Abstract
Bamboo mosaic virus (BaMV) satellite RNA (satBaMV) depends on BaMV for its replication and encapsidation. SatBaMV-encoded P20 protein is an RNA-binding protein that facilitates satBaMV systemic movement in co-infected plants. Here, we examined phosphorylation of P20 and its regulatory functions. Recombinant P20 (rP20) was phosphorylated by host cellular kinase(s) in vitro, and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and mutational analyses revealed Ser-11 as the phosphorylation site. The phosphor-mimic rP20 protein interactions with satBaMV-translated mutant P20 were affected. In overlay assay, the Asp mutation at S11 (S11D) completely abolished the self-interaction of rP20 and significantly inhibited the interaction with both the WT and S11A rP20. In chemical cross-linking assays, S11D failed to oligomerize. Electrophoretic mobility shift assay and subsequent Hill transformation analysis revealed a low affinity of the phospho-mimicking rP20 for satBaMV RNA. Substantial modulation of satBaMV RNA conformation upon interaction with nonphospho-mimic rP20 in circular dichroism analysis indicated formation of stable satBaMV ribonucleoprotein complexes. The dissimilar satBaMV translation regulation of the nonphospho- and phospho-mimic rP20 suggests that phosphorylation of P20 in the ribonucleoprotein complex converts the translation-incompetent satBaMV RNA to messenger RNA. The phospho-deficient or phospho-mimicking P20 mutant of satBaMV delayed the systemic spread of satBaMV in co-infected Nicotiana benthamiana with BaMV. Thus, satBaMV likely regulates the formation of satBaMV RNP complex during co-infection in planta.
INTRODUCTION
One of the most common post-translational modifications, phosphorylation/dephosphorylation, plays a fundamental role in the regulation of many protein activities in vivo by a balancing between the two processes. Plant cell kinases mediate viral protein phosphorylation (1–16). The functional implications of the phosphorylation of nonstructural viral proteins have been elaborated (1–4,8–10,14,17–33), and the kinases involved have been characterized to some extent (4,5,7,8,12,13,15,21,27,28,34,35). Cell-to-cell movement of viruses involves the formation of a ribonucleoprotein (RNP) complex that interacts with host protein(s) to increase the size exclusion limit of plasmodesmata and to potentiate its own transport into neighboring cells (36,37). Viral movement proteins (MPs) exist as phosphoproteins, and phosphorylation specifies their role in regulating viral movement (20,21,23,37,38). The best studied is the Ser/Thr phosphorylation of Tobacco mosaic virus (TMV) MP in vitro and in vivo (1,4,39,40). TMV MP is phosphorylated by cell wall-associated protein kinase (1,41) and protein kinase C-like enzymes (2). Plasmodesmata-associated protein kinase PAPK1 specifically phosphorylates the C-terminal residues of TMV MP that affect the movement capacity of the protein (4,12). TMV MP is phosphorylated by endoplasmic reticulum-associated kinase activity, and phosphorylation strongly inhibits the cell-to-cell movement and delays the systemic spread of TMV in Nicotiana tabacum (10). Membrane association of TMV MP is eliminated by phosphorylation (24). In Tomato mosaic virus (ToMV) MP, Ser-28 and -37 are the preferable contexts for phosphorylation by casein kinase 2 (CK2), CK2-like protein kinase or recombinant N. tabacum RIO kinase (3,6,7,25). Ser-37 phosphorylation profoundly affects the cell-to-cell movement of the viral genome and the subcellular distribution of ToMV-MP (18). Ser/Thr phosphorylation of MPs has also been reported in Apple chlorotic leaf spot virus (42), Turnip yellow mosaic virus (43), Potato leaf roll virus (44), Cucumber mosaic virus (45) and Brome mosaic virus (30). Potato virus A (PVA) coat protein (CP) is phosphorylated by a CK2-like enzyme in vivo and in vitro (8,9), and phosphorylation reduces the CP affinity for RNA in PVA to cause a defect in cell-to-cell movement. The recombinant and native triple gene block protein 1 (TGBp1) of Potato virus X are phosphorylated by activities in N. tabacum extracts, including distinct CK2 activity (15). These various findings make it clear that phosphorylation orchestrates the molecular interactions and functions of viral MPs during infection.
Bamboo mosaic virus (BaMV), a member of the genus Potexvirus, is a single-stranded, positive-sense RNA genome of 6.4 kb with five genes (46–48). The 5′-proximal gene codes for the replicase and the 3′-proximal gene codes for the CP. The CP gene is preceded by three partially overlapping TGB genes coding for three MPs referred to as TGBp1, 2 and 3. As in other potexviruses, in BaMV, the TGB-coded MPs, along with the CP, are essential for cell-to-cell and long-distance movement (49–51). Satellite RNA associated with BaMV (satBaMV) is the only satellite found in the potexvirus group. The satBaMV genome, a single-stranded, linear RNA molecule of 836 nt [excluding the poly(A) tail], encodes a single non-structural protein of 20 kDa (P20) (52) that is not required for satBaMV replication (53). The coding region is flanked by a leader of 159 nt and a 3′-terminal untranslated region of 129 nt. SatBaMV depends absolutely on BaMV for its replication and encapsidation; however, it has low similarity with the BaMV genome, except at the 5′ terminus (48,52). P20 binds to satBaMV and BaMV RNAs with a strong affinity through its arginine-rich motif (ARM) (54). P20 shares common biological properties with viral MPs (6,7,18,25,30,45,54–56) including RNA-binding activity (54), self-interaction, intracellular targeting and efficient cell-to-cell movement (57), and has implications in satBaMV movement during co-infection in plants (53,57,58). The MP properties of P20 suggest that the function of P20 might be regulated by a phosphorylation event during co-infection.
Here, we investigated phosphorylation of P20 in vitro and satBaMV co-infection in vivo. Comprehensive biochemical and molecular approaches verified Ser-11 as the phosphorylation site of P20. Mimicked phosphorylation of P20 abolished its self-interaction and dimerization, and inhibited interactions with its non-phosphorylated form and satBaMV RNA. Substantial circular dichroism (CD) modulation of the satBaMV RNA conformation upon interaction with only nonphospho-mimic recombinant P20 (rP20) indicated unstacking of bases and formation of stable satBaMV RNP complexes. Phospho-mimicking P20 enabled the translation of incompetent satBaMV RNA to translatable RNA. Moreover, the accumulation level of phospho-deficient and mimicking P20 mutants of satBaMV was greatly reduced in systemic leaves of Nicotiana benthamiana co-infected with BaMV. The collective findings in this study provide insight into the regulation of the satBaMV-P20 RNP complex by phosphorylation during satBaMV co-infection. To our knowledge, this is the first report of phosphorylation of a plant satellite RNA-encoded protein.
MATERIALS AND METHODS
BaMV and satBaMV
Plasmids pCB (GenBank accession number AF018156) and pCBSF4 (GenBank accession number NC003497) are the infectious clones of BaMV and satBaMV, driven by Cauliflower mosaic virus (CaMV) 35S promoter, respectively (50,53). The pBSF4 is also an infectious clone of satBaMV under the control of T7 promoter (53). Virions of BaMV-S were purified and viral RNA was extracted as described (59).
Analysis of P20 sequence
The P20 amino acid sequence was analyzed for phosphorylation sites in the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos) (60) and the molecular mass was determined by using EXPASY (http://www.expasy.ch/tools/pi_tool.html).
Site-directed mutagenesis
SatBaMV mutant constructs were generated by introducing point mutations at Ser (S)-11 and -14 by a two-step polymerase chain reaction (PCR)-mediated mutagenesis. Amino acids S11 and S14 or both were substituted by alanine (A). To mimic phosphorylation, S11 was substituted by aspartic acid (D). For each single and double mutation, appropriate primers were used (Table 1). The first set of PCR fragments was generated from pBSF4 as a template with the respective sense primer (S11A, S14A or S11D) and a common antisense primer (BS43). A second set of PCR fragments was amplified with a common sense primer (BS19), the first-step PCR fragment of each of the mutants as an antisense primer and pBSF4 as a template. The PCR fragments were digested with PstI and XbaI and cloned into pBSF4 after PstI–XbaI excision and ligation. All constructs were sequenced to confirm the nature of engineered mutations and the absence of second site mutations. The four satBaMV mutants were pBSF4-S11A; pBSF4-S14A; pBSF4-S11,14A and pBSF4-S11D. Plasmid pBSGFP was cloned by replacing the P20 open reading frame (ORF) in pBSF4 with the green florescent protein (GFP) gene at BstXI-EcoNI sites. All DNA manipulations followed standard cloning procedures (61). For translation control plasmid, pETGFP was generated. It contains ORF of GFP without any satBaMV-derived sequences. The ORF of GFP protein was amplified from pGFP (Clontech) by PCR with primer pair pET21-GFP-F and pET21-GFP-R (Table 1). The PCR product was then digested with NdeI and XhoI and ligated into NdeI/XhoI digested pET21b vector, which resulted in pET21b-GFP. The DNA fragment contained T7 promoter, GFP ORF and T7 terminator was amplified from pET21b-GFP by PCR using primer pair T7P and T7T (Table 1). After digestion with PstI and BamHI, the PCR product was cloned into PstI/BamHI digested pUC119 vector, which resulted in pETGFP.
Table 1.
Sequence of oligonucleotides
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| S11A | CGCCAGAGAGCGCGTGTC |
| S14A | CGCGTGTCGCCCAAATGA |
| S11/14A | CGCCAGAGAGCGCGTGTCGCCCAAATGA |
| S11D | CGCCAGAGAGACCGTGTCTCC |
| BS19 | TGCCTGCAGTAATACGACTCACTATAGAAAACTCACCGCAACGA |
| BS43 | GTCGACTCTAGAT(15) |
| pET21-GFP-F | GGAATTCCATATGGTGAGCA AGGGCGAGGA |
| pET21-GFP-R | CCGCTCGAGCTTGTAC AGCTCGTCCA TGC |
| T7P | AACTGCAGTAATACGACTCACTATA |
| T7T | CGGGATCCT(15)CAAAAAACCC CTCAAGACCC |
All primers were designed corresponding to the sequence of pBSF4 (52).
aBoldface letters indicate the amino substitution.
Expression and purification of recombinant P20
The P20 gene was PCR-amplified from pBSF4-S11A or pBSF4-S11D with appropriate primers (Table 1) and cloned into corresponding sites in the expression vector pET21b (Novagen); the mutant P20 expression constructs were pET-P20S11A and pET-P20S11D. The wild-type (WT) (54) and mutant constructs were each transformed into Escherichia coli strain BL21 (DE3) (Novagen), and the recombinant protein was expressed and purified as described (54) with the following modifications. After 3 h of isopropyl β-d-1-thiogalactopyranoside (IPTG) induction, E. coli cells were harvested, resuspended in 10 mM sodium phosphate buffer, pH 7.5, and disrupted by sonication. The inclusion bodies were recovered and solubilized in TKM buffer (50 mM Tris–HCl, pH 8, 50 mM KCl, 10 mM magnesium acetate, 1 mM DTT) containing 8 M urea on ice for 30 min. Denatured rP20 was purified by Ni-NTA affinity chromatography (Novagen) and renatured by step-wise dialysis against TKM buffer with lower concentrations of urea. Protein purity was analyzed by 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and the concentration was determined using a Bio-Rad protein assay kit (Bio-RAD).
In vitro transcription
Infectious satBaMV RNA transcripts were generated by linearizing pBSF4, pBSF4-S11A, pBSF4-S14A, pBSF4-S11,14A or pBSF4-S11D with XbaI (New England Biolabs) and transcription in vitro with T7 RNA polymerase and m7GpppG (New England Biolabs) as the cap analog as described (48,52,53). Likewise, satBaMV transcripts encoding GFP were generated from pBSGFP and control ETGFP transcript from pETGFP.
In vitro phosphorylation
One-month-old N. benthamiana plants were mechanically co-inoculated with 0.1 μg BaMV-S RNA and 0.1 μg satBaMV transcripts. Inoculated leaves were harvested at 5 days post-inoculation (dpi) and homogenized in protein extraction buffer (25 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 1 mM DTT, 1 mM PMSF) at 4°C. The homogenate was filtered through a miracloth (Calbiochem) and centrifuged at 3000g for 5 min, then the precipitate was resuspended in protein extraction buffer. Purified WT rP20 was incubated with freshly prepared N. benthamiana leaf protein extract, phosphoryl donor [γ-32P]ATP or [γ-32P]GTP (10 µCi/reaction) (Amersham Pharmacia), and 2 μl of 10X phosphorylation buffer (1 M Tris–HCl, pH 8, 1 M NaCl, 3 mM DTT, 0.5 M MgCl2) in the presence or absence of 100 mM EDTA in a total volume of 20 μl at 25°C for 30 min. rP20 was purified from the phosphorylation mixture by Ni-NTA affinity chromatography, resolved on 12.5% SDS–PAGE, electroblotted onto a polyvinylidene fluoride (PVDF) membrane (Immobilon-P, Millipore) and probed with rabbit polyclonal anti-P20 serum (57). Phosphorylation was detected by autoradiography.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
Purified WT rP20 or in vitro-phosphorylated rP20 was resolved on 12.5% SDS–PAGE, detected by ProteoSilver Plus Silver Staining (Sigma) and digested in-gel with sequencing-grade CNBr (Sigma). CNBr-digested native WT rP20 was included as a control. Peptides were analyzed by use of a MALDI-TOF MS (Bruker-Daltonics, Billerica, MA, USA) in positive ion reflector mode in a mass range of 1010–4200 (m/z). The molecular ions were assigned to P20 peptides according to the sequence and the CNBr cleavage specificity was detected by use of PAWS software (http://www.proteometrics.com).
Protoplast transfection and P20 metabolic labeling
N. benthamiana protoplasts were isolated and transfected as described (53). For each inoculation, 2 × 105 protoplasts were transfected with 1.0 μg BaMV-S RNA or co-transfected with 1.0 μg satBaMV transcripts. Uninfected protoplasts or protoplasts infected with BaMV alone were used as controls. Protoplasts were cultured with [33P]orthophosphate (200 µCi/ml) (Amersham Pharmacia) in electroporation buffer (10 mM HEPES, pH 7.2, 40 mM mannitol, 5 mM CaCl2, 100 mM KCl) under constant light for 20 h. Protoplasts were harvested, and labeled proteins were extracted in extraction buffer [20 mM Tris–HCl, pH 8, 10 mM EDTA, 10 mM benzamidine, 0.3% 2-mercaptoethanol (v/v), 1 mM phenylmethylsulfonyl fluoride (PMSF), 100 mM β-glycerophosphate, 1% Triton X-100 (v/v)] by intermittent vortexing for 1 h at 4°C. The lysate was centrifuged at 9000 g for 10 min, and the unincorporated [33P]orthophosphate was removed by use of a NAP-5 column (Amersham Pharmacia) equilibrated with immunoprecipitation buffer [50 mM Tris–HCl, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100 (v/v), 0.02% NaN3 (w/v)]. P20 was then immunoprecipitated with rabbit anti-P20 serum (54) at 4°C for 3 h, adsorbed to protein A agarose beads (Sigma) at 4°C for 3 h and centrifuged at 5000 g for 5 min. After washing in the immunoprecipitation buffer, the pellet was suspended in 30 µl of 1X sample buffer for 12.5% SDS–PAGE. Radiolabeled P20 was detected by autoradiography and immunoblotting.
Electrophoretic mobility shift assay
Affinity-purified recombinant WT, S11A or S11D rP20 was incubated with 5 ng WT satBaMV riboprobe and 2 U RNasin (Promega) in 15 µl RNA-binding buffer [10 mM HEPES, pH 7.4, 1 mM EDTA, 50 mM NaCl, 1 mM DTT, 1 mg/ml bovine serum albumin (BSA), 10% glycerol (v/v)] on ice for 30 min. Protein–RNA binding was terminated by adding gel loading buffer, and the samples were analyzed by 1% nondenaturing agarose gel electrophoresis in 0.5X TBE buffer (40 mM Tris-boric acid, 1 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0) at 150 V for 1 h at 4°C. The gel was dried and analyzed in a PhophorImager and quantified by use of ImageQuant v.5.2 (Molecular Dynamics, Sunnyvale, CA, USA).
CD spectroscopy
Conformational changes in satBaMV RNA transcripts after binding to rP20 were analyzed by CD on a Jasco J-715 spectropolarimeter (Tokyo, Japan) with a 0.1-cm path-length water-jacketed quartz cuvette. SatBaMV transcripts that were not pre-incubated with protein were included as controls. Temperature (4°C) was regulated by use of a Neslab RTE 110 water bath (Portsmouth, NH, USA) equipped with a microprocessor and a temperature sensor. Optical resolution, bandwidth and response time were 1 sec, 1.0 nm and 1 s, respectively. Five hundred nanograms of WT satBaMV RNA transcripts were incubated with 50 ng WT, S11A or S11D rP20 in PBS buffer (50 mM sodium phosphate, pH 7.0, 150 mM NaCl) on ice for 30 min. RNA incubated without any protein was included as a control. RNA ellipticity was recorded in the near-ultraviolet (UV) region (250–300 nm) in 0.5-nm steps with an average time of 2 s/step, and 200 000 points were acquired. CD was expressed in molar ellipticity and the final spectrum of each sample was the average of five consecutive scans; the spectra of only buffer were subtracted for baseline artifacts.
In vitro translation
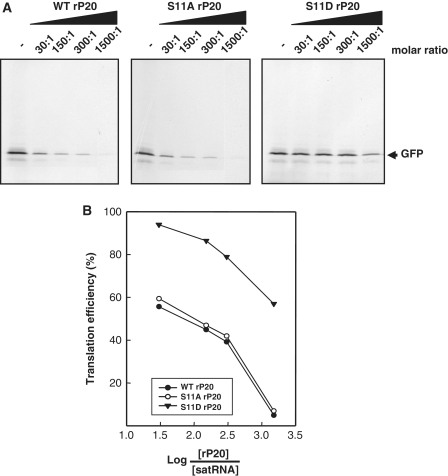
Cell-free translation of satBaMV transcripts (0.5 µg) was performed in wheat germ extract (WGE) (Promega) in the presence of 5 µCi of [35S]-Met as described by the manufacturer. For translation repression assay, 0.5 µg transcripts derived from pBSGFP were denatured at 95°C for 30 s and incubated with WT, S11A or S11D rP20 at different molar ratios on ice for 30 min. The transcripts were then translated in the WGE system in the presence of [35S]-Met as described earlier. Transcripts from pBSGFP not pre-incubated with rP20 or transcripts from pETGFP pre-incubated with rP20 served as a control. Translated GFP was resolved on 12.5% SDS–PAGE, detected by autoradiography and quantified as described above. Translation efficiency curve was plotted by percentage translation versus the molar ratio of log protein to RNA (62).
Protein overlay assay
An indicated amount of purified WT, S11A or S11D rP20 in 1 µl TKM buffer was dot-blotted onto a PVDF membrane in duplicate. Protein adsorption was verified by 0.1% Amido black (w/v) staining. Blots were blocked for 2 h in 1% BSA (w/v) in 140 mM NaCl, 10 mM Tris–HCl, pH 7.4, 2 mM EDTA, 0.1% Tween 20 (v/v) and 2 mM DTT at room temperature. [35S]-Met labeled WT, S11A and S11D were translated in vitro as described above. The blots were probed with either of the radiolabeled rP20 overnight at 4°C and washed in Tris-buffered saline buffer containing 0.05% Tween-20 (v/v). BSA was used as a control. Protein interactions were detected by autoradiography.
Chemical cross-linking
BaMV-S and WT or mutant satBaMV co-infected N. benthamiana leaf protein extracts were cross-linked with glutaraldehyde, and P20 species were immunodetected as described earlier (57).
Plant inoculation, RNA extraction and northern blot analysis
Plants of N. benthamiana were inoculated at the three leaf stages (∼4 weeks after seed germination), eight plants for each treatment. Each leaf was inoculated with 1 μg pCB (50) alone or with 1 μg of WT, pCBSF4 (50), or mutants of satBaMV. Total RNAs were extracted from leaf tissue as described by Verwoerd et al. (63). Accumulation of BaMV and satBaMV in the infected leaves at 10 dpi and 20 dpi were assayed by northern blot hybridization (53). Probes for detection of BaMV and satBaMV RNA were prepared from digoxigenin (DIG)-labeled in vitro transcripts from HindIII-linearized pBaHB (64) or EcoNI-linearized pBSHE (53), respectively. The hybridized membranes were immuno-detected with anti-DIG Fab fragments coupled to alkaline phosphatase (Roche) and visualized using a chemiluminescent substrate, CSPD (Amersham, UK). The banding signals were scanned by PhosphorImager (FUJIFILM LAS-4000) and quantified using Multi Gauge version 3.1 (FUJIFILM, Japan).
RESULTS
P20 is phosphorylated in vitro and in vivo
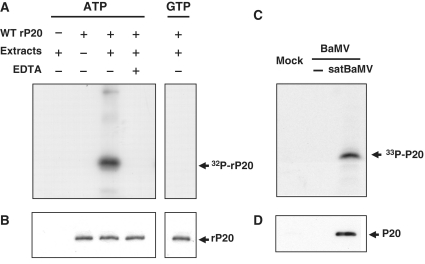
To determine whether satBaMV-encoded P20 is a substrate for host cellular kinases, we tested kinase activity with co-infected N. benthamiana leaf protein extract in vitro. rP20 was used as a substrate, immunoprecipitated and detected by autoradiography (Figure 1A). rP20 was phosphorylated by [γ-32P]-ATP but was not [γ-32P]-GTP used as a phosphoryl donor. Addition of EDTA inhibited phosphorylation, and no phosphorylation was detected in the absence of the substrate. Omission of plant extract in the reaction mix did not result in protein phosphorylation, which confirmed that the substrate was not auto-phosphorylated. rP20 was also phosphorylated with healthy N. benthamiana leaf protein extract used as the kinase source (data not shown). N. benthamiana protoplasts were co-inoculated with BaMV RNA and satBaMV RNA transcripts by electroporation with metabolically labeled [33P]orthophosphate. After 20-h post-inoculation (hpi), protoplast lysates were immunoprecipitated with polyclonal anti-P20 serum, resolved on 12.5% SDS–PAGE and analyzed by autoradiography (Figure 1C). Phosphorylation was detected only in the co-inoculated protoplasts and not in controls. The phosphorylated protein of 20 kDa detected was specifically recognized by the anti-P20 serum (Figure 1B and D), which confirmed the identity of phospho-P20. No protein was immunodetected by the pre-immune serum (data not shown).
Figure 1.
Phosphorylation of P20. (A) In vitro phosphorylation. Purified rP20 was co-incubated in protein extracts of BaMV and satBaMV co-infected N. benthamiana leaves in the absence or presence of 100 mM EDTA and [γ-32P]ATP or [γ-32P]GTP and analyzed by 12.5% SDS–PAGE and autoradiography. Reaction mixture lacking rP20 or leaf protein extract served as controls. (C) In vivo phosphorylation. N. benthamiana protoplasts were mock-inoculated, inoculated with BaMV viral RNA alone or co-inoculated with BaMV RNA and satBaMV RNA transcripts by electroporation and cultured with [33P]orthophosphate. After 20 h inoculation, protoplasts were harvested, lysed, total proteins were immunoprecipitated with anti-P20 serum, analyzed by 12.5% SDS–PAGE and detected by autoradiography. (B and D), Immunodetection of P20 phosphorylated in vitro and in vivo with anti-P20 serum.
S11 contributes to P20 phosphorylation in vivo
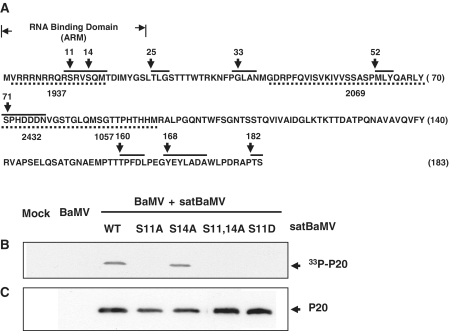
According to NetPhos prediction, P20 contains seven phosphorylation sites: Ser (S) residues 11, 14 and 71; Thr (T) residues 25, 160 and 182; and Tyr (Y) residue 168, which displayed high phosphorylation potential (>0.7) (Figure 2A). S11 and S14 are clustered within the RNA-binding domain of P20 (54). To confirm the phosphorylation targets, rP20 was phosphorylated in vitro, digested with CNBr and analyzed by MALDI-TOF MS in positive ion reflector mode. Native rP20 was included as a control. The mass spectra displayed the phosphorylation sites of rP20 which resides within the residues 2–15 of the ARM, an overlapping functional domain of P20 (57) (data not shown). Because of the close proximity of the residues, we could not precisely determine whether both residues were the targets for phosphorylation. To determine whether S11 and S14 are phosphorylated in vivo, we introduced mutations at S11 and S14 in an infectious satBaMV clone. One or both of the Ser residues were replaced by nonphosphorylatable alanine (S11A, S14A or S11,14A). To mimic phosphorylation in P20, S11 was replaced with an aspartic acid, and S14 was left unchanged (S11D). During satBaMV co-infection in N. benthamiana protoplasts, phosphorylation was determined by metabolic labeling and autoradiography (Figure 2B), and phospho-P20 was immunodetected (Figure 2C). We detected no such phosphorylation signal in mock- and BaMV-inoculated protoplast lysates. Phosphorylation of mutant S14A and WT P20 was similar. By contrast, S11 mutations (S11A, S11,14A and S11D) apparently abolished the phosphorylation, indicating that S11 is a potent target of phosphorylation.
Figure 2.
Phosphorylation sites of P20. (A) P20 sequence was analyzed by use of NetPhos 2.0 software and the predicted sites are shown. P20 sequence is represented by a one-letter amino acid code, and the number on right indicates the number of the last residue in that row. Seven phosphorylation sites—Ser (S) residues 11, 14 and 71; Thr (T) residues 25, 160 and 182; and Tyr (Y) residue 168 in the consensus sequence (solid upper line)—show high phosphorylation potential (>0.7). (B and C) Phosphorylation of satBaMV-encoded P20 in vivo. (B) BaMV RNA was co-inoculated with transcripts of wild-type (WT) or satBaMV mutants into N. benthamiana protoplasts and metabolically labeled with [33P]orthophosphate. Protoplast lysate was immunoprecipitated with anti-P20 serum and analyzed by 12.5% SDS–PAGE and autoradiography. Protoplasts that were mock-inoculated and inoculated with BaMV RNA alone served as a control. (C) Immunoblotting of P20 with anti-P20 serum.
Mimicked phosphorylation of rP20 affects satBaMV RNA binding
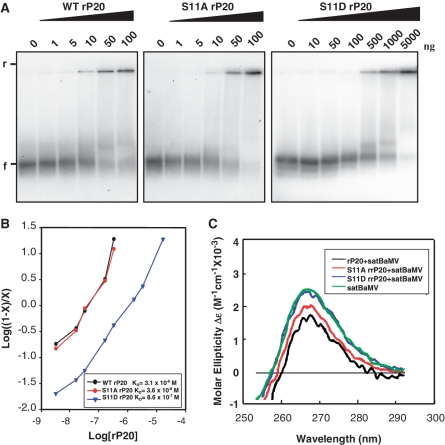
To test whether and to what extent S11 phosphorylation affects satBaMV RNA binding, we used the phospho-mimic (S11D) rP20 in electrophoretic mobility shift assay (EMSA; Figure 3A). As a control, we included the nonphospho-mimic (S11A) rP20. Mobility transition from free RNA to 50% RNA retardation occurred at 10 ng S11A rP20, which was comparable to that of WT rP20. Both proteins retarded almost all of the available RNA at 100 ng, which indicates that both nonphospho- and nonphospho-mimic P20 have no inhibitory effect on satBaMV RNA binding. In contrast, binding of S11D rP20 to the RNA probe was weak, and complete mobility shift required 50 times more protein than S11A rP20. From free riboprobe quantitative analysis, Hill coefficient for WT, S11A and S11D rP20 was determined as 0.97, 0.93 and 0.79, respectively. From linear regression analysis (Figure 3B), apparent dissociation constant (KD) for the WT, S11A and S11D RNP complexes was determined as 3.1 × 10−8 M, 3.6 × 10−8 M and 8.6 × 10−7 M, respectively.
Figure 3.
RNA binding activity of wild-type and mutant rP20. (A) EMSA. Indicated amounts of WT, S11A or S11D rP20 were co-incubated with 5 ng 32P-labeled WT satBaMV transcripts and resolved on 1% non-denaturing agarose gel electrophoresis in 0.5 X TBE buffer (40 mM Tris-boric acid, 1 mM EDTA, pH 8.0) at 150 V and 4°C for 1 h. The gel was analyzed by PhosphorImager. ‘f’ and ‘r’ indicate free and retarded satBaMV RNA, respectively. (B) EMSA data underwent Hill transformation by ImageQuant v.5.2. Data are plotted by log [(1 − x)/x] versus log rP20 concentration, where x is [RNA]/[RNA]total). The slope of best-fit equation determines the Hill coefficient. The dissociation constants (KD) for WT, S11A and S11D rP20 are 3.1 × 10−8 M, 3.6 × 10−8 M and 8.6 × 10−7 M, respectively. (C) Circular dichroism (CD) spectra of satBaMV RNA. In total, 500 ng of WT satBaMV transcripts were incubated with 50 ng WT, S11A or S11D rP20 in PBS buffer (50 mM sodium phosphate, pH 7.0, 150 mM NaCl) on ice for 30 min. SatBaMV transcripts not pre-incubated with rP20 were included as a control. RNA ellipticity was recorded in near-UV region (250–300 nm), and CD is expressed in molar ellipticity.
SatBaMV RNA undergoes conformational changes upon RNP complex formation
To monitor structural changes of satBaMV RNA, if any, upon interacting with phosphorylated P20, satBaMV RNA transcripts were pre-incubated with S11D rP20, and conformation was analyzed by CD spectroscopy (Figure 3C). We used RNA pre-incubated with WT or S11A rP20 and RNA without pre-incubation as controls. Free RNA transcripts showed intense positive ellipticity at 267 nm. On binding to WT or S11A rP20, satBaMV RNA underwent conformation change characterized by decreased molar ellipticity and a red shift. By contrast, RNA incubated with S11D rP20 did not undergo significant conformational change, which implies that the satBaMV RNA binds to phospho-mimicking P20 with less affinity than to nonphospho- or nonphospho-mimicking P20.
Phosphorylation inhibits self-interaction and dimerization of P20
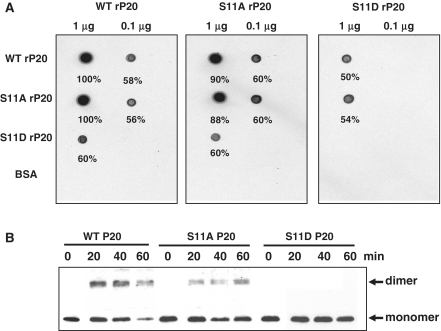
Protein interactions and oligomerization potential of P20 have a role in satBaMV movement (57). To assess whether phosphorylation affects the protein interactions of P20, we performed protein overlay and glutaraldehyde cross-linking assays. On overlay assay, WT, S11A or S11D rP20 was individually adsorbed onto a PVDF membrane and probed with [35S]-Met labeled rP20s, and the interaction was assessed by autoradiography (Figure 4A). Self-interaction of WT rP20 was strong (left panel). The S11 Ala mutation (S11A) inhibited the self-interaction of rP20 (middle panel) but to a lesser extent than the WT. The Asp mutation at S11 (S11D) completely abolished the self-interaction of rP20 (right panel). The interaction of S11D rP20 with both WT rP20 (left and right panels) and S11A rP20 (middle panel) was significantly inhibited compared to the WT and S11A rP20 interactions (left and middle panel). None of the rP20s interacted with BSA. For cross-linking assay, crude protein extracts from BaMV and satBaMV co-inoculated in N. benthamiana leaves were cross-linked at different time points, resolved on 12.5% SDS–PAGE and immunoprobed with anti-P20 serum (Figure 4B). Both WT and S11A P20 dimerized after 20 min, and dimerization of WT P20 was more efficient than that of S11A P20. S11D P20 did not dimerize under identical experimental conditions. Like the recombinant P20-His (57), P20 expressed in BaMV- and satBaMV-co-infected N. benthamiana leaves revealed dimerization potential; the possibility of dimerization resulting from the self-interaction of P20 or interaction between P20 and the helper virus or host protein, or both, cannot be excluded since P20 was derived from the crude plant protein extract.
Figure 4.
Protein interactions of wild-type and mutant rP20. (A) Overlay assay. Purified WT, S11A or S11D rP20 was dot-blotted onto a PVDF membrane. After blocking with 1% BSA (w/v) in 140 mM NaCl, 10 mM Tris–HCl, pH 7.4, 2 mM EDTA, 0.1% Tween 20 (v/v) and 2 mM DTT at room temperature, the membrane was overlayed at 4°C with 35[S]-Met-labeled WT, S11A or S11D translated in vitro. After washing the membrane in Tris-buffered saline buffer containing 0.05% Tween 20 (v/v), protein interactions were detected by autoradiography. Bovine serum albumin was used as a control. (B) Glutaraldehyde cross-linking assay. Total protein extracted from N. benthamiana leaves co-infected with BaMV and WT, S11A or S11D satBaMV was cross-linked with 0.025% glutaraldehyde (v/v) for indicated times at 30°C, resolved on 12.5% SDS–PAGE and immunodetected with anti-P20 serum.
Mimicking phospho-P20 in the RNP complex converts non-translatable to translatable satBaMV RNA
SatBaMV RNA binding of rP20 is highly cooperative and sequence preferential (54). To determine the translation efficiency of satBaMV RNA in the RNP complex containing phospho-mimicking P20, BSGFP, satBaMV RNA transcripts encoding GFP, were pre-incubated with S11D rP20, followed by translation and autoradiography (Figure 5A and B). The ETGFP transcripts from pETGFP without any satBaMV-derived sequences served as control. All tested RNAs were pre-incubated with WT or S11A rP20 and served as controls, along with non-pre-incubated RNA. Translation efficiency of the BSGFP RNA was progressively inhibited with increasing molar ratios of WT rP20 to RNA, and the RNA was non-translatable at 1500:1 molar ratio. Kinetics and suppression of BSGFP RNA translation by S11A rP20 were analogous to those of WT rP20. By contrast, S11D rP20 did not inhibit satBaMV translation at the same molar ratio of protein to RNA and failed to inhibit the translation even at 1500:1 molar ratio. The translation efficiency of control ETGFP was not significantly inhibited when transcripts were pre-inoculated with WT rP20, S11A or S11D rP20 at molar ratio 30:1–300:1 while slightly affected at 1500:1 molar ratio (data not shown).
Figure 5.
Translation repression of rP20. (A) SatBaMV transcripts (0.5 µg) encoding GFP were pre-incubated on ice with WT, S11A or S11D rP20 at indicated molar ratio for 30 min, translated in a wheat-germ extract system with [35S]-Met, resolved on 12.5% SDS–PAGE and detected by autoradiography. (B) Labeled GFP was quantified by ImageQuant v.5.2. Translation efficiency curve for each treatment in (A) was plotted by percentage translation versus molar ratio of log protein to RNA.
Phospho-deficient and phospho-mimicking P20 mutants of satBaMV move less efficiently than WT satBaMV in N. benthamiana
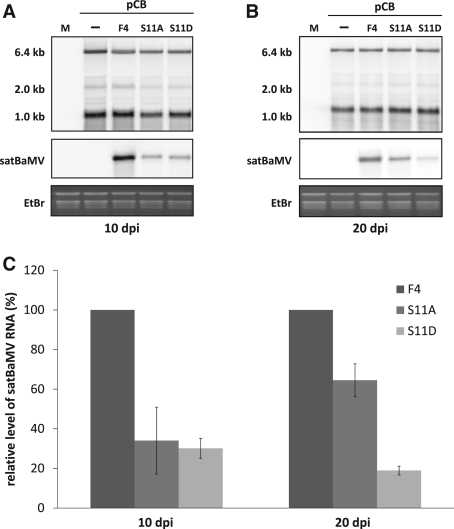
To analyze the biological function of P20 phosphorylation in satBaMV movement in planta, leaves of N. benthamiana were inoculated with pCB, or co-inoculated with WT pCBSF4 (50), pCBSF4-S11A or pCBSF4-S11D. The sytemic leaves at 10 dpi (the fifth leaf) and 20 dpi (the eighth leaf) were harvested for northern blot analysis. As shown previously, the presence of satBaMV slightly affected the accumulation level of BaMV in the systemic leaves of infected plants at 10 and 20 dpi (Figure 6A and B). To statistically analyze the results from three independent experiments, we set the level of WT F4 satBaMV as 100%. As shown in Figure 6A and C, the RNA level of S11A and S11D was reduced to about 30 and 25% of WT satBaMV at 10 dpi, respectively. For S11D, the RNA accumulation was even strongly reduced in the eighth systemic leaves at 20 dpi (Figure 6B and C). Accordingly, the protein level of S11A and S11D was also proportionately decreased (data not shown). These results show that the two mutants, S11A and S11D, delayed systemic movement in the co-infected N. benthamiana with BaMV.
Figure 6.
The accumulation level of wild-type (WT) and mutants of satBaMV in BaMV co-infected N. benthamiana. Three leaves of 1-month-old N. benthamiana plants were co-inoculated with 1 μg of BaMV infectious clone, pCB, or with 1 μg of WT (pCBSF4) and mutants (pCBSF4-S11A and pCBSF4-S11D) of satBaMV. The fifth and eighth systemic leaves were harvested at 10 and 20 days after inoculation (dpi), respectively. (A and B) Northern blot analysis of BaMV and satBaMV RNA from systemic leaves at 10 dpi (A) and 20 dpi (B). The accumulation level of BaMV and satBaMV was measured by hybridization with a specific riboprobe to detect positive-strand BaMV (64) and satBaMV RNA (53), respectively. Hybridization signals were detected in a PhosphorImager and were quantified by use of Multi Gauge, version 3.1 Ethidium bromide-agarose gel prior to blotting shows equal loading as revealed by an abundance of ribosomal RNA in each line. (C) Data show the average accumulation of satBaMVs from three independent experiments. Level of WT F4 was normalized to 100%.
DISCUSSION
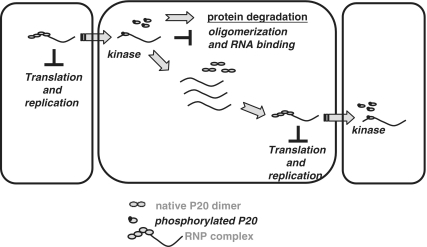
Phosphorylation of viral MP controls the spread of viruses (3,4,8–10,12,17,20,21,23,37,42–45,65). Our study demonstrates phosphorylation of a satellite RNA-encoded protein, P20, and elaborates its functional implications in satBaMV movement. SatBaMV-encoded P20 is phosphorylated in vitro and in vivo. Use of only ATP and not GTP as phosphorylation donor in the phosphorylation reaction suggested that kinase(s) other than CKII are involved in P20 phosphorylation, because the latter is unique in using both GTP and ATP as phosphate donors (6,7,9,66). CKII phosphorylates MPs of PVA (9), CaMV (67) and ToMV (7,45). Inhibition of P20 phosphorylation by EDTA suggests requirement of a divalent metal cation by the P20 kinase because it can essentially activate the enzyme through noncovalent interaction with ATP, which in turn serves as a true phosphate donor for kinases. Of the seven phosphorylation sites predicted in P20 by NetPhos algorithm, and verified by MALDI-TOF MS, the effect of Asp or Ala substitution at S11 in satBaMV infectious clones indicated that S11 is the favorable context for phosphorylation in co-infected plants. Retention of efficient replication of satBaMV mutants demonstrates that S11 is not involved in the satBaMV replication (Figures 2 and 6). Considering the stoichiometric and labile nature of the phosphoproteins, EMSA experiments further revealed that only WT and S11A rP20 bound to the satBaMV RNA in a cooperative mode. However, the high KD of S11D indicated that mimicked phosphorylation leads to less affinity of rP20 to satBaMV RNA. The weak RNA interaction with S11D rP20 is consistent with the fact that the addition of negative charges to P20 moiety by phosphorylation significantly modulates the protein and thereby the protein–protein and protein–RNA interactions. This finding agrees with the differential CD of satBaMV RNA upon binding to nonphospho-mimicked and phospho-mimicked rP20 (Figure 3C). Substantial modulation of satBaMV RNA conformation upon interacting with only WT and S11A rP20 is a consequence of strong protein interaction that leads to the unstacking of bases and stable satBaMV RNP complexes (68). Furthermore, inefficient translation inhibition by S11D alone, but not by WT or S11A rP20 at molar ratios identical to those of WT and S11A rP20, further confirms its weak satBaMV RNA binding (Figure 5). As suggested by the dissimilar regulating features of nonphospho- and phospho-mimicked rP20 in satBaMV RNA translation, phosphorylation of P20 in the satBaMV-P20 RNP complex may trigger the translatability of satBaMV RNA (Figure 5). From our findings, the events for P20 phosphorylation-regulated satBaMV co-infection can take place at two steps (Figure 7). At the early stage of satBaMV co-infection, a pool of satBaMV RNA in the primary infected cell is excluded from replication and interacts with the native P20 in a strong co-operative mode to form a stable satBaMV-P20 RNP complex, which renders the RNA translation incompetent. The RNP complex is facilitated and destined to plasmodesmata and then to the neighboring cells. P20 localized in the plasmodesmata (57) may be accessible to both plasmodesmata protein kinase(s) and cellular kinases. This suggestion is consistent with our data showing that P20 serves as a substrate for the host kinases in vitro and in vivo (Figures 1 and 2). During the plasmodesmal translocation of satBaMV RNP complex, phosphorylation of P20 leads to decrease in affinity of the protein to satBaMV RNA (Figure 3), thus freeing the satBaMV RNA for replication and translation in the newly infected cell. Dephosphorylated P20 or translated P20, upon binding to the newly synthesized satBaMV RNA, renders satBaMV translation incompetent. Hence, during satBaMV co-infection, P20 phosphorylation converts the translation-incompetent satBaMV to translatable satBaMV RNA (Figure 5), which is destined solely for cell-to-cell trafficking. Repression of nonphospho- or phospho-deficient P20 (WT or S11A rP20) by satBaMV translation is functionally analogous to that of other viral MPs (2,4,8,17,18,20,26). The negative interaction of phosphorylated P20 and satBaMV RNA may switch the satBaMV RNA function from movement to replication, and both mechanisms may operate concurrently. In the second event, as the P20 ARM encompasses S11 that is indispensable for the cell-to-cell spread of P20 (2), S11 phosphorylation may be vital for the transport of satBaMV-P20 RNP complex during co-infection. Phosphorylation may target P20 to the host degradation pathway, which might be the cause for P20 turnover at later stages of satBaMV co-infection (57). The functional relevance of P20 phosphorylation is supported by our mutational study of satBaMV in planta. Both phospho-deficient S11A and phospho-mimicking S11D mutants resulted in a significant reduction of accumulation of satBaMV RNA in the systemic leaves of co-infected N. benthamiana (Figure 6). P20 phosphorylation may therefore have control over satBaMV co-infection in a regulated manner and represent a host cell defense pathway.
Figure 7.
Model for P20 phosphorylation-regulated satBaMV co-infection. At the early stage of satBaMV co-infection, a pool of satBaMV RNA in the primary infected cell is excluded from replication and interacts with the native P20 in a strong co-operative mode to form a stable satBaMV-P20 RNP complex, which renders the RNA translation incompetent. The RNP complex is facilitated and destined to plasmodesmata and then to the neighboring cells. P20 localized in the plasmodesmata (57) may be accessible to the plasmodesmata kinase(s) and/or to other cellular kinases. S11 phosphorylation of P20 in the RNP complex prevents its dimerization and reduces the RNA-binding activity, thereby potentiating the dissociation of the RNP complex. The free RNA efficiently translates and/or replicates in the newly co-infected cell. Translated P20, on binding to the satBaMV RNA, represses the translation and regulation of satBaMV co-infection by continued P20 phosphorylation.
FUNDING
National Science Council (grant number NSC-93-2321-B-001-002); and Academia Sinica Investigator Award, Taipei, Taiwan. Funding for open access charge: Institute of Plant and Microbial Biology, Academia Sinica.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank M. Z. Fang for sequencing, T. N. Wen's assistance from Proteomics Core Lab (Institute of Plant and Microbial Biology, Academia Sinica) and Mr. Chih-Hao Chang for technical assistance. We also thank Douglas Platt for his careful reading and editing of this manuscript.
REFERENCES
- 1.Citovsky V. Probing plasmodesmal transport with plant viruses. Plant Physiol. 1993;102:1071–1076. doi: 10.1104/pp.102.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karpova OV, Ivanov KI, Rodionova NP, Dorokhov Yu L, Atabekov JG. Nontranslatability and dissimilar behavior in plants and protoplasts of viral RNA and movement protein complexes formed in vitro. Virology. 1997;230:11–21. doi: 10.1006/viro.1997.8472. [DOI] [PubMed] [Google Scholar]
- 3.Kawakami S, Padgett HS, Hosokawa D, Okada Y, Beachy RN, Watanabe Y. Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J. Virol. 1999;73:6831–6840. doi: 10.1128/jvi.73.8.6831-6840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waigmann E, Chen MH, Bachmaier R, Ghoshroy S, Citovsky V. Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 2000;19:4875–4884. doi: 10.1093/emboj/19.18.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atabekov JG, Rodionova NP, Karpova OV, Kozlovsky SV, Novikov VK, Arkhipenko MV. Translational activation of encapsidated potato virus X RNA by coat protein phosphorylation. Virology. 2001;286:466–474. doi: 10.1006/viro.2001.1013. [DOI] [PubMed] [Google Scholar]
- 6.Matsushita Y, Hanazawa K, Yoshioka K, Oguchi T, Kawakami S, Watanabe Y, Nishiguchi M, Nyunoya H. In vitro phosphorylation of the movement protein of tomato mosaic tobamovirus by a cellular kinase. J. Gen. Virol. 2000;81:2095–2102. doi: 10.1099/0022-1317-81-8-2095. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita Y, Ohshima M, Yoshioka K, Nishiguchi M, Nyunoya H. The catalytic subunit of protein kinase CK2 phosphorylates in vitro the movement protein of Tomato mosaic virus. J. Gen. Virol. 2003;84:497–505. doi: 10.1099/vir.0.18839-0. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov KI, Puustinen P, Merits A, Saarma M, Makinen K. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 2001;276:13530–13540. doi: 10.1074/jbc.M009551200. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov KI, Puustinen P, Gabrenaite R, Vihinen H, Ronnstrand L, Valmu L, Kalkkinen N, Makinen K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell. 2003;15:2124–2139. doi: 10.1105/tpc.012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karger EM, Frolova OY, Fedorova NV, Baratova LA, Ovchinnikova TV, Susi P, Makinen K, Ronnstrand L, Dorokhov YL, Atabekov JG. Dysfunctionality of a tobacco mosaic virus movement protein mutant mimicking threonine 104 phosphorylation. J. Gen. Virol. 2003;84:727–732. doi: 10.1099/vir.0.18972-0. [DOI] [PubMed] [Google Scholar]
- 11.Jakubiec A, Jupin I. Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res. 2007;129:73–79. doi: 10.1016/j.virusres.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JY, Taoka K, Yoo BC, Ben-Nissan G, Kim DJ, Lucas WJ. Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell. 2005;17:2817–2831. doi: 10.1105/tpc.105.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JY. Phosphorylation of movement proteins by the plasmodesmal-associated protein kinase. Methods Mol. Biol. 2008;451:625–639. doi: 10.1007/978-1-59745-102-4_42. [DOI] [PubMed] [Google Scholar]
- 14.Zayakina O, Arkhipenko M, Kozlovsky S, Nikitin N, Smirnov A, Susi P, Rodionova N, Karpova O, Atabekov J. Mutagenic analysis of potato virus X movement protein (TGBp1) and the coat protein (CP): in vitro TGBp1-CP binding and viral RNA translation activation. Mol. Plant Pathol. 2008;9:37–44. doi: 10.1111/j.1364-3703.2007.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modena NA, Zelada AM, Conte F, Mentaberry A. Phosphorylation of the TGBp1 movement protein of Potato virus X by a Nicotiana tabacum CK2-like activity. Virus Res. 2008;137:16–23. doi: 10.1016/j.virusres.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Makinen KM, Ivanov KI. Phosphorylation analysis of plant viral proteins. Methods Mol. Biol. 2008;451:339–359. doi: 10.1007/978-1-59745-102-4_24. [DOI] [PubMed] [Google Scholar]
- 17.Karpova OV, Rodionova NP, Ivanov KI, Kozlovsky SV, Dorokhov YL, Atabekov JG. Phosphorylation of tobacco mosaic virus movement protein abolishes its translation repressing ability. Virology. 1999;261:20–24. doi: 10.1006/viro.1999.9842. [DOI] [PubMed] [Google Scholar]
- 18.Kawakami S, Hori K, Hosokawa D, Okada Y, Watanabe Y. Defective tobamovirus movement protein lacking wild-type phosphorylation sites can be complemented by substitutions found in revertants. J Virol. 2003;77:1452–1461. doi: 10.1128/JVI.77.2.1452-1461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SH, Palukaitis P, Park YI. Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. EMBO J. 2002;21:2292–2300. doi: 10.1093/emboj/21.9.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waigmann E, Ueki S, Trutnyeva K, Citovsky V. The ins and outs of non-destructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 2004;23:195–250. [Google Scholar]
- 21.Lee JY, Lucas WJ. Phosphorylation of viral movement proteins–regulation of cell-to-cell trafficking. Trends Microbiol. 2001;9:5–8. doi: 10.1016/s0966-842x(00)01901-6. discussion 8. [DOI] [PubMed] [Google Scholar]
- 22.Puustinen P, Rajamaki ML, Ivanov KI, Valkonen JP, Makinen K. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J. Virol. 2002;76:12703–12711. doi: 10.1128/JVI.76.24.12703-12711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas WJ, Lee JY. Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell. Biol. 2004;5:712–726. doi: 10.1038/nrm1470. [DOI] [PubMed] [Google Scholar]
- 24.Fujiki M, Kawakami S, Kim RW, Beachy RN. Domains of tobacco mosaic virus movement protein essential for its membrane association. J. Gen. Virol. 2006;87:2699–2707. doi: 10.1099/vir.0.81936-0. [DOI] [PubMed] [Google Scholar]
- 25.Yoshioka K, Matsushita Y, Kasahara M, Konagaya K, Nyunoya H. Interaction of tomato mosaic virus movement protein with tobacco RIO kinase. Mol. Cells. 2004;17:223–229. [PubMed] [Google Scholar]
- 26.Trutnyeva K, Bachmaier R, Waigmann E. Mimicking carboxyterminal phosphorylation differentially effects subcellular distribution and cell-to-cell movement of Tobacco mosaic virus movement protein. Virology. 2005;332:563–577. doi: 10.1016/j.virol.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 27.Shapka N, Stork J, Nagy PD. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology. 2005;343:65–78. doi: 10.1016/j.virol.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Stork J, Panaviene Z, Nagy PD. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology. 2005;343:79–92. doi: 10.1016/j.virol.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Jakubiec A, Tournier V, Drugeon G, Pflieger S, Camborde L, Vinh J, Hericourt F, Redeker V, Jupin I. Phosphorylation of viral RNA-dependent RNA polymerase and its role in replication of a plus-strand RNA virus. J. Biol. Chem. 2006;281:21236–21249. doi: 10.1074/jbc.M600052200. [DOI] [PubMed] [Google Scholar]
- 30.Akamatsu N, Takeda A, Kishimoto M, Kaido M, Okuno T, Mise K. Phosphorylation and interaction of the movement and coat proteins of brome mosaic virus in infected barley protoplasts. Arch. Virol. 2007;152:2087–2093. doi: 10.1007/s00705-007-1038-6. [DOI] [PubMed] [Google Scholar]
- 31.Hafren A, Makinen K. Purification of viral genome-linked protein VPg from potato virus A-infected plants reveals several post-translationally modified forms of the protein. J. Gen. Virol. 2008;89:1509–1518. doi: 10.1099/vir.0.83649-0. [DOI] [PubMed] [Google Scholar]
- 32.Lewsey M, Surette M, Robertson FC, Ziebell H, Choi SH, Ryu KH, Canto T, Palukaitis P, Payne T, Walsh JA, et al. The role of the Cucumber mosaic virus 2b protein in viral movement and symptom induction. Mol. Plant Microbe Interact. 2009;22:642–654. doi: 10.1094/MPMI-22-6-0642. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez I, Martinez L, Rakitina DV, Lewsey MG, Atencio FA, Llave C, Kalinina NO, Carr JP, Palukaitis P, Canto T. Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol. Plant Microbe Interact. 2010;23:294–303. doi: 10.1094/MPMI-23-3-0294. [DOI] [PubMed] [Google Scholar]
- 34.Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EP. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J. Virol. 2006;80:6648–6656. doi: 10.1128/JVI.00173-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piroux N, Saunders K, Page A, Stanley J. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology. 2007;362:428–440. doi: 10.1016/j.virol.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 36.Lucas WJ. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology. 2006;344:169–184. doi: 10.1016/j.virol.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Verchot-Lubicz J, Ye CM, Bamunusinghe D. Molecular biology of potexviruses: recent advances. J. Gen. Virol. 2007;88:1643–1655. doi: 10.1099/vir.0.82667-0. [DOI] [PubMed] [Google Scholar]
- 38.Lewsey MG, Gonzalez I, Kalinina NO, Palukaitis P, Canto T, Carr JP. Symptom induction and RNA silencing suppression by the cucumber mosaic virus 2b protein. Plant Signal Behav. 2010;5:705–708. doi: 10.4161/psb.5.6.11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe Y, Ogawa T, Okada Y. In vivo phosphorylation of the 30-kDa protein of tobacco mosaic virus. FEBS Lett. 1992;313:181–184. doi: 10.1016/0014-5793(92)81440-w. [DOI] [PubMed] [Google Scholar]
- 40.Haley A, Hunter T, Kiberstis P, Zimmern D. Multiple serine phosphorylation sites on the 30 kDa TMV cell-to-cell movement protein synthesized in tobacco protoplasts. Plant J. 1995;8:715–724. doi: 10.1046/j.1365-313x.1995.08050715.x. [DOI] [PubMed] [Google Scholar]
- 41.Atabekov JG, Taliansky ME. Expression of a plant virus-coded transport function by different viral genomes. Adv. Virus Res. 1990;38:201–248. doi: 10.1016/s0065-3527(08)60863-5. [DOI] [PubMed] [Google Scholar]
- 42.Sato K, Yoshikawa N, Takahashi T, Taira H. Expression, subcellular location and modification of the 50 kDa protein encoded by ORF2 of the apple chlorotic leaf spot trichovirus genome. J. Gen. Virol. 1995;76 (Pt 6):1503–1507. doi: 10.1099/0022-1317-76-6-1503. [DOI] [PubMed] [Google Scholar]
- 43.Seron K, Bernasconi L, Allet B, Haenni AL. Expression of the 69K movement protein of turnip yellow mosaic virus in insect cells. Virology. 1996;219:274–278. doi: 10.1006/viro.1996.0246. [DOI] [PubMed] [Google Scholar]
- 44.Sokolova M, Prufer D, Tacke E, Rohde W. The potato leafroll virus 17K movement protein is phosphorylated by a membrane-associated protein kinase from potato with biochemical features of protein kinase C. FEBS Lett. 1997;400:201–205. doi: 10.1016/s0014-5793(96)01380-4. [DOI] [PubMed] [Google Scholar]
- 45.Matsushita Y, Yoshioka K, Shigyo T, Takahashi H, Nyunoy H. Phosphorylation of the movement protein of cucumber mosaic virus in transgenic tobacco plants. Virus Genes. 2002;24:231–234. doi: 10.1023/a:1015324415110. [DOI] [PubMed] [Google Scholar]
- 46.Lin NS, Huang TZ, Hsu YH. Infection of barely protoplasts with bamboo mosaic virus RNA. Bot. Bull. Acad. Sin. 1992;33:271–275. [Google Scholar]
- 47.Yang CC, Liu JS, Lin CP, Lin NS. Nucleotide sequence and phylogenetic analysis of a bamboo mosaic potexvirus isolate from common bamboo (Bambusa vulgaris McClure) Bot. Bull. Acad. Sin. 1997;38:77–84. [Google Scholar]
- 48.Lin NS, Lin BY, Lo NW, Hu CC, Chow TY, Hsu YH. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 1994;75 (Pt 9):2513–2518. doi: 10.1099/0022-1317-75-9-2513. [DOI] [PubMed] [Google Scholar]
- 49.Chang BY, Lin NS, Liou DY, Chen JP, Liou GG, Hsu YH. Subcellular localization of the 28 kDa protein of the triple-gene-block of bamboo mosaic potexvirus. J. Gen. Virol. 1997;78 (Pt 5):1175–1179. doi: 10.1099/0022-1317-78-5-1175. [DOI] [PubMed] [Google Scholar]
- 50.Lin MK, Chang BY, Liao JT, Lin NS, Hsu YH. Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 2004;85:251–259. doi: 10.1099/vir.0.19442-0. [DOI] [PubMed] [Google Scholar]
- 51.Lin MK, Hu CC, Lin NS, Chang BY, Hsu YH. Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the bamboo mosaic virus satellite RNA-mediated expression system. J. Gen. Virol. 2006;87:1357–1367. doi: 10.1099/vir.0.81625-0. [DOI] [PubMed] [Google Scholar]
- 52.Lin NS, Hsu YH. A satellite RNA associated with bamboo mosaic potexvirus. Virology. 1994;202:707–714. doi: 10.1006/viro.1994.1392. [DOI] [PubMed] [Google Scholar]
- 53.Lin NS, Lee YS, Lin BY, Lee CW, Hsu YH. The open reading frame of bamboo mosaic potexvirus satellite RNA is not essential for its replication and can be replaced with a bacterial gene. Proc. Natl Acad. Sci. USA. 1996;93:3138–3142. doi: 10.1073/pnas.93.7.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai MS, Hsu YH, Lin NS. Bamboo mosaic potexvirus satellite RNA (satBaMV RNA)-encoded P20 protein preferentially binds to satBaMV RNA. J. Virol. 1999;73:3032–3039. doi: 10.1128/jvi.73.4.3032-3039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu HT, Hsu YH, Bi IP, Lin NS, Chang BY. Biological functions of the cytoplasmic TGBp1 inclusions of bamboo mosaic potexvirus. Arch. Virol. 2004;149:1027–1035. doi: 10.1007/s00705-003-0254-y. [DOI] [PubMed] [Google Scholar]
- 56.Isogai M, Yoshikawa N. Mapping the RNA-binding domain on the Apple chlorotic leaf spot virus movement protein. J Gen. Virol. 2005;86:225–229. doi: 10.1099/vir.0.80493-0. [DOI] [PubMed] [Google Scholar]
- 57.Vijaya Palani P, Kasiviswanathan V, Chen JC, Chen W, Hsu YH, Lin NS. The arginine-rich motif of Bamboo mosaic virus satellite RNA-encoded P20 mediates self-interaction, intracellular targeting, and cell-to-cell movement. Mol. Plant Microbe Interact. 2006;19:758–767. doi: 10.1094/MPMI-19-0758. [DOI] [PubMed] [Google Scholar]
- 58.Palani PV, Chiu M, Chen W, Wang CC, Lin CC, Hsu CC, Cheng CP, Chen CM, Hsu YH, Lin NS. Subcellular localization and expression of bamboo mosaic virus satellite RNA-encoded protein. J. Gen. Virol. 2009;90:507–518. doi: 10.1099/vir.0.004994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin NS, Chen CC. Association of Bamboo mosaic virus (BaMV) and BaMV-specific electron-dense crystalline bodies with chloroplasts. Phytopathology. 1991;81:1551–1555. [Google Scholar]
- 60.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J, Fritsch F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 62.Kim SH, Kalinina NO, Andreev I, Ryabov EV, Fitzgerald AG, Taliansky ME, Palukaitis P. The C-terminal 33 amino acids of the cucumber mosaic virus 3a protein affect virus movement, RNA binding and inhibition of infection and translation. J. Gen. Virol. 2004;85:221–230. doi: 10.1099/vir.0.19583-0. [DOI] [PubMed] [Google Scholar]
- 63.Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin NS, Chai YJ, Huang TY, Chang TY, Hsu YH. Incidence of bamboo mosaic potexvirus in Taiwan. Plant Dis. 1993;77:448–450. [Google Scholar]
- 65.Rhee Y, Gurel F, Gafni Y, Dingwall C, Citovsky V. A genetic system for detection of protein nuclear import and export. Nat. Biotechnol. 2000;18:433–437. doi: 10.1038/74500. [DOI] [PubMed] [Google Scholar]
- 66.Li M, Lyon MK, Garcea RL. In vitro phosphorylation of the polyomavirus major capsid protein VP1 on serine 66 by casein kinase II. J. Biol. Chem. 1995;270:26006–26011. doi: 10.1074/jbc.270.43.26006. [DOI] [PubMed] [Google Scholar]
- 67.Chapdelaine Y, Kirk D, Karsies A, Hohn T, Leclerc D. Mutation of capsid protein phosphorylation sites abolishes cauliflower mosaic virus infectivity. J. Virol. 2002;76:11748–11752. doi: 10.1128/JVI.76.22.11748-11752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vilar M, Esteve V, Pallas V, Marcos JF, Perez-Paya E. Structural properties of carnation mottle virus p7 movement protein and its RNA-binding domain. J. Biol. Chem. 2001;276:18122–18129. doi: 10.1074/jbc.M100706200. [DOI] [PubMed] [Google Scholar]