Abstract
microRNAs (miRNAs) are a versatile class of non-coding RNAs involved in regulation of various biological processes. miRNA-122 (miR-122) is specifically and abundantly expressed in human liver. In this study, we employed 3′-end biotinylated synthetic miR-122 to identify its targets based on affinity purification. Quantitative RT-PCR analysis of the affinity purified RNAs demonstrated a specific enrichment of several known miR-122 targets such as CAT-1 (also called SLC7A1), ADAM17 and BCL-w. Using microarray analysis of affinity purified RNAs, we also discovered many candidate target genes of miR-122. Among these candidates, we confirmed that protein kinase, interferon-inducible double-stranded RNA-dependent activator (PRKRA), a Dicer-interacting protein, is a direct target gene of miR-122. miRNA quantitative-RT–PCR results indicated that miR-122 and small interfering RNA against PRKRA may facilitate the accumulation of newly synthesized miRNAs but did not detectably affect endogenous miRNAs levels. Our findings will lead to further understanding of multiple functions of this hepato-specific miRNA. We conclude that miR-122 could repress PRKRA expression and facilitate accumulation of newly synthesized miRNAs.
INTRODUCTION
MicroRNAs (miRNAs) are small conserved RNAs of ∼22 nt which negatively modulate gene expression in animals and plants, primarily through base paring to the 3′-untranslated region (UTR) of target messenger RNAs (mRNAs). This leads to mRNA cleavage and/or translation repression (1). miRNAs are primarily transcribed by RNA polymerase II as part of capped and polyadenylated primary transcripts (pri-miRNAs) that can be either protein-coding or non-coding. The primary transcript is cleaved by Drosha ribonuclease III enzyme to produce an ∼70-nt stem–loop precursor miRNA (pre-miRNA), which is further cleaved by the cytoplasmic Dicer ribonuclease to generate the mature miRNA. The mature miRNA is incorporated into an RNA-induced silencing complex (RISC), which recognizes target mRNAs through imperfect base pairing with the miRNA. Bioinformatic analysis predicts that each miRNA may regulate hundreds of target genes, suggesting that miRNAs may play a role in almost every biological pathway (2). Indeed, miRNAs have been implicated in the regulation of various cellular processes, including cell proliferation, apoptosis and stress responses (3–6).
One of the first clues of the existence of miRNAs in mammals came from studies on genetic alterations in woodchuck liver tumors. In 1989, a gene rearrangement of c-myc and an unusual transcript, named hcr, was described in one of these tumors. This transcript was characterized as liver specific, essentially non-coding, specifically nuclear and processed by endonucleases (7). Furthermore, hcr was proposed to be the precursor for miR-122. In the current understanding, the part of the hcr transcript encompassing the so-called ‘pri-miRNA’ is predicted to be processed to form a 66-nt long ‘pre-miRNA’, which presents a hairpin structure with 79% base pairing, and which will ultimately be cleaved by the endonuclease Dicer to form the mature miR-122 (8). Recent works on tissue-specific miRNAs has demonstrated miRNAs participation in tissue specification and cell lineage decisions (9–11). Among these tissue-specific miRNAs, miR-122 is one which is specifically expressed in adult liver and constitutes 70% of the total miRNA population (12–14). Recent studies showed that miR-122 could modulate lipid metabolism (15,16), hepatitis C virus (HCV) replication (17–19), apoptosis (20) and play a role in hepatocellular carcinoma (HCC) (21–23). To better understand the role of this liver-specific miRNA, the identification of the target genes of miR-122 is necessary.
In the present study, we employed 3′-end biotinylated synthetic miR-122 to identify its target genes based on affinity purification as described previously (24,25). Quantitative reverse transcriptase–polymerase chain reaction (RT–PCR) analysis of the affinity purified RNAs demonstrated a specific enrichment of several reported miR-122 targets, such as CAT-1 (13), ADAM17 (22) and BCL-w (20). By microarray, many candidate target genes of miR-122 were identified. We also verified that PRKRA, which showed a high of enrichment in affinity purification assay, was a new target gene of miR-122. Furthermore, the over-expression of miR-122 could facilitate the accumulation of newly synthesized miRNA.
MATERIALS AND METHODS
Cell lines and cultures
HepG2 and HeLa cell lines were cultured in DMEM (GIBCO BRL, Grand Island, NY, USA) containing 10% FBS with 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C with 5% CO2.
Affinity purification experiments
To identify mRNAs associated with miRNA-122, affinity purification experiments were performed as described previously (Supplementary Figure S1, see Supplementary Methods for details) (24,25). Synthetic miRNA-122 duplexes were produced carrying a biotin group attached to the 3′-end of the miRNA sense strand (TaKaRa, Dalian, China) and transfected into HepG2 cells. Cells were harvested 48 h after transfection. The isolated RNA was ready for downstream qRT–PCR or microarray analysis.
Real-time qRT–PCR for mRNA
Total RNA was isolated using TRI Reagent (Sigma-Aldrich, St Louis, USA). cDNA was generated by reverse transcription using 1 μg of total RNA and ImProm-IITM Reverse Transcription System (Promega, Madison, WI, USA). Quantitative real-time PCR was performed on the MX 3000PTM PCR Instrument (Stratagene, La Jolla, USA) using SYBR Premix EX TaqTM (TaKaRa). Forward (F) and reverse (R) primers used were as follows: GAPDH-F 5′-tcagtggtggacctgacctg-3′, GAPDH-R 5′-tgctgtagccaaattcgttg-3′; BCL-w -F 5′-tttggttcggctttatcagg-3′, BCL-w-R 5′-gaggactgcgagttccaaag-3′; CAT-1-F 5′-ggctgtcctctggtgagaag-3′, CAT-1-R 5′-ggccaccagatcaaaagtgt-3′; ADAM-17-F 5′-ctgtggtgcaaaagcagaaa-3′, ADAM-17-R 5′-tgccaaatgcctcatattca-3′; PRKRA-F 5′-acgaatacggcatgaagacc-3′, PRKRA-R 5′-tggaagggtcaggcattaag-3′; A20-F 5′-gagagcacaatggctgaaca-3′, A20-R 5′-tccagtgtgtatcggtgcat-3′.
Microarray hybridization
RNA from affinity purification experiments was linear amplified (Two-Cycle Eukaryotic Target Labeling Kit, P/N 900494, Affymetrix, Inc., Santa Clara, CA, USA) and analyzed on Affymetrix human u133 plus 2.0 microarrays, which include a set of human maintenance genes to facilitate the normalization and scaling of array experiments. This set of genes serves as a tool to normalize and scale the data prior to performing data comparisons.
miRNA target site analysis
Affymetrix probe set identifiers were mapped to Ensembl transcripts using the Ensembl Biomart system. The 3′-UTR sequences were extracted from Ensembl using Biomart utilities. UltraEdit software was used to search candidate 3′-UTR for seed matches (perfect Watson–Crick matches between the 6-mer from bases 2 to 7 of the miRNA from the 5′-end) with none or one of three kinds of extensions: (i) an A across from nucleotide 1 in the miRNA (seedM+t1A), (ii) an additional match between the site and nucleotide 8 in the miRNA (seedM+m8M) and (iii), the combination of (i) and (ii) (seedM+m8M+t1A). TargetScan software (www.targetscan.org) was used to search for conserved miR-122-binding sites.
Plasmid construction
For the expression of miR-122, miR-133 or miR-30a, genomic fragment of Homo sapiens miR-122, miR-133 or miR-30a precursor was amplified and cloned into pcDNA3.0 (Invitrogen, Carlsbad, CA, USA). PRKRA 3′-UTR segment was subcloned into the pGL3 Control vector (Promega) immediately downstream of the stop codon of the luciferase gene. PCR with the appropriate primers also generated inserts with point substitutions in the miRNA complementary sites. Wild-type and mutant inserts were confirmed by sequencing.
miRNA qRT–PCR
For miRNA detection, total RNA was polyadenylated by poly (A) polymerase (Ambion, Austin, USA). An amount of 50 μl polyadenylation reaction was set up with 10 μg total RNA and 1 μl (2U) poly (A) polymerase according to the manufacturer's protocol. After incubation at 37°C for 60 min, poly (A)-tailed total RNA was recovered by phenol/chloroform extraction and ethanol precipitation. RT reaction was performed using 1 μg poly (A) tailed total RNA and 1 μg RT primer [5′-gcgagcacagaattaatacgatcactatagg(t)18VN-3′] with 1 μl ImProm-IITM Reverse Transcriptase (Promega) according to the manufacturer's protocol. qPCR was performed as described in the method of Quantitect SYBR Green PCR Kit (Qiagen, Hilden, Germany) with Mx3000pTM (Stratagene) supplied with analytical software. One primer of miRNA amplification is miRNA specific (miR-122 5′-tggagtgtgacaatggtgtttg-3′; miR-16 5′-tagcagcacgtaaatattggcg-3′; miR-24 5′-tggctcagttcagcaggaacag-3′; miR-133 5′-tttggtccccttcaaccagctg-3′; miR-30a 5′-tgtaaacatcctcgactggaag-3′), and the other is a universal primer (5′-gcgagcacagaattaatacgac-3′). U6 snRNA levels were used for normalization (U6-F 5′-cgcttcggcagcacatatacta-3′; U6-R 5′-cgcttcacgaatttgcgtgtca-3′).
miRNAs, small interfering RNAs and transfection
The miR-122 duplex and PRKRA small interfering RNAs (siRNAs) were designed and synthesized by GenePharma (GenePharma, Shanghai, China). The sequences of miRNA duplex and siRNAs were presented in Supplementary Table S1. siRNAs and miRNAs were transfected using Lipofectamine 2000 (Invitrogen). In brief, cells were cultured in a six-well plate to 50% confluence. For each well, 5 μl siRNA (20 μM) or miRNA (20 μM) was added into 250 μl Opti-MEM medium (GIBCO BRL), 4 μl of Lipofectamine 2000 into 250 μl Opti-MEM medium and then mixed siRNA or miRNA with Lipofectamine 2000 after 5 min incubation. After 20 min, the mixture was added to cells and incubated for 6 h before replacing the medium. Total RNA and protein were prepared 48 or 72 h after transfection and were used for qRT–PCR or western blotting analysis.
Western blot
Total cell lysate was prepared in 1× SDS buffer. Proteins at the same amount were separated by SDS–PAGE and transferred onto PVDF membranes. After probing with anti-PRKRA (10771-1-AP; Protein Tech Group, Inc., Chicago, USA) or anti-β-actin antibody (Beijing Zhongshan Biotechnology, Beijing, China) and incubating with proper secondary antibody, antigen–antibody complex was visualized by enhanced chemiluminescence's reagents Supersignal (Pierce, Rochford, IL, USA).
Luciferase reporter assay
Adherent HepG2 cells were grown in DMEM with 10% FBS to 80–90% confluency in 24-well plates. Cells were co-transfected with 100 ng of firefly luciferase reporter vector containing the PRKRA 3′-UTR (named pGL3-PRKRA-3′-UTR) or PRKRA mutant (named pGL3-PRKRA-3′-UTRmut) and 8 ng of the control vector containing Renilla luciferase, pRL-TK (Promega), in a final volume of 0.5 ml using Lipofectamine 2000 (Invitrogen). Firefly and Renilla luciferase activities were measured consecutively using the Dual-luciferase assays (Promega) 48 h after transfection.
Statistic analysis
All data are presented as means ± SD. Differences were assessed by two-tailed Student's t-test using Excel software. P < 0.05 was considered to be statistically significant.
RESULTS
Affinity purification of miR-122 target mRNAs
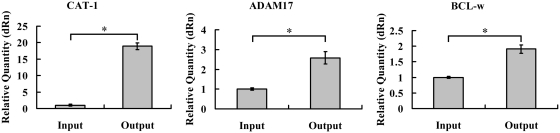
To identify mRNA targets of miR-122, we employed an affinity-based target-identification procedure in which miR-122 is synthesized with a 3′-biotin group allowing for subsequent purification with streptavidin (Supplementary Figure S1). This technique has been previously verified for affinity purification of miRNA targets in Drosophila melanogaster cells and human neuronal cells (24,25). We validated the technique using a biotin-tagged miR-122 targeting several reported targets such as CAT-1 (13), ADAM17 (22) and BCL-w (20). Affinity purification experiments in HepG2 cells resulted in a 2- to 19-fold enrichment of these endogenous targets (Figure 1).
Figure 1.
Affinity purification with biotin-tagged miR-122 from human hepatoma HepG2 cells and quantitative RT–PCR for its endogenous target CAT-1, ADAM17 and BCL-w. HepG2 cells were treated with cell lysis buffer 48 h after transfection. Data were normalized to the level of GAPDH mRNA. Results of the mean of triplicate quantitative PCR assays with standard deviation of the mean are presented. *P < 0.05.
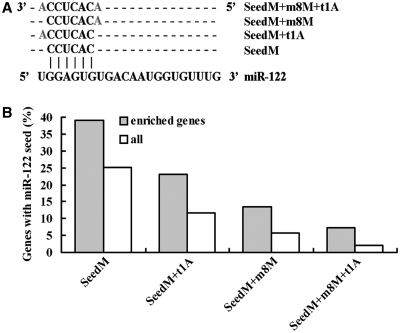
To identify more targets of miR-122, RNAs from affinity purification experiment were linear amplified and analyzed on microarrays. Microarray results showed that 1474 genes were enriched >2-fold by 3′-biotin modified miR-122 (output) compared with total RNA (input) (Supplementary Table 2). We examined the 3′-UTRs for the presence of the 6-nt sequence CACTCC, which is the reverse complement of the nucleotides 2–7 seed in the mature miR-122 sequence. About 39% of the enriched mRNAs having at least one miR-122 recognition sequence, while the seed match frequency in all annotated human 3′-UTRs was only 25%, implying that a significant pool of the enriched mRNAs correspond to direct miR-122 targets in the liver (P < 0.001, Figure 2B and Supplementary Table S3). Detailed studies indicate that the presence of extended seed matches increases the likelihood that a given message is regulated by a miRNA (26,27). We therefore examined the identified seed matches for the presence of an A anchor corresponding to the 5′ most nucleotide of miR-122, as well as for an extended match to base 8 of miR-122 as described by Lewis et al. (26). Both the 7-nt and the 8-nt seed matches were significantly enriched in our microarray data (Figure 2Aand B; Supplementary Table S3). Moreover, about one-third of the conserved targets predicted by TargetScan software showed >2-fold of enrichment by biotin modified miR-122 (38 out of 124, Supplementary Table S2).
Figure 2.
A significant enrichment of miR-122 targets by biotin-tagged miR-122. (A) Different seed types of miR-122. (B) The occurrence of miR-122 6-, 7- or 8-nt seed sequence matches in biotin-miR-122 enriched mRNAs (grey columns) and in all human annotated genes (white columns).
The GO analysis revealed that the enriched genes were over represented in gene categories involved in cellular functions associated with the regulation of transcription and RNA splicing, as well as development and cell cycle (Supplementary Table S4 and See Supplementary Table S5 for details).
Interaction between miR-122 and the 3′-UTR of PRKRA mRNA
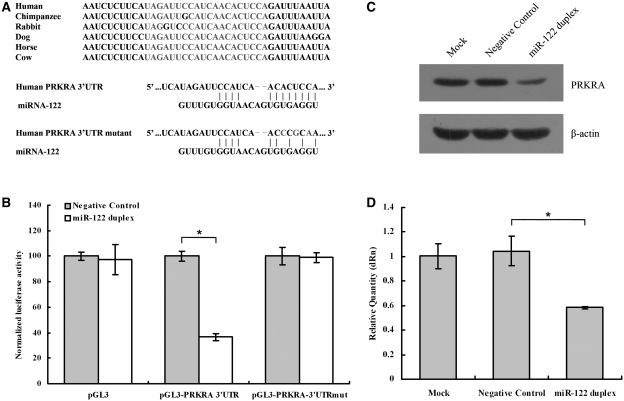
In previously mentioned affinity purification experiment, PRKRA showed a high-level enrichment (∼45-fold) and harbored one putative binding site for miR-122 in its mRNA's 3′-UTR which was conserved across various species (Figure 3A). To investigate the potential miRNA:mRNA interaction, the human PRKRA 3′-UTR was subcloned after the firefly luciferase open reading frame (ORF) and cotransfected into HepG2 cells with the miR-122 duplex, which could mimic miR-122 molecule. miR-122 duplex could lead to a 60% decrease of relative luciferase activity compared with Negative Control transfected cells (Figure 3B). An analogous reporter with point substitutions disrupting the target sites (as illustrated, Figure 3A) was also contransfected with miR-122 duplex. There was no decrease of relative luciferase activity in miR-122 duplex transfected cells compared with Negative Control transfected cells (Figure 3B).
Figure 3.
miR-122 inhibits PRKRA expression. (A upper panel) The target site of miR-122 in PRKRA 3′-UTR is conserved among mammalian species. (A lower panel) Predicted duplex formation between miR-122 and the targeted PRKRA 3′-UTR. The PRKRA 3′-UTR mutant is identical with the wild-type except that its three point substitutions disrupting pairing to miR-122 seed. (B) pGL3-PRKRA-3′-UTR reporter plasmid in which the luciferase-coding sequence had been fused to the 3′-UTR of PRKRA was cotransfected into HepG2 cells with Negative Control (grey columns) or miR-122 duplex (white columns). Luciferase activity was normalized relative to a simultaneously transfected Renilla expression plasmid. 3′-UTR-Mut indicates the introduction of alterations into the seed complementary sites shown in Figure 3A. Results of the mean of quadruplicate assays with standard deviation of the mean are presented. *P < 0.05. (C) Western blots of PRKRA from HepG2 cells. Cells were transfected with Negative Control or miR-122 duplex. Cells were harvested 48 h later, and 30 μg of whole-cell lysate was added into each lane. A β-actin antibody was used in a reprobing as a loading control. (D) Real-time RT–PCR of PRKRA in HepG2 cells transfected with the Negative Control or miR-122 duplex for 48 h. Data were normalized to the level of GAPDH mRNA. Results of the mean of triplicate qPCR assays with standard deviation of the mean are presented. *P < 0.05.
PRKRA is a potential target of miR-122
To learn whether miR-122 can affect endogenous PRKRA, we examined the impact of this miR-122 on PRKRA protein expression in cultured cells. Western blot from protein extracts obtained from the HepG2 cells revealed dramatically reduction in PRKRA protein level after miR-122 duplex transfection (Figure 3C). In contrast, negative control-transfected cells showed no reduction in PRKRA protein level. Because inhibition of expression by miRNA may also be mediated by mRNA degradation (28,29), we examined whether the PRKRA mRNA levels might be affected by miR-122. Figure 3D showed the PRKRA mRNA level was dramatically reduced by miR-122 duplex (42% reduction). Taken together, affinity purification experiment, luciferase data, immunoassay and real-time qRT–PCR assay provided strong evidences that PRKRA is a target of miR-122.
miR-122 facilitates accumulation of newly synthesized miRNA through downregulate PRKRA
PRKRA (PACT) cDNA was cloned by virtue of its interaction with PKR (30). PRKRA is a human cellular protein that heterodimerizes with PKR through its double-stranded RNA-binding domains (dsRBDs) and activates PKR pathway in vitro and in vivo in the absence of dsRNA. In contrast to PRKRA, the human immunodeficiency virus transactivating response RNA-binding protein (TRBP), another dsRBD protein with 42% identity to PRKRA, has an inhibitory effect on PKR (31). The opposite effects of PRKRA and TRBP on PKR activity are mediated by the C-terminal dsRBDs, which are devoid of detectable dsRNA-binding properties (31). PRKRA and TRBP have been demonstrated recently to interact with RNase III Dicer, a key enzyme involving in miRNA maturation, through their C-terminal dsRBDs (32–35). By using siRNA against TRBP, Chendrimada et al. (32) found that TRBP knockdown reduced endogenous miRNA levels in cell culture, whereas Haase et al. (33) saw no such reduction.
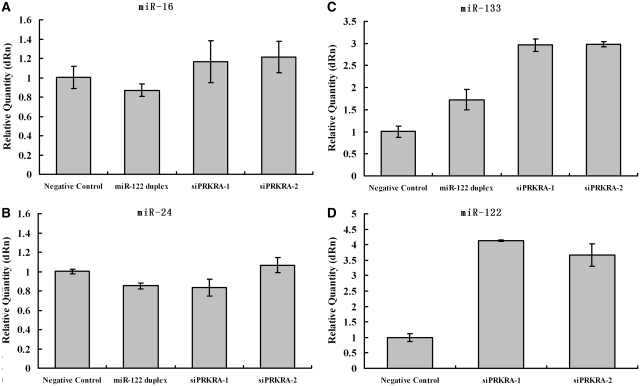
To assess the potential role of PRKRA in miRNA pathway, we transfected HeLa cells with miR-122 duplex or siRNAs against PRKRA. Via miRNA-RT–qPCR, we found the endogenous levels of miR-16 and miR-24 changed only slightly after the depletion of PRKRA (Figure 4A and B). This supports the previous observation that mature miRNA is highly stable in cells and therefore the changes in the steady-state level of mature miRNA may be difficult to detect (36–38). Then we cotransfected miR-133 expressing vector (pcDNA3.0-miR133) with miR-122 duplex or siRNAs against PRKRA into HeLa cells. Results from miRNA-RT–qPCR indicated that miR-122 duplex, siPRKRA-1 and siPRKRA-2 could facilitate accumulation of newly synthesized miR-133 (Figure 4C). We also found that PRKRA siRNAs could also facilitate accumulation of newly synthesized miR-122 when HeLa cells were cotransfected with miR-122 expressing vector (pcDNA3.0-miR122) (Figure 4D). Similar results were observed in HepG2 cell line (data not shown). To monitor PKR activity, we analysis the expression of Tnfaip3 (A20) which could be induced by PKR activation through NF-κB pathway (39,40). Real time qRT–PCR results showed that downregulation of PRKRA by siRNA has no effect on A20 expression (Supplementary Figure S3). Our findings that PRKRA knockdown facilitated accumulation of newly synthesized miRNA support the hypothesis that PRKRA and TRBP have opposite effects on miRNA maturation just like the situation in PKR pathway (Figure 5).
Figure 4.
miR-122 facilitates the accumulation of newly synthesized miRNA through regulating PRKRA. (A and B) miRNA-RT–qPCR detected the expression of miR-16 and miR-24. HeLa cells were transfected with the Negative Control siRNA, miR-122 duplex, siPRKRA-1 or siPRKRA-2 for 48 h. (C) miRNA-RT–qPCR detected the expression of miR-133. HeLa cells were cotransfected with miR-133 expressing vector (pcDNA3.0-miR133) and Negative Control siRNA, miR-122 duplex, siPRKRA-1 or siPRKRA-2 for 48 h. (D) The same experiment as described in (C), except for cotransfecting HeLa cells with miR-122 expressing vector (pcDNA3.0-miR122) and Negative Control siRNA, siPRKRA-1 or siPRKRA-2 for 48 h. Total RNA was poly-A tailed, reverse transcripted and then real-time PCR tested. Data were normalized to the level of U6 RNA. Results of the mean of triplicate quantitative PCR assays with standard deviation of the mean are presented.
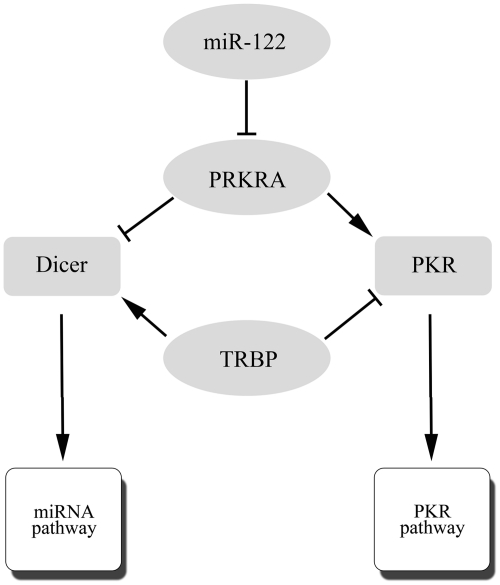
Figure 5.
The regulation of miRNA maturation and PKR pathway by TRBP and PRKRA. PRKRA and TRBP involved in the regulation of PKR pathway and the maturation of miRNAs through interacting with PKR or Dicer, respectively.
DISCUSSION
The present study shows that PRKRA could be enriched by 3′-biotin modified miR-122. It is also shown that PRKRA is negatively regulated by miR-122 via a specific target site within the 3′-UTR. Moreover, we showed that miR-122 facilitates accumulation of newly synthesized miRNA through downregulating PRKRA expression. The identification of miR-122 as an important regulator of miRNA maturation emphasizes an essential role of this liver-specific miRNA.
Sophisticated mechanisms regulating RNA may explain the gap between the great complexity of cellular functions and the limited number of primary transcripts. Regulation by miRNAs underscores this possibility, as each miRNA is believed to bind directly to many mRNAs to regulate their translation or stability and thereby control a wide range of activities. Despite their biological importance, determining the targets of miRNAs is a major challenge. Attention has largely been laid on computational predictions of targets based on the observation that many miRNAs can recognize their targets by binding to motifs in the 3′-UTR sequences complementary to bases 2–8 of the miRNA (the seed region) (41). Whereas these algorithms have been instrumental in many studies of individual miRNA:mRNA interactions, unbiased approaches to study miRNA target recognition are important to discover new features of miRNAs. In this study, we used a direct affinity-based procedure to isolate target mRNAs bound by miR-122. These results will help us to understand the function of this liver-specific miRNA. While preparing this manuscript one group has published a paper demonstrating miR-122 targeting CUTL1 (CUX1) during liver development, which is also enriched by biotin-tagged miR-122 (Supplementary Table S2) (42).
The affinity-based target-identification procedure was performed following Orom et al. (24,25) with some modification. The non-specific binding of RNAs to streptavidin–agarose beads was unavoidable even we washed the beads four times with cell lysis buffer. Therefore, GAPDH with no putative target site of miR-122 was chosen to serve as an internal control. If one gene is the target of miR-122, the output would contain more mRNA/GAPDH than the Input. To validate our results, we also normalize mRNA with β-actin (no putative target site of miR-122 in its 3′-UTR). Results from real-time PCR showed that miR-122's known target gene had enrichment in Ouput when normalized to GAPDH or β-actin (Figure 1 and Supplementary Figure S4).
miRNAs are generated by a two-step processing pathway. Primary miRNAs are processed to pre-miRNAs by Drosha. These pre-miRNAs are cleaved by Dicer to generate mature miRNAs (43–45). In human cells, Drosha exists as part of a protein complex called the Microprocessor complex, which contains the dsRNA-binding protein DGCR8 (also called Pasha). DGCR8 is essential for Drosha activity and capable of binding single-stranded fragments of the pri-miRNA that are required for proper processing (46,47). Dicer also exists as part of a protein complex which contains two dsRNA-binding proteins, TRBP and PRKRA (32–35). It was reported that TRBP knockdown reduced endogenous miRNA levels in cell culture (32). Here, we found that PRKRA knockdown by miR-122 duplex or siRNAs against PRKRA could facilitate accumulation of newly synthesized miRNA. Thus dsRNA-binding protein such as DGCR8, TRBP and PRKRA could affect the miRNAs processing pathway by interacting with Drosha or Dicer.
However, our finding that PRKRA is a negative regulator of miRNA processing is in direct conflict with the finding of Lee et al. (34). They demonstrated that the reduction of PRKRA protein results in reduced miRNA accumulation. The contradictory results probably reflect the different methodologies. Lee et al. depleted PRKRA by RNAi from a HeLa cell line that expresses pri-miR-30a from the tetracycline-inducible promoter. After incubation with siRNA, the cell line was exposed to doxycycline, the derivative of tetracycline, for induction of pri-miR-30a. By using northern blotting on miR-30a, they found that reduction of PRKRA protein levels results in reduced miRNA accumulation. While we co-transfected siRNA and miRNA-expressing vector into HeLa cell line. By using miRNA quantitative-RT–PCR, we found that PRKRA siRNAs could facilitate the accumulation of miRNA. Moreover, we co-transfected PRKRA siRNAs and miR-30a-expressing vector (the specific miRNA investigated by Lee et al.) into HeLa cell line. Forty-eight hours after transfection, we employed miRNA quantitative-RT–PCR to detect miR-30a expression and found that PRKRA siRNAs could also facilitate the accumulation of miR-30a (Supplementary Figure S5).
An interesting paradox exists between TRBP and PRKRA, which shares 42% amino acid sequence identity. TRBP inhibits the dsRNA-activated PKR, whereas PRKRA activates this protein—both using their C-terminal domains for these functions (31). TRBP and PRKRA could interact with Dicer, raising the intriguing possibility that TRBP and PRKRA modulate PKR differentially in response to dsRNAs in the cytoplasm. For instance, it would be disadvantageous to the cell to have miRNA precursors activating PKR, as this may ultimately lead to cell death. Thus, our findings suggest that TRBP and PRKRA might be responsible for keeping the balance between PKR pathway and pre-miRNA processing (Figure 5).
In conclusion, miR-122, a hepato-specific miRNA, inhibits the expression of Dicer-interacting PRKRA gene by binding to the 3′-UTR of PRKRA mRNA and thereby facilitates the accumulation of newly synthesized miRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary material, Supplementary tables, Supplementary figures 1–5.
FUNDING
Chinese State Key Projects for Basic Research (2010CB912801, 2007CB914601); partially by grants from Chinese Key Project for the Infectious Diseases (2008ZX10002-016); and Chinese National Natural Science Foundation (30870529, 30873008). Funding for open access charge: Chinese State Key Projects for Basic Research (2010CB912801).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 3.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl Acad. Sci. USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dresios J, Aschrafi A, Owens GC, Vanderklish W, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl Acad. Sci. USA. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moroy T, Etiemble J, Bougueleret L, Hadchouel M, Tiollais P, Buendia MA. Structure and expression of hcr, a locus rearranged with c-myc in a woodchuck hepatocellular carcinoma. Oncogene. 1989;4:59–65. [PubMed] [Google Scholar]
- 8.Chang J, Provost P, Taylor JM. Resistance of human hepatitis delta virus RNAs to dicer activity. J. Virol. 2003;77:11910–11917. doi: 10.1128/JVI.77.22.11910-11917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 13.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 14.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl Acad. Sci. USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 16.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 18.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 21.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 23.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta V, Huang X, Patel RC. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology. 2003;315:283–291. doi: 10.1016/s0042-6822(03)00589-0. [DOI] [PubMed] [Google Scholar]
- 32.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 37.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 39.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donze O, Deng J, Curran J, Sladek R, Picard D, Sonenberg N. The protein kinase PKR: a molecular clock that sequentially activates survival and death programs. EMBO J. 2004;23:564–571. doi: 10.1038/sj.emboj.7600078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 43.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 44.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 45.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 47.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.