atg10 and atg18a-2 showed enhanced resistance to G. cichoracearum. Furthermore, enhanced disease resistance in atg2 was correlated with increased accumulation of transcripts of defense-related genes, such as PR1.1 Recently, Lenz et al. and Lai et al. reported that atg5, atg7, atg10 and atg18a mutants exhibited enhanced susceptibility to B. cinerea,21,23 indicating that autophagy positively affects defense response toward necrotrophic pathogen.
Accumulating evidence indicates that autophagy plays important role in plant defense response and disease resistance. However, there are still some debate about the role of autophagy towards pathogens. Here we showed atg18a-2 also displayed enhanced disease resistance to powdery mildew, indicating that autophagy plays a negative role in disease resistance to biotrophic pathogens.
Acknowledgments
We thank ABRC for providing the atg18a-2 T-DNA insertion line. This work was supported by grants from National Basic Research Program of China (2011CB100700) and the National Natural Science Foundation of China (30771168).
References
- 1.Wang Y, Nishimura MT, Zhao T, Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04669.x. In press. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 5.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 7.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 8.Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimoto K, Takano Y, Sakai Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010;584:1350–1358. doi: 10.1016/j.febslet.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42:535–546. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- 15.Qin G, Ma Z, Zhang L, Xing S, Hou X, Deng J, et al. Arabidopsis AtBECLIN 1/AtATG6/AtVPS30 is essential for pollen germination and plant development. Cell Res. 2007;17:249–263. doi: 10.1038/cr.2007.7. [DOI] [PubMed] [Google Scholar]
- 16.Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12a and ATG12b loci. Plant J. 2010;62:483–493. doi: 10.1111/j.1365-313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- 19.Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- 20.Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 21.Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011;66:818–830. doi: 10.1111/j.1365-313X.2011.04546.x. [DOI] [PubMed] [Google Scholar]
- 22.Lenz HD, Vierstra RD, Nurnberger T, Gust AA. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal Behav. 2011;6:1040–1042. doi: 10.4161/psb.6.7.15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai Z, Wang F, Zheng Z, Fan B, Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66:953–968. doi: 10.1111/j.1365-313X.2011.04553.x. [DOI] [PubMed] [Google Scholar]


