Abstract
Physiological and pathophysiological stress attenuates global translation via phosphorylation of eIF2α. This in turn leads to the reprogramming of gene expression that is required for adaptive stress response. One class of cellular messenger RNAs whose translation was reported to be insensitive to eIF2α phosphorylation-mediated repression of translation is that harboring an Internal Ribosome Entry Site (IRES). IRES-mediated translation of several apoptosis-regulating genes increases in response to hypoxia, serum deprivation or gamma irradiation and promotes tumor cell survival and chemoresistance. However, the molecular mechanism that allows IRES-mediated translation to continue in an eIF2α-independent manner is not known. Here we have used the X-chromosome linked Inhibitor of Apoptosis, XIAP, IRES to address this question. Using toeprinting assay, western blot analysis and polysomal profiling we show that the XIAP IRES supports cap-independent translation when eIF2α is phosphorylated both in vitro and in vivo. During normal growth condition eIF2α-dependent translation on the IRES is preferred. However, IRES-mediated translation switches to eIF5B-dependent mode when eIF2α is phosphorylated as a consequence of cellular stress.
INTRODUCTION
A sizeable proportion of cellular messenger RNAs (mRNAs) has been shown to be translated by a cap-independent mechanism using an Internal Ribosome Entry Site (IRES) element (1–3). Many mRNAs that contain IRES encode proteins that play important roles in cell growth, proliferation, differentiation and the regulation of apoptosis. The IRES mechanism is utilized preferentially during conditions when normal cap-dependent translation initiation is compromised and in fact represents a critical survival ‘switch’ during oncogenesis (3,4). Translational control by internal initiation thus represents a novel and unique regulatory mechanism that is critical for cell survival; however, the mechanistic understanding of cellular IRES-mediated translation is missing.
Initiation of translation in eukaryotes is a complex and highly regulated process. Briefly, eukaryotic mRNAs contain a m7G cap structure at the 5′-end that is essential for their translation. Cap-dependent initiation requires interaction of initiation factor eIF4E (the cap-binding protein) and its partners eIF4A and eIF4G with the 5′-end of the mRNA, which is then followed by recruitment of ribosomal subunits, recognition of the AUG start codon and commencement of polypeptide chain elongation (1,5). In contrast, IRES are thought to recruit 40S ribosomal subunit directly to the vicinity of the initiation codon, thereby bypassing the requirement for the 5′ cap-structure and eIF4E binding. With the exception of some viral RNAs, there is no evidence to suggest that the steps following 40S recruitment to the mRNA differ between the cap- and IRES-dependent translation initiation mechanisms of eukaryotic mRNAs.
Cells respond to physiological and pathophysiological stress by phosphorylation of α subunit of eIF2 which results in an attenuation of global protein synthesis. eIF2 is required for the formation of ternary complex and delivery of Met-RNAiMet into the P-site of the ribosome to initiate protein synthesis (6). Phosphorylation of eIF2α leads to reduced availability of ternary complex, and a concomitant decrease in global translation rates (6). However, translation of some mRNAs (such as ATF4 and GCN2) continues (or is even enhanced) under conditions that cause eIF2α phosphorylation since their translation is controlled by upstream open reading frames, a setting in which low levels of ternary complex promote translation of the downstream open reading frame (6). Continuous translation of several IRES-containing mRNAs during conditions of phosphorylated eIF2α and reduced ternary complex availability has also been reported. For example, translation of vascular endothelial growth factor (VEGF) was substantially increased during tumor hypoxia (4), while the IRES-mediated translation of X-chromosome linked Inhibitor of Apoptosis (XIAP) sustained during ER stress (7), or was increased in response to serum starvation, low-dose gamma irradiation or glucose deficiency (8–11). It is not clear, however, how IRES-mediated translation of cellular mRNAs proceeds under these conditions, and what is the precise molecular mechanism that allows cellular IRES to function in an eIF2α-independent manner. We have used the XIAP IRES element to address this question.
XIAP is a prototype member of the Inhibitor of Apoptosis (IAP) protein family, which binds to and inhibits key caspases involved both in the initiation and execution steps of apoptosis (12). There is now overwhelming evidence for a role for the IAP genes, and XIAP in particular, in cancer (13). Targeting of XIAP is a promising therapeutic opportunity in the treatment of cancer and is currently in clinical trials (14–17). XIAP is encoded by two distinct mRNAs that differ in their 5′-UTRs; the major, shorter 5′-UTR promotes a basal level of XIAP expression under normal growth conditions, while the less abundant, longer 5′-UTR contains an IRES element and supports cap-independent translation during stress (11). Importantly, the IRES-mediated upregulation of XIAP in response to irradiation, IL-6 treatment or cellular transformation enhances cell survival, confirming that IRES-dependent translation of XIAP is critically involved in mediating tumor cell survival (8,9,18,19). In this report we have extensively characterized the XIAP IRES by toeprinting assay for its function in vitro. We demonstrate that XIAP IRES is able to form translation-competent initiation complexes in a cap-independent manner, both in untreated and poly I:C treated rabbit reticulocyte lysate (RRL), which induces eIF2α phosphorylation. Furthermore, we describe the mechanism by which translation initiation proceeds on cellular IRES when eIF2α is phosphorylated. We identify an eIF5B-dependent switch that promotes IRES-mediated translation during conditions of pathophysiological stress both in vitro and in vivo. Our data demonstrate that the eIF2α-dependent pathway is utilized for IRES-dependent translation initiation during normal growth conditions, whereas cellular stresses that inactivate eIF2α by phosphorylation cause IRES-dependent translation to switch to an eIF5B-dependent mode.
MATERIALS AND METHODS
Constructs, cell culture and transfection
The hepatitis C virus (HCV) IRES construct described previously (20,21), which contains both streptotag aptamer and a unique toeprint primer-binding site, was obtained as a gift from Dr Peter Lukavsky. The XIAP IRES construct was generated by replacing the HCV IRES with the XIAP IRES sequence (171 nt) along with 38 nt of coding region and T7 promoter sequences. Similarly, the human β-globin 5′-UTR (50 nt along with 248 nt coding sequence; derived from clone 6663141, Open Biosystems) was used to replace the HCV IRES. The PPT mutant and SC mutants were created by site-directed mutagenesis of the XIAP IRES construct using primers listed in Supplementary Table S1. All plasmids were verified by restriction digest and sequencing.
HEK293T cells were maintained in standard conditions in Dulbecco’s modified Eagle’s medium (DMEM) and transfection of small interfering RNAs (siRNAs) was performed using Lipofectamine RNAimax (Invitrogen). Briefly, cells were seeded at a density of 2.5 × 105 cells/well in 6-well plates and simultaneously transfected with 20 nM of Stealth RNAi™ siRNAs targeting eIF5B or non-targeting siRNA control (Invitrogen). Seventy-two hours post-transfection, eIF2α phosphorylation was induced by poly I:C transfection (25 μg/ml) using Lipofectamine 2000 (Invitrogen). Eighteen hours after poly I:C transfection cells were harvested in RIPA buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% (vol/vol) NP40, 0.05% (wt/vol) SDS, 0.5% (wt/vol) Deoxycholic acid] for western blot analysis, or in RNA lysis buffer [15 mM Tris (pH 7.4), 15 mM MgCl2, 300 mM NaCl, 1% (vol/vol) Triton-X100, 100 U/ml RNAsin and 0.1 mg/ml Cycloheximide (CHX)] for polysome profiling.
In vitro transcription
DNA templates used for the synthesis of RNAs for the toeprinting assays and streptomycin affinity chromatography were amplified by polymerase chain reaction (PCR) from the above-mentioned constructs. The 5′ primers incorporated the T7 promoter sequence to allow for RNA transcription; the reverse primer included streptotag aptamer sequence and 51 T residues (Supplementary Table S1) which were added to the end of the PCR products to provide poly-A tail for the transcribed RNA. In vitro transcription was performed using the Megashortscript and mMessage mMachine kit (Ambion) was used to generate capped versions of RNA. The newly-synthesized RNA was purified by size exclusion chromatography (22) or ethanol precipitation.
Toeprinting assay
Toeprinting was performed as described by Locker and Lukavsky (21), with some modifications. Briefly, the RRL (Green Hectares) was treated with RNAsein (Promega) and Guanosine 5′-[β,γ-imido]triphosphate (GMP-PNP) for 5 min at 30°C. RRL was treated first with poly I:C & adenosine triphosphate (ATP) at 37°C for 20 min if the toeprinting assay was performed in the presence of poly I:C. Subsequently, RNA, ATP and guanosine triphosphatecDNA (GTP) were added, as indicated, and the reactions were incubated at 30°C for further 5 min. The reaction volume was brought to 40 μl by the addition of toeprinting buffer [20 mM Tris–HCl (pH 7.6), 100 mM KOAc, 2.5 mM MG(OAc)2, 5% (wt/vol) sucrose, 2 mM DTT and 0.5 mM spermidine] and incubated at 30°C for 3 min. Subsequently, 5 pmol of toeprinting primer (5-CTCGATATGTGCATCTGTA-3) (5′-end labeled with IRDye™800) was added and reaction was incubated on ice for 10 min. A quantity of 1 mM dNTPs, 5 mM Mg(OAc)2 and 1 μl of avian myeloblastosis virus reverse transcriptase (Promega) were added to the reaction and the final volume was brought to 50 μl by toeprinting buffer. Primer extension was allowed to occur for 45 min at 30°C. The cDNA products were purified by phenol:chloroform extraction and analyzed on a standard 6% sequencing gel using a model 4200 IR2 sequence analyzer (LI-COR, Lincoln, Nebraska, USA). The concentrations of the toeprinting assay components are as follows unless otherwise specified: RRL, 15 μl; GMP-PNP, 1.7 mM; ATP, 1.82 mM; GTP, 1.8 mM; RNA, 800 ng; poly I:C, 150 ng/ml.
Western blot analysis
Cells were lysed in RIPA buffer for 30 min at 4°C, followed by centrifugation at 13 000 rpm to remove debris. Equal amounts of protein were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), transferred to nitrocellulose membranes and probed with antibodies against GST-XIAP (23), β-actin (Abcam), phospho-eIF2α (Abcam), eIF2α (Abcam) and eIF5B (Protein Technology). Membranes were then incubated with species-specific horseradish peroxidase-conjugated secondary antibody (Cell Signaling) followed by detection with ECL substrate (Pierce), and quantified using Odyssey densitometry software (LI-COR).
Polysome profiling and quantitative reverse transcription–PCR
HEK293T cells from two 6-well plates per condition were lysed in RNA lysis buffer and polysomal profiling was performed as described (11). Gradients were fractionated using ISCO gradient fraction collector machine. The fractions (1 ml) were spiked with 100 ng of an in vitro transcribed chloramphenicol acetyltransferase (CAT) RNA, to ensure technical consistency in RNA isolation. RNA was isolated as described (11) and cDNA was generated from equal volumes of RNA using the qScript™ reverse transcription kit (Quanta Biosciences). Quantitative PCR was performed on a Realplex2 real-time thermocycler with PerfeCTa™ SYBR® Green FastMix™ (Quanta Biosciences) using primers specific to IRES containing (longer UTR) and non-IRES (shorter UTR) mRNA of XIAP, CAT or β-actin (Supplementary Table S2).
RNA-streptomycin affinity chromatography
Untreated RRL of 4 ml (Green Hectares) was incubated with 150 ng/ml poly I:C, 1 mM ATP, 10 μl ribonuclease inhibitor (Promega) and 0.1 mg/ml CHX at 37°C for 20 min. Binding buffer of 12 ml [20 mM Tris (pH 7.6), 10 mM MgCl2, 120 mM KCl, 8% sucrose, 2 mM dithiothreitol] containing EDTA-free protease inhibitor cocktail (Roche) and 1 mM puromycin was mixed with RRL and the reaction was incubated at 37°C for 10 min. An in vitro transcribed, uncapped strepto-tagged XIAP IRES RNA, 1 mM GTP and 40S and 60S ribosomal subunits purified from HeLa cells were added to the RRL and further incubated for 10 min at 37°C to form the 80S initiation complex. RNA-dihydrostreptomycin affinity chromatography was performed as described (20,21) and the isolated initiation complex was analyzed by Tris/Borate/EDTA (TBE) agarose gel electrophoresis and a toeprinting assay. The negative control experiment was performed in the same manner using 1 ml RRL and 3 ml binding buffer but XIAP IRES RNA was omitted.
RESULTS
In vitro characterization of the XIAP IRES initiation complex
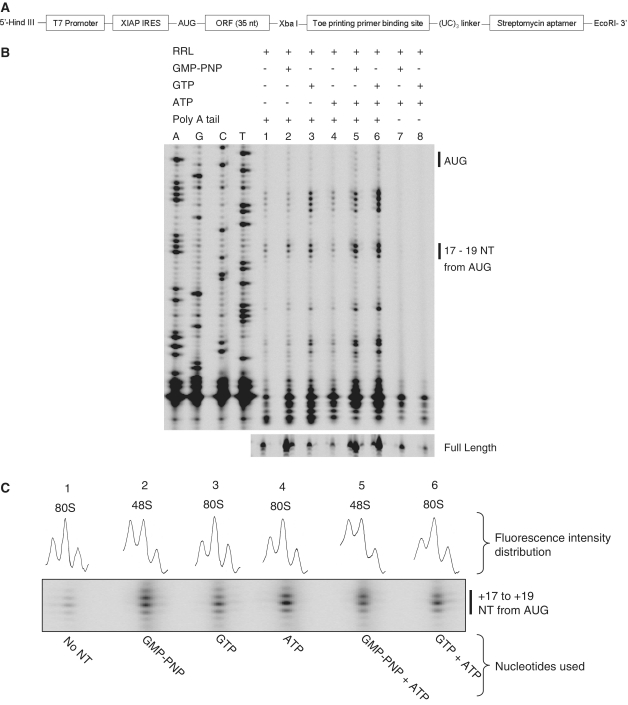
Stress-induced translation of XIAP mRNA is driven by an IRES element (24); however, the mechanism of XIAP IRES function is not known. The ability of the XIAP IRES to nucleate the formation of initiation complex was determined using toeprinting assay on an in vitro transcribed uncapped XIAP IRES RNA (Fig. 1A). After XIAP IRES initiation complexes were allowed to form in RRL, reverse transcription of the mRNA-ribosome complex typically yielded toeprints +17 to +19 nt downstream of AUG (Fig. 1B, lanes 1–6), which is a hallmark of ribosome recruitment to the AUG and stable ribosome–RNA complex formation (25–27). The ability of XIAP IRES to recruit ribosomes was impaired in the absence of a poly A tail (Figure 1B, lanes 7 and 8) which is in accordance with previously published reports (28). XIAP IRES initiation complexes could be further differentiated depending on the distribution of fluorescence intensities between +17 to +19 nt (Fig. 1C) as described previously (27). The type of initiation complex observed was dependent on the type of guanine nucleotide treatment. When RRL was pretreated with GMP-PNP or GMP-PNP and ATP, a 48 S initiation complex was formed (characterized by toeprint distribution +17 ≥ +18 > +19; Figure 1C, lanes 2 and 5). In contrast, 80S complex was observed in GTP, ATP or GTP + ATP treated, or untreated RRL (characterized by toeprint distribution +17<+18> +19; Figure 1C, lanes 1, 3, 4 and 6). Importantly, reverse transcription of XIAP IRES RNA in the absence of RRL, GMP-PNP, GTP or ATP did not yield any toeprints in the +17 to +19 range (Supplementary Figure S1A), strongly indicating that the +17 to +19 toeprints are not due to hindrance by the structure of the mRNA during reverse transcription, but rather are the result of specific ribosome recruitment to the XIAP IRES. We will refer to these toeprints as 40S leading edge toeprints.
Figure 1.
Initiation complex formation on uncapped XIAP IRES RNA. (A) Schematic diagram of the DNA construct of the streptomycin aptamer-tagged XIAP IRES RNA used for analysis of initiation complexes by toeprinting and affinity purification. (B) XIAP IRES initiation complexes were formed on the in vitro transcribed RNA with or without poly-A tail in RRL pre-treated with GMP-PNP or GTP and/or ATP. Subsequently, initiation complexes were analyzed by toeprinting. (C) A magnified portion of the toeprinting gel showing toeprints from +17 to +19 downstream of the AUG with the distribution of fluorescence intensities indicated. XIAP IRES initiation complexes 48S, 80S or 48S + 80S were determined as described by Shirokikh et al. (27).
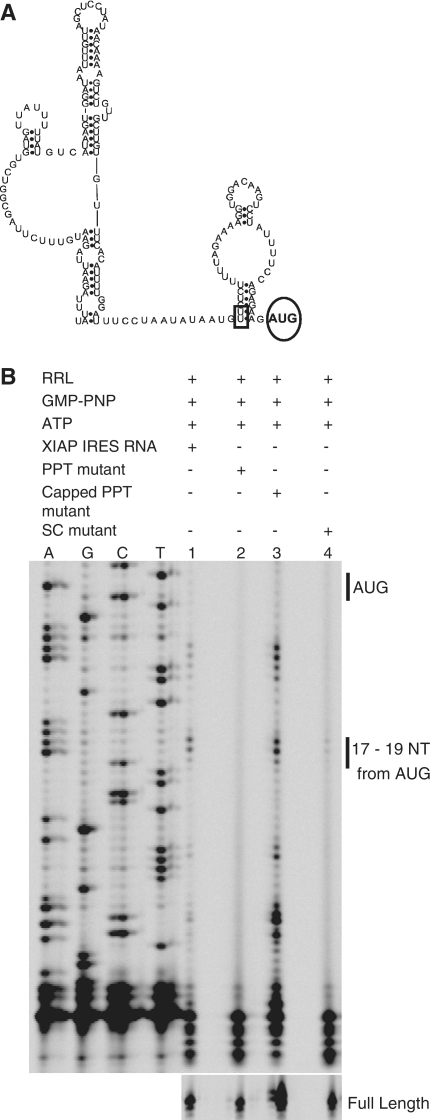
To further authenticate the formation of 40S leading edge toeprints on the XIAP IRES, additional control experiments were performed. First, the initiation codon AUG was mutated to AAC (highlighted by a circle in Figure 2A; referred to as Start Codon, SC, mutant). The 40S leading edge toeprints were not observed with the SC mutant (Figure 2B, lane 4). These data confirm that XIAP IRES initiation complexes are formed on the authentic initiation AUG codon. Second, the substitution of UU to AA in the polypyrimidine tract (PPT) of XIAP IRES (highlighted by a rectangle in Figure 2A) was shown to result in a complete loss of XIAP IRES activity in the bicistronic reporter assay (24). Toeprinting assay was therefore performed using the same PPT mutant. The absence of 40S leading edge toeprints indicates the inability of the XIAP IRES PPT mutant to form an initiation complex (Fig. 2B, lane 2). Strikingly, the PPT mutant was able to form an initiation complex once a m7G cap was added to the 5′-end of the in vitro transcribed RNA (Figure 2B, lane 3). Finally, the 48S initiation complex was detected only on m7G-capped human β-globin RNA, and not in the absence of the cap (Supplementary Figure S1B). Together, these data confirm that the in vitro conditions established in the RRL system do not allow for random recruitment of the ribosome to uncapped RNA; importantly, either a 5′ m7G cap or a functional IRES element are stringent requirements for ribosome recruitment and initiation complex formation.
Figure 2.
XIAP IRES initiation complexes are IRES dependent. (A) Secondary structure model of the XIAP IRES (54) with the introduced mutations within the IRES indicated. The UU sequence of the polypyrimidine tract was mutated to AA in the PPT mutant (indicated with a rectangle); the initiation codon AUG was replaced with AAC in the SC mutant (indicated with a circle). (B) Toeprinting analysis was carried out to determine the ability of PPT, capped PPT and SC XIAP IRES mutants to form initiation complexes in RRL pre-treated with GMP-PNP and ATP.
In addition to the +17 to + 19 toeprints we also observed toeprints at +8 to +11, +29 to +30 and +35 to +39 nt downstream of the AUG in XIAP IRES RNA. It was suggested previously that such toeprints could represent ‘ribosomal jumps’ (29). However, the addition of cycloheximide (which freezes the ribosome) to RRL did not eliminate these toeprints (Supplementary Figure S1C, lane 4), thus ruling them out as ‘ribosomal jumps’. Furthermore, these additional toeprints were absent from the PPT or SC mutants of XIAP IRES RNA, which are unable to recruit the ribosome (Figure 2B lanes 2 and 4). These observations suggest that ribosome recruitment to the AUG induces overall structural rearrangement of XIAP IRES RNA, which results in additional toeprints at +8 to +11, +29 to +30 and +35 to +39 nt downstream of AUG.
Formation of the XIAP IRES initiation complex is not dependent on eIF2α
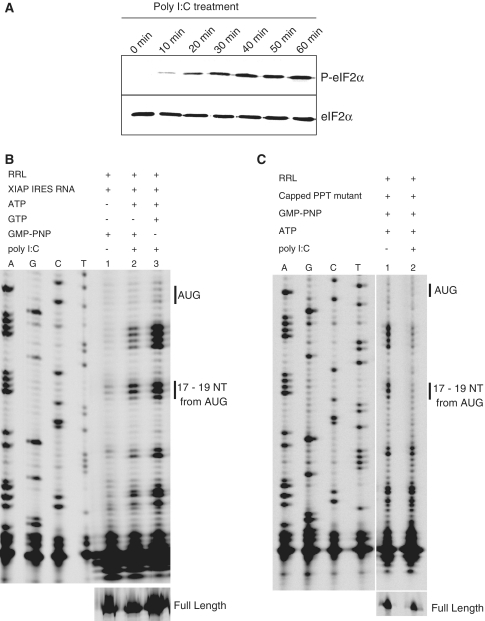
Poly (I)-poly(C) (poly I:C) treatment of RRL activates double stranded RNA (dsRNA)-dependent protein kinase (PKR), which leads to phosphorylation of eIF2α and subsequent attenuation of cap-dependent protein translation (30). Treatment of RRL with poly I:C (150 ng/μl) for 5 to 8 min results in a complete shut-off of protein synthesis (30,31).We wished to use this system to study XIAP IRES initiation under conditions of cellular stress. Sustained phosphorylation of eIF2α was observed in RRL following poly I:C treatment (Figure 3A), which resulted in the inhibition of translation as indicated by the inability of the capped XIAP IRES PPT mutant to form the 40S leading edge toeprint (Figure 3C, lane 2). Strikingly, the uncapped wt XIAP IRES RNA was able to form an initiation complex in the poly I:C-treated RRL (Figure 3B, lanes 2 and 3), and in fact the fluorescence intensity of the 40S leading edge toeprints was enhanced by poly I:C treatment (Figure 3B, lanes 2 and 3). These data indicate that XIAP IRES-dependent translation initiation proceeds in poly I:C treated RRL, whereas cap-dependent translation initiation is inhibited. Reverse transcription of XIAP IRES RNA in the absence of RRL, GMP-PNP, GTP and ATP, but in the presence of poly I:C, did not show any toeprints +17 to +19 nt downstream of AUG (Supplementary Figure S2). This result confirms that these toeprints are not due to XIAP IRES RNA structure changes that may be potentially caused by poly I:C treatment, which could hinder the reverse transcriptase.
Figure 3.
eIF2α phosphorylation-independent formation of the XIAP IRES initiation complexes. (A) RRL was treated with poly I:C (to induce eIF2α phosphorylation) and ATP for the indicated times; western blot analysis was performed to confirm phosphorylation of eIF2α. (B) Toeprinting assay of XIAP IRES initiation complexes was performed in the presence of GMP-PNP or GTP in RRL pre-treated with poly I:C and ATP. (C) The ability of the capped PPT mutant to form initiation complexes in poly I:C treated and untreated RRL was determined by toeprinting assay as in (B).
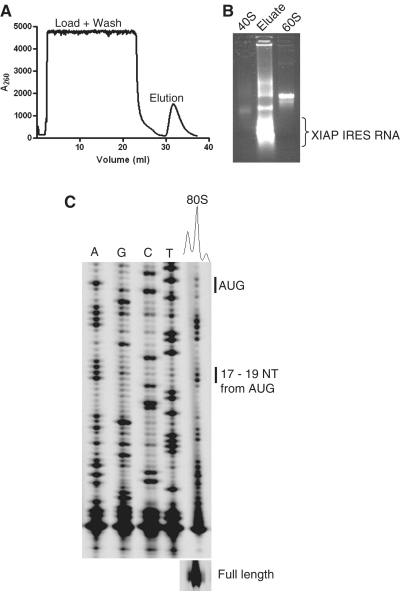
We further wished to confirm the nature of the observed 40S leading edge toeprints, and therefore used a streptomycin–RNA affinity chromatography approach (20,21) to isolate and characterize initiation complex formed on the XIAP IRES RNA. An in vitro transcribed uncapped strepto-tagged XIAP IRES RNA was incubated with poly I:C and cycloheximide-treated RRL. In order to achieve higher yield of XIAP IRES RNA ribosome complex, the nuclease untreated RRL was supplemented with purified 40S and 60S from HeLa cells. Following initiation complex formation, strepto-tagged XIAP IRES RNA was captured on dihydrostreptomycin resin, washed and eluted using streptomycin solution (Figure 4A). The eluate was then analyzed by TBE agarose gel electrophoresis. The presence of 40S and 60S RNA and free XIAP IRES RNA in the eluate confirmed that the 80S was indeed formed on XIAP IRES RNA in poly I:C treated RRL (Figure 4B). Toeprinting analysis of the eluate further confirmed the presence of 80S initiation complexes (Figure 4C). In contrast, the 80S initiation complex could not be isolated in a control reaction (Supplementary Figure S3A). These data indicate that XIAP IRES 80S initiation complex was purified specifically using XIAP IRES RNA affinity chromatography.
Figure 4.
Isolation of XIAP IRES initiation complexes formed in poly I:C-treated RRL. RRL was supplemented with purified 40S and 60S subunits from HeLa cells and subsequently treated with poly I:C, ATP and cycloheximide. Initiation complexes were formed on strepto-tagged XIAP IRES RNA and affinity purified using streptomycin affinity chromatography as described by Locker et al. (20). (A) A typical profile of RNA/initiation complex elution from the dihydrostreptomycin column. (B) Agarose gel electrophoresis analysis of the eluate from the streptomycin column. (C) Without further purification 5μl of eluate was analyzed by toeprinting assay for XIAP IRES initiation complexes.
An eIF5B-dependent mode of XIAP IRES-mediated translation initiation
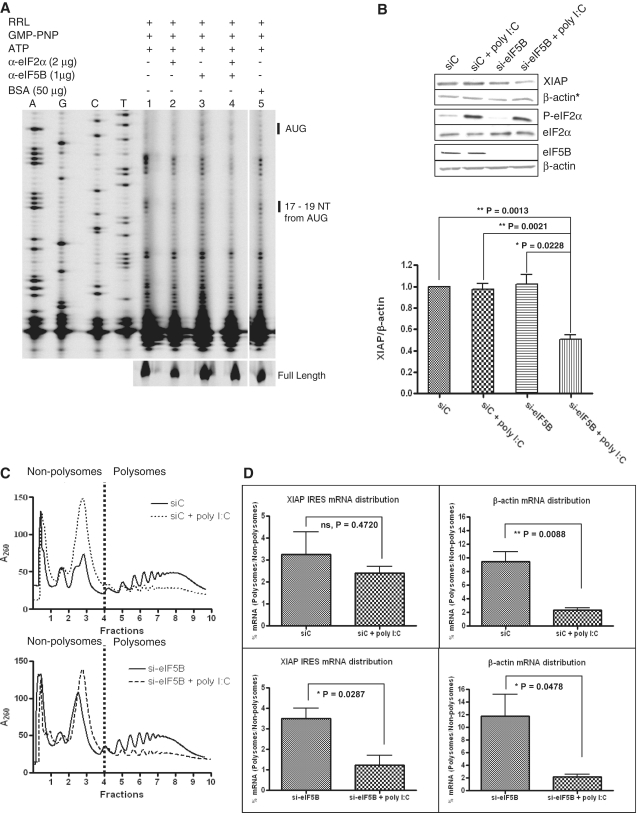
We have shown that the XIAP IRES is capable of recruiting ribosomes and forming translation initiation complex even when eIF2α is inactivated by phosphorylation. Previous reports identified a bacterial-like mode of translation initiation on viral IRES [HCV, (32); CSFV, (31)] that utilizes eIF5B when eIF2α is inactivated by phosphorylation. This prompted us to investigate if eIF5B could also be used by a cellular IRES when eIF2α is phosphorylated. Initiation factors eIF2α and/or eIF5B were thus immuno-inactivated in RRL prior to performing the toeprinting assay on the XIAP IRES RNA. We observed that initiation complex formation was partially inhibited when either eIF2α or eIF5B were immuno-inactivated; however, XIAP IRES initiation complex formation was completely abrogated when both eIF2α and eIF5B were immuno-inactivated (Figure 5A).
Figure 5.
Translation initiation on XIAP IRES RNA switches to an eIF5B-dependent mode when eIF2 is inactivated. (A) A quantity of 5 μl of RRL was incubated with the anti-eIF2α and/or anti-eIF5B antibodies at 4°C overnight. Following the immuno-inactivation of the indicated initiation factors, the toeprinting assay of the XIAP IRES initiation complex was performed. (B) HEK293T cells were transiently transfected with siRNA targeting eIF5B or non-targeting control siRNA and poly I:C as indicated. Western blot analysis (upper panel) was performed to verify eIF5B knockdown and phosphorylation of eIF2α, and the levels of endogenous XIAP protein. XIAP levels are represented as the ratio of XIAP to β-actin (lower panel; n = 4, mean ± SEM). The actin blot used for XIAP normalization is indicated with an asterisk. (C) Polysome profiling of HEK293T cells transiently transfected with siRNA targeting eIF5B or non-targeting control siRNA and poly I:C as indicated. Non-polysome (translationally inactive) and polysome (translationally active) fractions are separated by broken vertical lines. Representative profile of three independent experiments is shown. (D) Individual fractions were probed for the presence of β-actin and IRES-containing endogenous XIAP mRNAs by qRT–PCR. Percent distribution of specific mRNAs relative to the spiked CAT RNA control across the gradient was determined. The amount (%) of specific mRNA present in polysomes relative to non-polysomes quantifies the change in translation efficiency (n = 3, mean ± SEM).
In order to extend our in vitro data, we wished to verify the alternative role of eIF2α and eIF5B in translation initiation on XIAP IRES in cells. HEK293T cells were transiently transfected with siRNA targeting either eIF5B (si-eIF5B) or non-targeting control siRNA (siC). A substantial reduction of eIF5B expression was achieved 72 h following siRNA transfection (Figure 5B, lower panel), at which point poly I:C transfection for an additional 18 h was used to induce eIF2α phosphorylation. The levels of endogenous XIAP protein were determined by western blot analysis (Figure 5B). As anticipated from our in vitro results, XIAP protein levels remained unchanged by either eIF2α phosphorylation or reduced eIF5B expression. However, XIAP protein levels were decreased by 50% when eIF5B expression decreased and eIF2α was phosphorylated simultaneously (Figure 5B).
We have shown recently that the translation of XIAP is alternatively regulated by two 5′-UTRs: a 323 nucleotide UTR (shorter) or a 1700 nucleotide UTR (longer). The longer UTR contains the IRES element and is used preferentially during stress, whereas translation mediated by the shorter UTR is cap dependent (11). To directly measure the effect of eIF2α phosphorylation and eIF5B reduction on the translation of the endogenous XIAP IRES-containing mRNA, polysome profiling was performed to determine the association of XIAP IRES containing mRNA with translating polysomes. HEK293T cells were transfected with eIF5B-specific siRNA and subsequently transfected with poly I:C, and the polysomal distribution of mRNAs was analyzed by density gradient centrifugation (Figure 5C). Western blot analysis was performed to verify eIF5B knockdown and phosphorylation of eIF2α in the lysates prepared for polysome profiling (Supplementary Figure S4A). Treatment of cells with poly I:C resulted in an attenuation of cap-dependent translation due to eIF2α phosphorylation, as demonstrated by a loss of high molecular weight polysomes and an increase in the non-polysomal peaks (Figure 5C). Surprisingly, we did not observe significant decrease in high molecular weight polysomes when eIF5B levels were reduced by siRNA. Although, eIF5B is known to play a role in ribosomal subunit joining (33), Pestova et al. (31) reported that eIF5B is dispensable to form elongation competent 80S initiation complex on CSFV IRES. This suggests that eIF5B is not essential for ribosomal subunit joining. The quantitative RT–PCR analysis of individual fractions showed that the endogenous IRES-containing XIAP mRNA remains in polysomes even when eIF2α is phosphorylated (Figure 5D, Supplementary Figure S4C). In contrast, β-actin and non-IRES XIAP (shorter-UTR) mRNAs shifted toward non-polysomes upon eIF2α phosphorylation (Figure 5D, Supplementary Figure S4C). Similarly, the reduced expression of eIF5B did not affect the association of IRES-containing XIAP mRNA with polysomes (Figure 5D, Supplementary Figure S4C). In contrast, the IRES-containing XIAP mRNA was shifted toward non-polysomes from polysomes when eIF5B expression was reduced and eIF2α was phosphorylated. However, steady-state levels of XIAP IRES mRNAs were not significantly affected by poly I:C transfection, reduced expression of eIF5B or by combination of both (Supplementary Figure S4B). These data indicate that XIAP IRES-dependent translation relies on eIF5B when eIF2α is phosphorylated. Together, these observations support a model in which translation initiation mediated by the XIAP IRES switches from an eIF2-dependent mode to an eIF5B-dependent mode when the availability of ternary complex decreases due to eIF2α phosphorylation.
DISCUSSION
XIAP is a potent IAP that interacts directly with caspases 3, 7 and 9 to block cell death (34). Cell survival during pathophysiological stress conditions depends on the abundance and activity of XIAP protein, which is determined at least partially by the expression mediated by its IRES. Interestingly, several groups reported that the IRES-mediated translation of XIAP is sustained during pathophysiological stress (8–11,35), despite the fact that these conditions lead to phosphorylation of eIF2α and a consequent reduction in the levels of available ternary complex which is required for translation of both capped and uncapped mRNAs. Here, we present the first functional characterization of cellular IRES in vitro and explain the mechanism by which translation initiation on XIAP IRES occurs during pathophysiological stress condition when eIF2α is phosphorylated. We show that XIAP IRES-dependent translation initiation switches to an eIF5B-dependent mode to avoid attenuation due to eIF2α phosphorylation.
We have used a toeprinting assay in RRL to show that the XIAP IRES recruits ribosome in the absence of a m7G cap and thus supports bona fide cap-independent translation. Since the validity of the XIAP IRES has been questioned in several recent reports (36,37) our data add unequivocal evidence to support the authenticity of the XIAP IRES. Previously it was shown that translation of cellular IRES is enhanced by the presence of poly-A tail in a cell-free translation system (28). Indeed, this is also true for the XIAP IRES, as we observed impaired ability of the IRES to form an initiation complex in the absence of poly-A-tail. In vitro initiation on the XIAP IRES forms either a 48S or a 80S initiation complex depending on available guanine nucleotides. These two types of initiation complexes were revealed by determining distribution of fluorescence intensities of the 40S leading edge toeprints (Figure 1). Toeprinting assays performed in cycloheximide-treated RRL yielded enhanced intensity of the 40S leading edge toeprints (Supplementary Figure S1C, compare lanes 1 and 4). This indicates that a high proportion of XIAP IRES RNAs harbor an elongation-competent 80S initiation complex, which can be inhibited by cycloheximide treatment, and provides evidence that the initiation complex formed on uncapped XIAP IRES RNA is indeed translation competent. We have shown previously that in vivo XIAP IRES activity is abolished by point mutations within the PPT of XIAP IRES (24). We have expanded this observation here in an in vitro toeprinting assay and demonstrate that mutations within the PPT abolished the ability of uncapped XIAP IRES to form an initiation complex probably due to alteration in the secondary structure and/or IRES trans-acting factors (ITAFs) binding site. Interestingly, the formation of the 40S leading edge toeprints was restored by the addition of a 5′ m7G cap to the PPT mutant IRES RNA, indicating that the recruitment of the ribosome and the subsequent formation of the initiation complex is sequence-specific and requires a functional IRES (Figure 2).
Published toeprinting analyses performed in RRL using HCV IRES, capped human β-globin or capped Hsp70 RNAs obtained 40S leading edge toeprints +16 to +18 nt downstream of the AUG (20,21,27,29). Using a capped human β-globin RNA, we have also observed 40S leading edge toeprints +16 to +18 nt downstream of the AUG (Supplementary Figure S1B). In contrast, toeprinting analysis of XIAP IRES RNA or the capped PPT mutant of XIAP IRES identified 40S leading edge toeprints +17 to +18 nt downstream of the AUG, for both 48S and 80S initiation complexes. It was suggested that changes in the structural organization of the 40S subunit within the initiation complex result in a change in the distribution of fluorescence intensities of 40S leading edge toeprints (27,29). Therefore, our data suggest that the conformation of 40S subunit in either the 48S or 80S initiation complexes formed on the XIAP IRES or capped PPT mutant is different from that formed on HCV IRES, or capped non-IRES RNAs.
Having established a faithful in vitro system to study XIAP IRES-dependent translation, we sought to dissect the mechanism that allows the translation of cellular IRES-containing mRNAs during conditions of cellular stress, which are typified by phosphorylated eIF2α. Several lines of evidence suggest that translation of IRES-containing mRNAs may continue even in the presence of phosphorylated eIF2α. For example, the IRES-mediated translation of VEGF and p120 catenin is resistant to the global translation attenuation due to eIF2α phosphorylation in the hypoxic environment of the inflammatory breast tumor (38,39). Similarly, enhanced translation of XIAP was observed in hypoxic breast cancer cell lines (40–42) in which the eIF2α remains phosphorylated due to higher abundance and activity of PKR (43). In addition, glucose deficiency induces phosphorylation of eIF2α and inhibition of protein synthesis but the XIAP mRNA can bypass this translational block (10). Our group has shown previously that the IRES-containing variant of XIAP mRNA is preferentially translated during serum deprivation which also leads to eIF2α phosphorylation (11). Here, we confirm these observation in an in vitro system and show that XIAP IRES forms an initiation complex in poly I:C treated RRL where eIF2α is phosphorylated (Figure 3). Using a RNA affinity chromatography approach we have isolated 80S initiation complex formed on the XIAP IRES RNA in poly I:C-treated RRL and show that the 80S initiation complex can be formed without the participation of eIF2 (Figure 4). Importantly, translation of the endogenous XIAP mRNA was not decreased in poly I:C-transfected HEK293T cells as determined by western blot analysis and polysome profiling (Figure 5). This indicates that the IRES-mediated translation of XIAP is indeed resistant to the global translation attenuation imparted by eIF2α phosphorylation. Interestingly, only the 48S but not the 80S initiation complex could be formed and isolated from RRL treated with GMP-PNP (Figure 1B, lane 2; Supplementary Figure S3B). Under these conditions eIF2α is not inactivated by phosphorylation and therefore our data suggest that XIAP IRES RNA preferentially utilizes an eIF2α-dependent translation initiation mechanism under normal conditions when eIF2 is fully active, whereas translation initiation on XIAP IRES RNA switches to an alternative, eIF2α-independent mode when the availability of ternary complex is reduced.
A few eIF2-independent modes of translation initiation have been proposed recently for the translation of viral RNAs. eIF2D (44), Ligatin, and MCT-1/DENR (45) have all been reported to deliver tRNA into the P-site of ribosome during HCV IRES-mediated translation. An eIF2-independent mode of translation initiation on CSFV IRES has been reported in which eIF5B, an eukaryotic homologue of bacterial IF2, delivers tRNA into the P-site of the ribosome to form a translation-competent initiation complex (31). Similar switch from an eukaryotic to a bacterial-like mode of translation initiation has been reported for the HCV IRES when eIF2α is inactivated by phosphorylation (32). The ability of HCV and CSFV IRES to recruit and place the 40S subunit in the vicinity of initiation codon in a scanning-independent manner allows them to utilize an eIF5B dependent mode of translation initiation (31,32). We report for the first time that a cellular IRES-containing mRNA utilizes a similar, eIF2α-independent mode of translation. A stable complex between the purified ribosome and XIAP IRES RNA could not be detected by a gel-shift assay performed in the absence of ITAFs or by a toeprinting assay performed in the absence of RRL (data not shown). However, the XIAP IRES initiation complex could be formed in RRL treated with hippuristanol (a small molecule inhibitor of eIF4A), indicating that eIF4A-dependent ribosome scanning is not required and the ribosome is likely placed in the vicinity of the AUG codon to form an initiation complex (Supplementary Figure S5). These observations strongly suggest the possibility that an eIF5B-dependent pathway operates on the XIAP IRES. Our in vivo data confirm this model, since we find that expression of XIAP protein decreases only when eIF5B expression is reduced and eIF2α is phosphorylated (Figure 5). This is in contrast to the cap-dependent translation of β-actin mRNA, which is decreased when eIF2α is phosphorylated (Figure 5). Thus we show that like viral IRES, a cellular IRES can operate in an eIF5B-dependent manner when the availability of ternary complex decreases due to eIF2α phosphorylation. We wished to extend our observations to other cellular IRES-containing mRNAs, and therefore assessed the polysomal distribution of mRNAs that encode other regulators of apoptosis, namely Bcl-xL, cIAP1, Apaf-1 and p97/DAP5. Surprisingly, we find that translation of these mRNAs is unable to utilize eIF5B when global translation is attenuated due to phosphorylation of eIF2α (data not shown), suggesting that not all cellular IRES operate in an eIF5B dependent mode.
Chemoresistance of many cancer cell types have been correlated with overexpression of XIAP (17,46–49). In fact, XIAP has been validated as a potent therapeutic target and various anti-XIAP strategies for cancer treatment are currently being tested in clinical trials (50). The specificity of these therapeutics for tumor cells is desirable, since XIAP deficiency in non-cancerous cells can lead to disorders such as X-linked lymphoproliferative disease (51,52). It is tempting to propose that specific targeting of the eIF5B-dependent mode of XIAP IRES-mediated translation may allow a reduction of XIAP levels only in cancer cells which express high levels of eIF2 (53). Therefore, it is crucial to understand the process of IRES-mediated translation initiation of XIAP during pathophysiological stress. The mechanism proposed in this report is the first step toward understanding the process of ribosome recruitment and initiation complex formation on XIAP IRES during pathophysiological stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (operating grant, MOP-89737). Funding for open access charge: CIHR MOP-89737.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Stephen Baird for assistance with LI-COR’s 4200 IR2 sequence analyzer and e-Seq software, Fahad Shamim for technical assistance, Faraz Farooq and members of the laboratory for their insightful discussion of the project. We are grateful to Dr. Peter Lukavsky for the gift of strepto-tagged HCV IRES construct, and to Dr. Jerry Pelletier for the gift of hippuristanol. MH is the CHEO Volunteer Association Endowed Scholar.
REFERENCES
- 1.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell. Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 2.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 4.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell. Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ron D, Harding HP. eIF2α phosphorylation in cellular stress responses and disease. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. New York: Cold Spring Harbor Laboratory Press; 2007. pp. 345–368. [Google Scholar]
- 7.Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcik M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 2004;279:17148–17157. doi: 10.1074/jbc.M308737200. [DOI] [PubMed] [Google Scholar]
- 8.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–375. doi: 10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holcik M, Yeh C, Korneluk RG, Chow T. Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene. 2000;19:4174–4177. doi: 10.1038/sj.onc.1203765. [DOI] [PubMed] [Google Scholar]
- 10.Muaddi H, Majumder M, Peidis P, Papadakis AI, Holcik M, Scheuner D, Kaufman RJ, Hatzoglou M, Koromilas AE. Phosphorylation of eIF2alpha at serine 51 is an important determinant of cell survival and adaptation to glucose deficiency. Mol. Biol. Cell. 2010;21:3220–3231. doi: 10.1091/mbc.E10-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley A, Jordan LE, Holcik M. Distinct 5′ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res. 2010;38:4665–4674. doi: 10.1093/nar/gkq241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- 13.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat. Rev. Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 14.Carter BZ, Mak DH, Morris SJ, Borthakur G, Estey E, Byrd AL, Konopleva M, Kantarjian H, Andreeff M. XIAP antisense oligonucleotide (AEG35156) achieves target knockdown and induces apoptosis preferentially in CD34+38- cells in a phase 1/2 study of patients with relapsed/refractory AML. Apoptosis. 2011;16:67–74. doi: 10.1007/s10495-010-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean E, Jodrell D, Connolly K, Danson S, Jolivet J, Durkin J, Morris S, Jowle D, Ward T, Cummings J, et al. Phase I trial of AEG35156 administered as a 7-day and 3-day continuous intravenous infusion in patients with advanced refractory cancer. J. Clin. Oncol. 2009;27:1660–1666. doi: 10.1200/JCO.2008.19.5677. [DOI] [PubMed] [Google Scholar]
- 16.Schimmer AD, Estey EH, Borthakur G, Carter BZ, Schiller GJ, Tallman MS, Altman JK, Karp JE, Kassis J, Hedley DW, et al. Phase I/II trial of AEG35156 X-linked inhibitor of apoptosis protein antisense oligonucleotide combined with idarubicin and cytarabine in patients with relapsed or primary refractory acute myeloid leukemia. J. Clin. Oncol. 2009;27:4741–4746. doi: 10.1200/JCO.2009.21.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamm I, Kornblau SM, Segall H, Krajewski S, Welsh K, Kitada S, Scudiero DA, Tudor G, Qui YH, Monks A, et al. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin. Cancer Res. 2000;6:1796–1803. [PubMed] [Google Scholar]
- 18.Yamagiwa Y, Marienfeld C, Meng F, Holcik M, Patel T. Translational regulation of X-linked inhibitor of apoptosis protein by interleukin-6: a novel mechanism of tumor cell survival. Cancer Res. 2004;64:1293–1298. doi: 10.1158/0008-5472.can-03-2517. [DOI] [PubMed] [Google Scholar]
- 19.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 20.Locker N, Easton LE, Lukavsky PJ. Affinity purification of eukaryotic 48S initiation complexes. RNA. 2006;12:683–690. doi: 10.1261/rna.2227906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locker N, Lukavsky PJ. A practical approach to isolate 48S complexes: affinity purification and analyses. Methods Enzymol. 2007;429:83–104. doi: 10.1016/S0076-6879(07)29005-6. [DOI] [PubMed] [Google Scholar]
- 22.Lukavsky PJ, Puglisi JD. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004;10:889–893. doi: 10.1261/rna.5264804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Feng Q, Kim JM, Schneiderman D, Liston P, Li M, Vanderhyden B, Faught W, Fung MF, Senterman M, et al. Human ovarian cancer and cisplatin resistance: possible role of inhibitor of apoptosis proteins. Endocrinology. 2001;142:370–380. doi: 10.1210/endo.142.1.7897. [DOI] [PubMed] [Google Scholar]
- 24.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell. Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 25.Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 26.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 27.Shirokikh NE, Alkalaeva EZ, Vassilenko KS, Afonina ZA, Alekhina OM, Kisselev LL, Spirin AS. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res. 2010;38:e15. doi: 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell. 2004;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- 30.Price NT, Welsh GI, Proud CG. Phosphorylation of only serine-51 in protein synthesis initiation factor-2 is associated with inhibition of peptide-chain initiation in reticulocyte lysates. Biochem. Biophys. Res. Commun. 1991;176:993–999. doi: 10.1016/0006-291x(91)90380-p. [DOI] [PubMed] [Google Scholar]
- 31.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 33.Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 2007;27:2384–2397. doi: 10.1128/MCB.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holcik M, Korneluk RG. XIAP, the guardian angel. Nat. Rev. Mol. Cell. Biol. 2001;2:550–556. doi: 10.1038/35080103. [DOI] [PubMed] [Google Scholar]
- 35.Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, Zhu X, Jordan LE, Scheuner D, Kaufman RJ, et al. eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 2010;285:17098–17111. doi: 10.1074/jbc.M110.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, Logg CR. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl Acad. Sci. USA. 2008;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saffran HA, Smiley JR. The XIAP IRES activates 3′ cistron expression by inducing production of monocistronic mRNA in the betagal/CAT bicistronic reporter system. RNA. 2009;15:1980–1985. doi: 10.1261/rna.1557809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell. Biol. 2009;11:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 39.Silvera D, Schneider RJ. Inflammatory breast cancer cells are constitutively adapted to hypoxia. Cell Cycle. 2009;8:3091–3096. doi: 10.4161/cc.8.19.9637. [DOI] [PubMed] [Google Scholar]
- 40.Aird KM, Ding X, Baras A, Wei J, Morse MA, Clay T, Lyerly HK, Devi GR. Trastuzumab signaling in ErbB2-overexpressing inflammatory breast cancer correlates with X-linked inhibitor of apoptosis protein expression. Mol. Cancer Ther. 2008;7:38–47. doi: 10.1158/1535-7163.MCT-07-0370. [DOI] [PubMed] [Google Scholar]
- 41.Aird KM, Ghanayem RB, Peplinski S, Lyerly HK, Devi GR. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol. Cancer Ther. 2010;9:1432–1442. doi: 10.1158/1535-7163.MCT-10-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Z, Liang YJ, Chen ZS, Wang XH, Ding Y, Chen LM, Fu LW. Overexpression of Survivin and XIAP in MDR cancer cells unrelated to P-glycoprotein. Oncol Rep. 2007;17:969–976. [PubMed] [Google Scholar]
- 43.Kim SH, Forman AP, Mathews MB, Gunnery S. Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene. 2000;19:3086–3094. doi: 10.1038/sj.onc.1203632. [DOI] [PubMed] [Google Scholar]
- 44.Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter BZ, Gronda M, Wang Z, Welsh K, Pinilla C, Andreeff M, Schober WD, Nefzi A, Pond GR, Mawji IA, et al. Small-molecule XIAP inhibitors derepress downstream effector caspases and induce apoptosis of acute myeloid leukemia cells. Blood. 2005;105:4043–4050. doi: 10.1182/blood-2004-08-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 48.Pardo OE, Lesay A, Arcaro A, Lopes R, Ng BL, Warne PH, McNeish IA, Tetley TD, Lemoine NR, Mehmet H, et al. Fibroblast growth factor 2-mediated translational control of IAPs blocks mitochondrial release of Smac/DIABLO and apoptosis in small cell lung cancer cells. Mol. Cell. Biol. 2003;23:7600–7610. doi: 10.1128/MCB.23.21.7600-7610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schimmer AD, Welsh K, Pinilla C, Wang Z, Krajewska M, Bonneau MJ, Pedersen IM, Kitada S, Scott FL, Bailly-Maitre B, et al. Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell. 2004;5:25–35. doi: 10.1016/s1535-6108(03)00332-5. [DOI] [PubMed] [Google Scholar]
- 50.Flygare JA, Fairbrother WJ. Small-molecule pan-IAP antagonists: a patent review. Expert. Opin. Ther. Pat. 2010;20:251–267. doi: 10.1517/13543770903567077. [DOI] [PubMed] [Google Scholar]
- 51.Marsh RA, Madden L, Kitchen BJ, Mody R, McClimon B, Jordan MB, Bleesing JJ, Zhang K, Filipovich AH. XIAP deficiency: a unique primary immunodeficiency best classified as X-linked familial hemophagocytic lymphohistiocytosis and not as X-linked lymphoproliferative disease. Blood. 2010;116:1079–1082. doi: 10.1182/blood-2010-01-256099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 53.Schneider RJ, Sonenberg N. Translational control in cancer development and progression. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. New York: Cold Spring Harbor Laboratory Press; 2007. pp. 401–431. [Google Scholar]
- 54.Baird SD, Lewis SM, Turcotte M, Holcik M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664–4677. doi: 10.1093/nar/gkm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.