Abstract
The Arabidopsis thaliana F-box protein MAX2 has been discovered in four separate genetic screens, indicating that it has roles in leaf senescence, seedling photosensitivity, shoot outgrowth and seed germination. Both strigolactones and karrikins can regulate A. thaliana seed germination and seedling photomorphogenesis in a MAX2-dependent manner, but only strigolactones inhibit shoot branching. How MAX2 mediates specific responses to both classes of structurally-related signals, and the origin of its dual role remains unknown. The moss Physcomitrella patens utilizes strigolactones and MAX2 orthologs are present across the land plants, suggesting that this signaling system could have an ancient origin. The seed of parasitic Orobanchaceae species germinate preferentially in response to strigolactones over karrikins, and putative Orobanchaceae MAX2 orthologs form a sub-clade distinct from those of other dicots. These observations suggest that lineage-specific evolution of MAX2 may have given rise to specialized responses to these signaling molecules.
Key words: karrikins, strigolactones, F-box protein, seed germination, photomorphogenesis, parasitic weeds, mycorrhiza, moss, axillary branching
A Role for MAX2 in Responses to Karrikins
Delaying germination allows plants to defer the critical stage of seedling establishment until environmental conditions are suitable, maximizing the chances of a successful transition to maturity. Physiological seed dormancy is an adaptive mechanism that prevents germination until particular endogenous and external conditions are met. Periodic bushfires provide a brief opportunity for plants to capitalise on the reduced competition for light, nutrients and water. As such, dormant seed of many plants from diverse families will germinate when exposed to smoke.1 Intensive efforts over recent years have led to the identification of a key bioactive compound in smoke, the butenolide 3-methyl-2H-furo[2,3-c]pyran-2-one, now known as karrikinolide or KAR1.2 Several related compounds have since been discovered in smoke, which together comprise a small family known as karrikins.3 Karrikins are potent germination stimulants, acting at concentrations as low as 1 nM.2 They also enhance post-germination responses to light, potentially enhancing seedling survival in the post-fire environment.4
Although Arabidopsis thaliana is not associated with fire-prone environments, the discovery that it is highly sensitive to karrikins unlocked an array of resources for identifying the genetic mechanisms of karrikin perception, signaling and response.5 Recently we initiated a screen for Arabidopsis mutants incapable of responding to karrikins. Two karrikin insensitive (kai) mutants, kai1-1 and kai1 2, exhibited increased seed dormancy that could not be recovered by the application of KAR1. Both mutants exhibited additional phenotypes: elongated hypocotyls, increased axillary shoot branching, reduced inflorescence height, delayed leaf senescence and curling of the leaf margins. As many of these phenotypes are observed in the Arabidopsis mutant more axillary growth 2 (max2), we sequenced the MAX2 gene in the kai1-1 and kai1-2 mutants and discovered that each carried frameshift alleles of MAX2. Additional max2 alleles also conferred increased seed dormancy and insensitivity to KAR1, supporting our conclusion that MAX2 is required for karrikin responses.6
Multiple Functions for MAX2
MAX2 has been implicated in several aspects of plant development, having been identified independently in screens for delayed leaf senescence (ore9) and light hyposensitivity (pps). However, MAX2 is most recently renowned for its role in responses to strigolactones, a class of plant-synthesized, carotenoid-derived hormones that inhibit shoot branching,7–9 influence root architecture,10–12 and promote the germination of parasitic weeds.13,14 Strigolactones also promote hyphal branching in arbuscular mycorhizal fungi.15 Two carotenoid-cleavage dioxygenases, CCD7/MAX3 and CCD8/MAX4, as well as a cytochrome P450, MAX1, are involved in strigolactone biosynthesis in Arabidopsis. All of the more axillary growth mutants share an increased shoot branching phenotype, but while max1, ccd7/max3 and ccd8/max4 branching can be restored to wild type levels by the supply of exogenous strigolactones, max2 cannot, suggesting that MAX2 is specifically required for strigolactone perception or signal transduction. Mutants in orthologous genes in rice, petunia and pea have demonstrated that this pathway for strigolactone control of shoot branching is conserved in higher plants.7,16,17
It is remarkable that MAX2 mediates responses to both strigolactones and karrikins, despite these growth regulators being produced in different manners and having distinct known ecological roles. Karrikins and strigolactones are partially similar in structure, having a butenolide ring in common (Fig. 1). We found that in Arabidopsis both compounds are capable of promoting seed germination, enhancing photomorphogenesis, and regulating a common set of early transcriptional response markers.4,5 However, karrikins are completely ineffective as inhibitors of shoot branching, being unable to rescue the max3 and max4 phenotypes.6 This fact demonstrates that while karrikins and strigolactones both signal through MAX2, the two classes of compounds are not equivalent and, at least in some developmental stages, there must be a means to perceive each of them distinctly. Indeed, while max2 mutants have increased seed dormancy and abnormally long hypocotyls, the strigolactone-deficient mutants max1, ccd7/max3 and ccd8/max4 do not, indicating that the loss of strigolactone signaling per se is probably not responsible for these aspects of the max2 phenotype.6 Thus, considering also its role in the regulation of leaf senescence and photomorphogenesis, MAX2 is a fundamentally important protein with several distinct functions in plant development.
Figure 1.
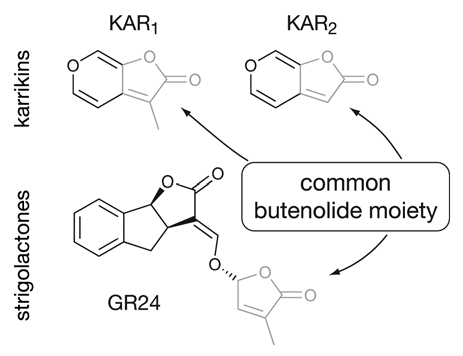
Chemical structure of two bioactive karrikins, KAR1 and KAR2, and of the synthetic strigolactone GR24. Note that both classes of compounds have a butenolide ring in common (grey) which is required for activity.
Implications of MAX2 Conservation among Land Plants
Although strigolactones were originally identified as germination stimulants of Striga spp. and then as promoters of mycorrhizae formation,18 strigolactone signaling systems are not limited to parasitic or mycorrhizal species, and appear to have an ancient origin among land plants. For example, it was recently demonstrated that strigolactones are produced and have developmental roles in the moss Physcomitrella patens, which unlike other bryophytes is not mycorrhizal.19 Mutation of the moss ortholog of CCD8 resulted in a reduction of strigolactone levels in the surrounding media. The Ppccd8Δ mutants exhibited a variety of growth phenotypes including earlier spore germination, increased branching of chloronemata and larger colonies due to continued elongation and branching of caulonemata. Interestingly, the growth of Ppccd8Δ was insensitive to colony density, at the cost of reproductive capacity, suggesting that strigolactones may have a role in quorum sensing. It is also notable that expression of the PpCCD7 ortholog in moss displayed strigolactone-responsive feedback inhibition in a similar manner to that seen for CCD7 in angiosperms.19–21
As a close homolog of MAX2 is present in the P. patens genome (45% identity) and at least some features of the angiosperm strigolactone biosynthesis pathway appear to be conserved, we hypothesize that PpMAX2 is required for moss responses to strigolactones, as it is in higher plants. This can readily be tested, as P. patens is amenable to homologous recombination.22 It would also be highly interesting to determine if karrikins can influence moss spore germination or development, as do strigolactones. If they do not, this could indicate that the capacity to recognize karrikins emerged after the development of a strigolactone signaling mechanism, or that a karrikin response pathway was lost in bryophytes while a strigolactone pathway was maintained.
In a survey of post-fire dynamics of bryophyte colonization in Tasmania, it was noted that the colonizing species were not among those bryophytes common to adjacent, unburnt areas,23 suggesting a fire-adapted trait may be in play. Examples of bryophyte spore banks and spore dormancy have also been described,24 setting up a line of inquiry that parallels angiosperm fire ecology. Can smoke, or specifically karrikins, activate spore germination of fire-following bryophytes as it can for the seed of many angiosperms, or does another post-fire cue such as light or nutrient availability influence recolonization patterns?
Parasitic weeds of the Orobanchaceae and Orobanche have a significant negative impact on agricultural yields in Africa and western Asia; millions of farmers already face substantial crop loss from these weeds, and as infestations continue to spread and intensify the economic cost is expected to be billions of USD.25 The seed of these parasitic weeds can lie dormant in the soil for up to 14 years, waiting to detect a nearby host. Extraordinary sensitivity to strigolactones exuded by a host root induces the parasitic weed seed to germinate, extend toward the host root, and form a haustorium connection to the host's phloem and/or xylem.26 There has been one report that a purified smokewater fraction containing KAR1 can promote germination of several parasitic weed species, including Striga hermonthica and Orobanche minor;27 however, three independent labs have been unable to detect a germination response with chemically-synthesized KAR1 applied to S. hermonthica, O. minor, O. aegyptica or O. crenata seed.5,28 Therefore we consider germination of these parasitic weeds to be a strigolactone-specific response. As we have shown that promotion of Arabidopsis thaliana seed germination by strigolactones and karrikins requires MAX2, we hypothesize that MAX2 orthologs are components of the highly sensitive germination response to strigolactones in parasitic weeds.
The transcriptomes of several parasitic weeds—Striga hermonthica, Orobanche aegyptica and Triphysaria versicolor—are currently being surveyed with deep-sequencing methods by the Parasitic Plant Genome Project (PPGP, ppgp.huck.psu.edu/). In addition to an assembled MAX2 ortholog sequence from Orobanche aegyptica (contig OrAe0GB1_43412), we have combined contigs from multiple library assemblies from PPGP to generate putative full-length MAX2 ortholog sequences from S. hermonthica and T. versicolor. These three putative parasitic plant MAX2 sequences have 58% identity to full-length Arabidopsis thaliana MAX2, and 73% identity among the highly conserved C-terminal 200 amino acids. These sequences form a monophyletic clade within the larger MAX2 family, representatives of which are present across the land plant groups (Fig. 2). It would be highly interesting to determine whether MAX2 polymorphisms in these parasitic species, or the loss of a karrikin receptor, have led to the strigolactone-specific germination response. We anticipate that a better understanding of the MAX2-dependent seed germination mechanism will lead to more sophisticated approaches for controlling Striga and Orobanche spp. infestation.
Figure 2.
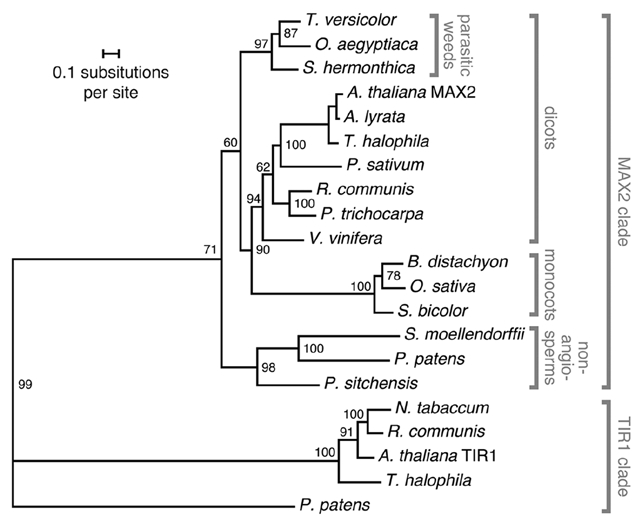
Maximum likelihood (ML) phylogeny of MA X2 orthologs from land plants. Sequences were obtained from GenBank (www.ncbi.nlm.nih.gov) and the Parasitic Plant Genome Project (ppgp.huck.psu.edu), using the Arabidopsis thaliana MA X2 amino acid sequence as a query. Full-length sequences were aligned using MA FFT (mafft.cbrc.jp/alignment/software/), and regions of poor alignment were removed manually with PFAAT.41 The phylogeny was constructed using RAxML BlackBox.42 The tree was rooted on a clade comprising Arabidopsis thaliana TIR1 and its orthologs. Numbers indicate branch support (100 bootstrap replicates). GenBank protein IDs for sequences used to generate the tree are: Arabidopsis thaliana, 18406017 (MA X2) and 254028670 (TIR1); Arabidopsis lyrata, 297824229; Thellungiella halophila, 312282253 and 312281471; Pisum sativum, 89329716; Ricinus communis, 255575295 and 255559322; Nicotiana tabacum, 254028670; Populus trichocarpa, 224128748; Vitis vinifera, 302143426; Oryza sativa cv. Japonica, 297724489; Sorghum bicolor, 242092018; Physcomitrella patens, 168039586 and 168062926; Selaginella moellendorffii, 302816439; and Picea sitchensis, 148906666. The Brachypodium distachyon sequence was derived from the locus identifier Bradi1g49120.1 (db.brachypodium.org). For Orobanche aegyptiaca, the protein sequence was derived from PPGP contig OrAe0GB1_43412. For Triphysaria versicolor and Striga hermonthica, sequences were assembled from multiple PPGP sequences to generate a contig that encoded a putative full-length protein.
MAX2 in Action
MAX2 encodes an F-box protein with C-terminal leucine-rich repeats (LRR). While some classes of F-box proteins are highly prone to birth and death in genomes, MAX2 is not; it is one of 20 ‘evolutionarily highly conserved’ F-box genes with only one copy present in the Arabidopsis thaliana, Oryza sativa and Populus trichocarpa genomes (Fig. 2).29,30 The F-box refers to an N-terminal motif which mediates interactions with the other core components of the SCF class of E3 ubiquitin-protein ligase complexes: Skp1 (ASK1 in Arabidopsis), Cullin1 and Rbx1. While these core subunits provide the ubiquitin ligase activity, the F-box subunit is thought to confer specificity for target proteins. Among the 692 F-box proteins in Arabidopsis, MAX2 belongs to a subfamily of 33 members with similar F-box-LRR domain organization.30 A number of members of this subfamily have already been found to have key roles in phytohormone signaling and plant growth regulation. Examples include the auxin receptors TIR1,31,32 AFB1, AFB2, AFB3 and AFB5;33,34 the jasmonate coreceptor COI1;35,36 the negative regulators of ethylene signaling EBF1 and EBF2;37,38 SLOMO, which has roles in auxin homeostasis and lateral organ initiation at the shoot meristem;39 and the regulators of plant growth and lateral root formation VFB1, VFB2, VFB3 and VFB4/SKIP2.40
As with other F-box proteins involved in hormone signaling, it is unsurprising that MAX2 has such a pleiotropic range of functions. Like its newfound roles in karrikin perception and seed dormancy, it is not unlikely that additional functions for MAX2 will be discovered in the future. Identifying the protein targets of SCFMAX2 is a key area of future research that will shed light on this issue. An important distinction from other characterized F-box proteins, such as TIR1, is that MAX2 is involved in the response to at least two distinct classes of compounds. As we noted previously in reference 6, the strigolactoneindependent phenotypes of max2 suggest that there may be a novel endogenous signal that max2 mutants can no longer perceive. Thus our results raise the possibility that MAX2 has an even broader importance to signal transduction networks in plants than has previously been considered, with its celebrated role in strigolactone responses being only one of many.
References
- 1.Roche S, Koch J, Dixon K. Smoke enhanced seed germination for mine rehabilitation in the southwest of western Australia. Restor Ecol. 1997;5:191–203. [Google Scholar]
- 2.Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. A compound from smoke that promotes seed germination. Science. 2004;305:977. doi: 10.1126/science.1099944. [DOI] [PubMed] [Google Scholar]
- 3.Flematti GR, Goddard-Borger ED, Merritt DJ, Ghisalberti EL, Dixon KW, Trengove RD. Preparation of 2H-furo[2,3-c]pyran-2-one derivatives and evaluation of their germination-promoting activity. J Agric Food Chem. 2007;55:2189–2194. doi: 10.1021/jf0633241. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DC, Flematti GR, Riseborough JA, Ghisalberti EL, Dixon KW, Smith SM. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:7095–7100. doi: 10.1073/pnas.0911635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson DC, Riseborough JA, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, et al. Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 2009;149:863–873. doi: 10.1104/pp.108.131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2011;108:8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17:1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 9.Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 10.Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta. 2011;233:209–216. doi: 10.1007/s00425-010-1310-y. [DOI] [PubMed] [Google Scholar]
- 11.Koltai H. Strigolactones are regulators of root development. New Phytologist. 2011;190:545–549. doi: 10.1111/j.1469-8137.2011.03678.x. [DOI] [PubMed] [Google Scholar]
- 12.Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 2011;155:721–734. doi: 10.1104/pp.110.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dor E, Yoneyama K, Wininger S, Kapulnik Y, Yoneyama K, Koltai H, et al. Strigolactone deficiency confers resistance in tomato line SL-ORT1 to the parasitic weeds Phelipanche and Orobanche spp. Phytopathology. 2011;101:213–222. doi: 10.1094/PHYTO-07-10-0184. [DOI] [PubMed] [Google Scholar]
- 14.Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta. 2007;225:1031–1038. doi: 10.1007/s00425-006-0410-1. [DOI] [PubMed] [Google Scholar]
- 15.Dor E, Joel DM, Kapulnik Y, Koltai H, Hershenhorn J. The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta. 2011;234:419–427. doi: 10.1007/s00425-011-1452-6. [DOI] [PubMed] [Google Scholar]
- 16.Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, et al. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51:1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- 17.Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, et al. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth and flower development. Plant Cell. 2005;17:746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X, Yoneyama K, Yoneyama K. The strigolactone story. Annu Rev Phytopathol. 2010;48:93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- 19.Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, et al. Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development. 2011;138:1531–1539. doi: 10.1242/dev.058495. [DOI] [PubMed] [Google Scholar]
- 20.Dun E, Hanan J, Beveridge C. Computational Modeling and Molecular Physiology Experiments Reveal New Insights into Shoot Branching in Pea. Plant Cell. 2009;21:3459. doi: 10.1105/tpc.109.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, et al. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 2006;142:1014–1026. doi: 10.1104/pp.106.087676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer DG, Zrÿd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 23.Duncan D, Dalton P. Recolonization by bryophytes following fire. J Bryology. 1982;12:53–63. [Google Scholar]
- 24.During H. New frontiers in bryology and lichenology—Diaspore banks. Bryologist. 2001;104:92–97. [Google Scholar]
- 25.Parker C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci. 2009;65:453–459. doi: 10.1002/ps.1713. [DOI] [PubMed] [Google Scholar]
- 26.Cardoso C, Ruyter-Spira C, Bouwmeester HJ. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 2011;180:414–420. doi: 10.1016/j.plantsci.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Daws MI, Pritchard HW, van Staden J. Butenolide from plant-derived smoke functions as a strigolactone analogue: Evidence from parasitic weed seed germination. South African Journal of Botany. 2008;74:116–120. [Google Scholar]
- 28.Chiwocha SDS, Dixon KW, Flematti GR, Ghisalberti EL, Merritt DJ, Nelson DC, et al. Karrikins: a new family of plant growth regulators in smoke. Plant Science. 2009;177:252–256. [Google Scholar]
- 29.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 30.Xu G, Ma H, Nei M, Kong H. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA. 2009;106:835–840. doi: 10.1073/pnas.0812043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 32.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 33.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, et al. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 2006;142:542–552. doi: 10.1104/pp.106.085969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, et al. The Arabidopsis CORONATINE INSENSITIVE1 Protein Is a Jasmonate Receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, et al. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell. 2007;19:509–523. doi: 10.1105/tpc.106.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 39.Lohmann D, Stacey N, Breuninger H, Jikumaru Y, Muller D, Sicard A, et al. SLOW MOTION Is Required for Within-Plant Auxin Homeostasis and Normal Timing of Lateral Organ Initiation at the Shoot Meristem in Arabidopsis. Plant Cell. 2010;22:335–348. doi: 10.1105/tpc.109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwager KM, Calderon-Villalobos LIA, Dohmann EMN, Willige BC, Knierer S, Nill C, et al. Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell. 2007;19:1163–1178. doi: 10.1105/tpc.105.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caffrey D, Dana P, Mathur V, Ocano M, Hong EJ, Wang Y, et al. PFAAT version 2.0: A tool for editing, annotating and analyzing multiple sequence alignments. BMC Bioinformatics. 2007;8:381. doi: 10.1186/1471-2105-8-381. 2007; 8: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A, Hoover P, Rougemont J. A rapid boot-strap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]


