Abstract
The L1 (LINE 1) retrotransposable element encodes two proteins, ORF1p and ORF2p. ORF2p is the L1 replicase, but the role of ORF1p is unknown. Mouse ORF1p, a coiled-coil-mediated trimer of ∼42-kDa monomers, binds nucleic acids and has nucleic acid chaperone activity. We purified human L1 ORF1p expressed in insect cells and made two findings that significantly advance our knowledge of the protein. First, in the absence of nucleic acids, the protein polymerizes under the very conditions (0.05 M NaCl) that are optimal for high (∼1 nM)-affinity nucleic acid binding. The non-coiled-coil C-terminal half mediates formation of the polymer, an active conformer that is instantly resolved to trimers, or multimers thereof, by nucleic acid. Second, the protein has a biphasic effect on mismatched double-stranded DNA, a proxy chaperone substrate. It protects the duplex from dissociation at 37°C before eventually melting it when largely polymeric. Therefore, polymerization of ORF1p seemingly affects its interaction with nucleic acids. Additionally, polymerization of ORF1p at its translation site could explain the heretofore-inexplicable phenomenon of cis preference—the favored retrotransposition of the actively translated L1 transcript, which is essential for L1 survival.
INTRODUCTION
The non-LTR L1 clade of autonomously replicating retrotransposons is ubiquitous in eukaryotic genomes (1,2) and is now the dominant transposable element in most mammalian lineages where L1 activity has generated ∼40% of their genomic DNA (e.g. ref. 3). L1 elements remain active in most modern mammalian species, including humans (4,5). They were sufficiently deleterious in early hominids to have been subject to negative selection (6), and may well still be in modern humans for they are a significant source of genetic diversity, defects and rearrangements (5,7–10), and perhaps a source of neuronal cell mosaicism (11).
Despite their profound effects, little is known about the regulation or biochemistry of L1 replication [most recently reviewed by Martin (12)]. L1 elements contain a 5′ untranslated region (UTR), which has a poorly understood regulatory role, two protein-encoding regions, ORF1 and ORF2, and a 3′UTR of unknown function. The L1 encoded proteins, ORF1p and ORF2p, are essential for retrotransposition in a cell culture-based assay (13).
ORF2p is the L1 replicase (13–15), catalyzing a reaction similar to the template primed reverse transcription (TPRT) reaction first described for the L1-like non-LTR retrotransposon R2 (16–18). This reaction relies on nicked genomic DNA to prime cDNA synthesis of the retrotransposon transcript, and has been partially recapitulated using extracts or ribonucleoprotein (RNP) particles isolated from cell cultures transfected with L1 retrotransposition vectors (19–21).
In contrast, the role of ORF1p in retrotranposition is largely unknown. Human and mouse embryonic cell tumors that synthesize L1 products contain ORF1p-containing RNPs, presumed intermediates of L1 retrotransposition as they also contain L1 transcripts (and other constituents) (22,23). However, ORF1p is homologous to neither retroviral gag proteins nor any other protein (12,24). Thus, the published literature offers few clues to its function.
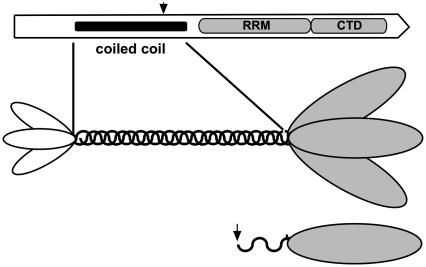
Nonetheless, studies on mouse ORF1p in vitro, (12,24), and more recently on the human protein (25), revealed structural and biochemical properties that are undoubtedly related to its role in vivo. Purified mouse ORF1p expressed in insect cells is a trimer of the ORF1-encoded 42-kDa primary sequence (monomer) (26), which is mediated by a coiled-coil domain in the N-terminal half of the protein (27). Figure 1 shows schematic versions of the ORF1p primary sequence and trimer.
Figure 1.
Schematic representation of ORF1p. The top diagram shows the location of the major domains in the ORF1p of an active L1Pa1 element (the L1.3 member of the Ta-1 subfamily, respective refs 4 and 38). The amino acids corresponding to the predicted coiled-coil domain (41), the RNA recognition motif (RRM, ref. 25), and the C-terminal domain (CTD, ref. 30) are shown in Supplementary Figure S1. The arrow indicates the start of the N-terminal deletion mutant, M128p (see text and Supplementary Data). The middle diagram depicts the trimer modeled on the relative sizes of the N-terminal region, coiled-coil domain, and C-terminal half of the mouse protein as revealed by atomic force microscopy (26). The areas of the ovals corresponding to the N-terminal region and C-terminal half are proportional to their masses. The bottom diagram depicts the M128p protein monomer.
The C-terminal half of the mouse and human proteins contains highly conserved residues, some of which have correlated with the activity of the purified protein or when examined using cell culture- or L1 RNP-based assays (13,20,25,28–30). However, high-affinity nucleic acid binding and nucleic acid chaperone activities in vitro, both of which have been correlated with retrotransposition, are properties of the trimer, not the isolated C-terminal region of the protein, which cannot form trimers (12,25,28,31,32).
Additionally, strong conservation of a coiled-coil domain in ORF1p throughout L1 evolution from fish to mammals (Walser, J.-C. and Furano, A.V., unpublished data, 24,33) suggests that the trimer configuration of the C-terminal halves imbues them with properties essential for retrotransposition. Thus, a coiled-coil domain persists even though it is far more variable than any other region of ORF1p, and is often subject to adaptive evolution and even complete replacement (23,24,33–35).
Nonetheless, trimer formation per se does not ensure retrotransposition. For example, a single amino acid change in the coiled-coil of the mouse protein, which drastically reduces retrotransposition, does not impair trimer formation (32). Furthermore, coiled-coil mutations in human ORF1p that have no discernible effect on the trimer can completely destroy retrotransposition (Callahan, K., Perez-Gonzalez, C. and Furano, A.V., unpublished data).
Although the full-length human protein is also a trimer, essentially all of the structural and biochemical work on this protein has been carried out on the monomeric carboxy-terminal half of the protein (25). Here, we examined the properties of purified full-length human ORF1p trimers expressed in insect cells and made two novel observations that significantly advance our knowledge of this protein:
First, ORF1p trimers readily polymerize when present in molar excess of nucleic acid under the very conditions (0.05 M NaCl) that support high affinity (∼1 nM) nucleic acid binding. Second, ORF1p polymerization seemingly affects the nature of its interaction with nucleic acid. Thus, while the protein protects mismatched duplexes from dissociation (melting), at sufficiently high molar excess and largely polymeric, the protein melts the duplex.
Although earlier studies noted the propensity of ORF1p to aggregate (23,36), our findings show that ORF1p polymers are an active conformer of the protein. Additionally, ORF1p polymerization at its site of translation, where it could preferentially bind its transcript, could account for a heretofore-inexplicable feature of L1 biology, namely cis preference—the strongly favored retrotransposition of the actively translated L1 transcript over non-L1 transcripts, a property critical for the survival of L1 elements (33,37).
MATERIALS AND METHODS
ORF1p expression constructs
The ORF1 sequence was derived from the L1.3 element (38), an active member of the most recently evolved Ta-1 subfamily of the human-specific L1Pa1 family (4). In every case, we inserted the tobacco etch virus protease site (TEV) 5′ to the ORF1 sequence. We could thereby cleave the affinity tag with the TEV protease, leaving just a glycine at the N-terminus of the purified ORF1p. For expression in Escherichia coli: The GST affinity tag was fused to the N-terminus of TEV-ORF1p (GST-TEV-ORF1p) in pGEX-5X (GE Healthcare). We generated an N-terminal deletion mutant of ORF1p by deleting its first 127 residues (Stratagene QuikChange deletion protocol) to generate M128p (arrow in Figure 1; residues 128–338, Supplementary Figure S1 and Supplementary Data). We fused thioredoxin (Trx) and a hexa-histidine (HIS) affinity tag to the N-terminus of TEV-M128p (Trx-HIS-TEV-M128p) in the pET32a vector (Novagen). For expression in insect cells, we fused HIS to the N-terminus of TEV-ORF1p (HIS-TEV-ORF1p) in the pFastBac1 vector (Invitrogen) for baculovirus infection.
Purification of ORF1p and M128p from E. coli
During this purification and the one described in the next section, we identified the ORF1p- or M128p-containing fractions by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Both full-length GST-TEV-ORF1p and Trx-HIS-TEV-M128p were expressed in Rosetta cells (EMD Chemicals) grown at 19°C for 16 h after induction with 2 mM IPTG. Trx-HIS-TEV-M128p was purified as described in the ‘Purification of human ORF1p from sf9 cells’ section.
We purified GST-TEV-ORF1p at 4°C as follows: We resuspended the cell pellet from 1 l of cells in 200 ml GST lysis buffer [10 mM Na phosphate, pH 7.2, 650 mM NaCl, 2.5 mM DTT and 0.5 mM ethylenediaminetetraacetic acid (EDTA)] followed by sonication. We centrifuged the lysate at 24k g for 30 min and incubated the supernatant with 1.5 ml glutathione beads (EMD Chemicals) for 1 h. We washed the slurry in 100 ml GST lysis buffer for 5 min, loaded the beads into a BioRad Glass Econo-column (1.5 × 10 cm) and eluted GST-TEV-ORF1p with 40 ml lysis buffer supplemented with 10 mM glutathione collecting 3 ml fractions for analysis.
We pooled the GST-TEV-ORF1p-containing fractions and incubated them with 650 nM TEV protease during overnight dialysis against 2 l Tris storage buffer (20 mM Tris–HCl, pH 8.0, 0.5 M NaCl, 1 mM EDTA, 10% w/v glycerol, 5 mM DTT). We further purified the protein as described in the ‘Purification of human ORF1p from sf9 cells’ section.
Purification of ORF1p from Sf9 cells
We generated the recombinant baculovirus from HIS-TEV-ORF1p-FastBac1 using the Bac-to-Bac® baculovirus expression system (Invitrogen). Sf9 cells were infected in suspension at 27°C at a multiplicity of infection of three virions/cell, harvested 72 h post infection and stored at −80°C.
We purified HIS-TEV-ORF1p at 4°C as follows: We resuspended the cell pellet from 1 liter of cells in 250 ml 100 mM Tris–Cl, pH 8.0, 1 mM β-mercaptoethanol, 0.5 M KCl, 2% NP-40 and after 30 min on ice sonicated the lysate. We centrifuged the lysate at 24k g for 30 min, incubated the supernatant with 1 ml Ni-NTA HIS bind resin beads (Novagen) for 1 h and then centrifuged the slurry at 3k g for 5 min. We washed the beads twice with 2.5 ml buffer E (20 mM Tris, 0.5 M NaCl, 5 mM DTT) supplemented with 40 mM imidazole, loaded them into a column as described in the previous section and eluted HIS-TEV-ORF1p from the Ni-NTA resin with 10 ml Buffer E that contained 400 mM imidazole collecting 1 ml fractions. We pooled the ORF1p-containing fractions and dialyzed them overnight against 1 liter Tris storage buffer in the presence of 650 nM TEV protease.
We further purified ORF1p (∼5 mL) on a HiLoad 26/60 Superdex-200 size exclusion column (GE Healthcare) that had been equilibrated in Tris storage buffer. We concentrated the protein by applying the pooled fractions to a 1-ml HiLoad heparin column (GE Healthcare) in Tris storage buffer and eluted the protein with 2 M NaCl in Tris storage buffer. We pooled the ORF1p-containing fractions and dialyzed the eluate against Tris storage buffer overnight. We determined the protein concentration by A280 nm in a 1-cm cell using the following calculated extinction coefficients: 1 mg/ml ORF1p =0.635 absorbance units; 1 mg/ml M128p = 0.723 absorbance units. Protein samples were stored in aliquots at −80°C.
Protein cross-linking
We dialyzed ORF1p or M128p against HEPES storage buffer (20 mM HEPES, pH 7.5, 10% w/v glycerol, 0.5 M NaCl); the proteins were stored at −80°C. The protein (added last) was diluted into 10–20-μl reactions at room temperature in oligonucleotide binding buffer (20 mM HEPES, pH 7.5, 10% w/v glycerol), adjusted to the desired concentration of NaCl (0.05–0.5 M) and various oligonucleotides as indicated in the Figures. After various times (immediately to 20–30 min) we added 0.1 vol of 10 or 0.5 mM ethylene glycobis(succinimidylsuccinate) (EGS, Pierce Biotechnology) freshly made in dimethyl sulfoxide (DMSO, Sigma). After 30–60 min at room temperature we quenched the reaction with 1/10th volume of 1 M Tris–Cl, pH 8.0, subjected the samples to SDS–PAGE, and stained the gels with either colloidal Coomassie blue (PageBlue, Fermentas) or silver stain (Pierce).
Oligonucleotides
We radiolabeled oligonucleotides (Table 1 – Midland Certified Reagent Company) with γ-[32P]-ATP (Perkin-Elmer) using T4 polynucleotide phosphorylase (Optikinase, Affymetrix) and removed unincorporated radiolabel by chromatography on a G50 spin column (GE Healthcare). We prepared radioactive duplex DNA by annealing 100 nM [32P]-oligonucleotide with 125 nM of its unlabeled complement in 0.05 M NaCl. We stored oligonucleotides at −20°C.
Table 1.
Oligonucleotide sequences
| N | Name | L | sequence |
|---|---|---|---|
| 1 | d120_c | 120 | AGCGAGTTGATGTTAGACTGTGTACTTTTTGCGAGTTGATGTTAGACTGTGTACTTTTTA AGCGAGTTGATGTTAGACTGTGTACTTTTTGCGAGTTGATGTTAGACTGTGTACTTTTTA |
| 2 | d60_c | 60 | AGCGAGTTGATGTTAGACTGTGTACTTTTTGCGAGTTGATGTTAGACTGTGTACTTTTTA |
| 3 | d29 | 29 | AAAAAGTACACAGTCTAACATCAACTCGC |
| 4 | d29_c | 29 | GCGAGTTGATGTTAGACTGTGTACTTTTT |
| 5 | d29_cmm | 29 | GCGAGTTGACGTCAGACCGTGCACTTTTT |
| 6 | d25_c | 25 | GAGTTGATGTTAGACTGTGTACTTT |
| 7 | d20_c | 20 | GTTGATGTTAGACTGTGTAC |
| 8 | d15_c | 15 | TTGATGTTAGACTGT |
| 9 | d10_c | 10 | TTGATGTTAG |
| 10 | d5_c | 5 | TTGAT |
| 11 | d53_pT-b | 53 | GAGAGATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCAGAGC |
| 12 | d58_pT-b | 58 | GAGAGATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCAGAGC |
| 13 | d60_orf1 | 60 | GATCATGCAGGACAGTCGGATCGCAACCTGATTTACTGTGTCATATAGTACGTGATTCAG |
| 14 | r60_U | 60 | UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUU |
| 15 | r30_3′utr-a | 30 | ACCUGCACAAUGUGCACAUGAAAAAAAAAA |
| 16 | r26_3′utr | 26 | ACCUGCACAAUGUGCACAUGAAAAAA |
| 17 | r22_3′utr | 22 | ACCUGCACAAUGUGCACAUGAA |
Each oligonucleotide has a prefix (d or r, for DNA or RNA) followed by its length and, in some cases, additional identifying information. Oligonucleotides 1–10 are based on the d29 (or its complement d29_c) oligonucleotide, which are identical to the ones called 29linear and c29linear respectively by Martin and Bushman (45); likewise, d29_cmm is identical to the cmm29linear (45), which when paired with d29 results in four mismatched positions (underlined). Oligonucleotide 2, d60_c, is a dimer of d29_c with an additional 5′ and 3′ A. d120_c (oligonucleotide 1) is a dimer of d60_c. Oligonucleotides 6–10 are each subsets of the d29_c sequence. Oligonucleotides 11–14 are arbitrary DNA sequences, and gel shift assays like those shown in Figure 6 showed little difference between these and d60_c in their ability to form shifted products with ORF1p (data not shown). The sequence of oligonucleotides 15–17 corresponds to positions 5980–6000 of the 3′ UTR of L1.3 (38) followed respectively by 10, 6 and 2 A's.
Filter-binding assay
We modified a two-membrane filter-binding assay (39) as follows: We gridded a 47-mm zeta-probe GT (Bio-Rad) membrane into 12 equal squares (using a pre-inked stamp) and after soaking it in binding buffer (see below) layered a presoaked 47-mm diameter 0.45-micron nitrocellulose membrane on it. We placed the filter layer (nitrocellulose filter up) on a fritted glass filter support that had been inserted into a side-arm flask connected to house vacuum. This apparatus was not only far easier to assemble than a traditional dot-blot apparatus but unlike the latter provided a uniform vacuum across the membrane. Binding reactions (prepared on ice) contained 0.2-nM single-stranded or 0.1-nM double-stranded [32P]-oligonucleotides in binding buffer [20 mM Tris, 10% glycerol (w/v), 0.5 mM MgCl2, 0.1 mM DTT and 100 μg/ml bovine serum albumin (BSA)). We added 1/10th volume of the appropriately diluted ORF1p in 0.5 M NaCl (final [NaCl] = 0.05 M) and incubated the reaction at 37°C for 1 h. We applied duplicate 5-μl samples of the binding reaction to the membrane layer under vacuum and washed it with 2 ml binding buffer containing 0.05 M NaCl. We exposed both dried membranes to Fujifilm BAS-MS Phosphor Imager screens and scanned them on a Fuji FLA-5000 series Image Analyzer (Fuji Medical Systems). We used Image Gauge software (version 3.0, Fuji Medical Systems) for quantitation. Fraction bound (FB) = [radioactivity bound to nitrocellulose / (radioactivity bound to nitrocellulose + radioactivity bound to zeta probe)]. We plotted the fraction bound as a function of protein concentration using KaleidaGraph version 4.1. We fit the data points using the following equation for a logistic function: y (fraction bound) = m1 + (m2 − m1)/(1 + (X/m3)^m4), where X = [ORF1p] added; m1 = [ORF1p] at maximal ligand bound; m2 = fraction ligand bound at 0 [ORF1p], m3 = [ORF1p] where half of the ligand is bound, i.e. [ORF1p0.5FB]; m4 = slope of the binding curve plotted as a function of the natural log of [ORF1p] (40).
Electrophoretic mobility shift competition assay
We added radioactive nucleic acids to gel shift buffer (20 mM HEPES, 0.5 mM MgCl2, 200 μg/ml BSA, 20 mM DTT) and added ORF1p to dilution buffer (20 mM HEPES, 0.5 mM MgCl2, 0.5 M NaCl) on ice. After equilibrating the [32P]-nucleic acid samples for 1 min at 30°C, we added ORF1p such that the final concentration of NaCl in the binding reaction was 0.05 M and then incubated the reactions for 20 min at 30°C. For competition experiments we added non-radioactive nucleic acids either with, or 10 min after, addition of the protein. We obtained the same results either way (data not shown). After adding loading buffer (15% glycerol, 0.05 M NaCl and bromophenol blue dye) we electrophoresed the samples for 65 min on 6% polyacrylamide gels in 20 mM HEPES, 10 mM sodium acetate buffer at 200 V at 4°C.
Melting assay
We mixed radioactive duplex (1 nM) in binding buffer (see ‘Filter-binding assay’ section) with ORF1p to a final concentration of 0.05 M NaCl on ice. After 5 min at 37°C we added 10 μl of the reaction to 10 μl of ice-cold stop mix (0.2% SDS, 3% glycerol, 400 ng/μl tRNA,and bromophenol blue) and resolved duplex and single-stranded oligonucleotides by electrophoresis for 1.5 h at 4°C as described for the gel shift experiments. We determined the radioactivity of the dried gels using a Phosphor Imager as described in the ‘Filter-binding assay’ section. Fraction single-stranded DNA = [radioactive single-stranded DNA / (radioactive single-stranded DNA + radioactive double stranded DNA)] − (fraction single-stranded DNA at 0 protein). We plotted the fraction of single-stranded DNA as a function of protein concentration.
Annealing assay
Unless, stated otherwise, we mixed 2 nM of d29 and 1.8 nM [32P]-d29_c oligonucleotides (Table 1) in binding buffer with ORF1p at a final concentration of 0.05 M NaCl, and after 5 min at 37°C treated the reactions as described in the ‘Melting assay’ section. Fraction double-stranded DNA = [radioactive double-stranded DNA/(radioactive double-stranded DNA + radioactive single-stranded DNA)]. We plotted the fraction of double-stranded DNA as a function of protein concentration without subtracting the value for 0 protein.
RESULTS
ORF1p expression in E. coli and baculovirus-infected insect cells
We found that expression of human ORF1p in E. coli produces trimers consisting of both full-length and N-terminal truncated monomers. The truncated monomers result from initiation of translation at conserved methionines near the C-terminus of the coiled-coil domain (Supplementary Figure S1 and Supplementary Data).
In contrast, expression in baculovirus-infected sf9 insect cells produced trimers that contain only full-length human ORF1p monomers (see Figure 2A, lanes 14 and 15 and Supplementary Figure S2 and Supplementary Data). Sedimentation equilibrium at 4.0°C and sedimentation velocity at 20.0°C showed that ORF1p sediments as a single species with a molecular mass of 122 kDa in 0.5 M NaCl (Supplementary Figure S2 and Supplementary Data), which corresponds to a trimer of the predicted molecular weight of the primary gene product (monomer).
Figure 2.
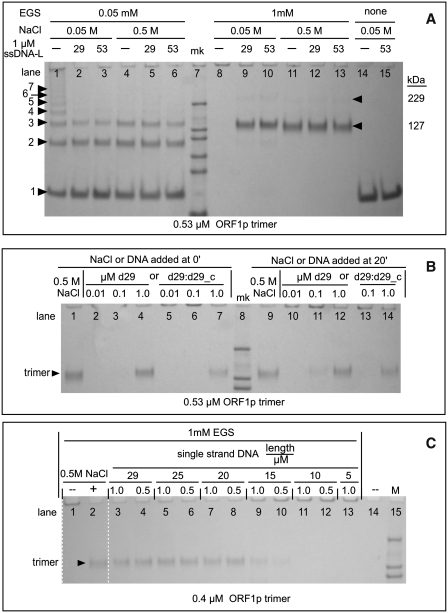
ORF1p polymers—effect of oligonucleotides and NaCl. Cross-linking with EGS was carried out as described in the ‘Materials and Methods’ section. (A) Cross-linked ORF1p products generated by 0.05 mM and 1.0 mM EGS at 0.05 and 0.5 M NaCl in the absence and presence of a molar excess of oligonucleotides. The 29-mer single-stranded DNA is d29_c; the 53-mer is d53_pT-b (Table 1). The numbers to the left of the arrowheads at lane 1 indicate the estimated number of monomers in each species. The estimated kDa's for bands 1–7 are respectively: 41, 85, 127, 163, 216, 229 and 298. The kDa of the marker bands (mk) are: 200, 116, 97, 66, 55, and 37. Estimated kDa >200 are extrapolated values and therefore only approximations. The arrowheads to the right of lane 13 indicate the position of the trimer and a putative dimer of the trimer (trimer2). (B) Effect of time of addition of oligonucleotide or 0.5 M NaCl on cross-linked products generated by 1 mM EGS. The reactions with oligonucleotides were in 0.05 M NaCl. The single-stranded DNA is d29; the double-stranded DNA is a duplex of d29 and d29_c (Table 1). The marker bands are 200, 116 and 97 kDa. One millimolar EGS was added immediately after NaCl or oligonucleotides were added to the reactions electrophoresed in lanes 9–14. (C) Effect of oligonucleotide length on ORF1p polymerization. Cross-linking was with 1 mM EGS. The oligonucleotides used in these experiments are all of the dN_c set shown in Table 1 where N is the length (L) of the oligonucleotide. Each oligonucleotide was tested at 1 and 0.5 µM, as indicated in the major header (length/µM) for lanes 3–13 in reactions that contained 0.05 M NaCl. The marker bands are 200, 116 and 97 kDa.
We constructed an N-terminal deletion mutant of ORF1p that retains 2.5 heptads of the predicted 13 heptads (41) of the coiled-coil (M128p, residues 128–338, Figure 1; Supplementary Figure S1 and Supplementary Data). Sedimentation velocity and sedimentation equilibrium analysis of this protein shows that M128p forms a strongly temperature-dependent trimer (Supplementary Figure S3). These results showed that the Kd for the monomer–trimer equilibrium is ∼3.4 μM at 4°C but ∼35 μM at 20°C (Supplementary Data). Thus, M128p would be largely monomeric in the experiments reported here, which were carried out at >20°C.
ORF1p forms polymers
While purifying ORF1p, we found that although ORF1p trimers were soluble in 0.5 M NaCl they immediately aggregated into a sedimentable form (∼17k g) at <0.1 M NaCl in the absence of nucleic acid (data not shown). However, preliminary experiments also showed that ORF1p bound nucleic acids maximally at 0.05 M NaCl, and not at all at 0.5 M NaCl (data not shown).
Therefore, before determining the parameters of nucleic acid interaction with ORF1p in 0.05 M NaCl, we examined the nature of the ORF1p aggregate and its relationship to the soluble form by reacting the protein under various conditions with the bifunctional chemical cross-linking reagent, ethylene glycobis(succinimidylsuccinate) (EGS). Preliminary experiments have shown that EGS was the most efficient of various bifunctional reagents in producing completely cross-linked ORF1p trimers (data not shown).
Figure 2A (lanes 14 and 15) shows that ORF1p that had been incubated in 0.05 M NaCl, in the absence or presence of nucleic acid, migrated as the expected ∼40-kDa monomer after being denatured and subject to PAGE under denaturing conditions.
However, if the protein in 0.5 M NaCl was first cross-linked with 1 mM EGS and then subjected to denaturing PAGE almost all of the protein was recovered in a band that migrated with the molecular weight of the trimer (lane 11). The results in lane 4 showed that this band was indeed a trimer. Here, partial cross-linking of ORF1p in 0.5 M NaCl with 0.05 mM EGS, which would reveal the constituent components of a trimer, did generate the expected mixture of monomers, dimers and trimers. Thus, in 0.5 M NaCl, ORF1p is largely a trimer.
In contrast, carrying out this pair of cross-linking reactions in 0.05-M NaCl produced dramatically different results. Incomplete cross-linking of ORF1p (lane 1) generated an ascending series of discrete multiples of the monomer that extended to the top of the gel. This result would be expected if the ORF1p trimers had polymerized into large aggregates. The results of lane 8 indicate that these polymers must be quite large because, after complete cross-linking, no protein was recovered on the gel.
To determine if nucleic acids affected the polymerization of ORF1p trimers, we compared the effect of a molar excess of nucleic acids on the cross-linked products obtained after partial or complete cross-linking. Lanes 2 and 3, and 9 and 10, of Figure 2A show that ORF1p is largely a trimer in 0.05 M NaCl in the presence of an excess of either a 29-mer or a 53-mer (d29_c, d53_pT-b respectively, Table 1). As ORF1p does not bind nucleic acids in 0.5 M NaCl we would expect the results in lanes 5 and 6 to not materially differ from those in lane 4, or those in lanes 12 and 13, to differ from lane 11.
Figure 2B shows the important result that ORF1p polymers are not denatured or inactive, but can bind nucleic acids. First, lanes 1–7 show the results of reactions carried out similarly to those shown in Figure 2A. Herein ORF1p was incubated with either 0.5 M NaCl (lane 1) or varying concentrations of single-stranded (lanes 2–4) or duplex (lanes 5–7) DNA for 20 min and then cross-linked with 1 mM EGS. As expected, ORF1p trimers were recovered from both the 0.5 M NaCl reaction, and those oligonucleotide-containing reactions that contained a sufficient concentration of DNA.
However, lanes 9–14 show that essentially the same results were obtained if 0.5 M NaCl or the oligonucleotides were added to ORF1p after it had been incubated for 20 min in just 0.05 M NaCl. By this time, all of the ORF1p would have polymerized (e.g. Figure 2A, lane 8). The addition of either 0.5 M NaCl (lane 9) or DNA (lanes 10–14) yields the same amounts of trimer as the reactions displayed in lanes 1–7. Thus, these additions depolymerized the pre-formed polymer, which occurred very rapidly for we added EGS immediately after adding the NaCl or oligonucleotides. Other experiments (data not shown) had shown that cross-linking is >90% complete within 5 min.
Although a perfectly matched duplex DNA both prevents polymerization (lane 7) and depolymerizes the polymer (lane 14), it is not as effective as the single-stranded nucleic acid. In contrast, a mismatched duplex is as efficient as the single-stranded oligonucleotide (data not shown). The mismatched duplex (a hybrid of d29 and d29_cmm, Table 1) was the same one used for the experiments shown in Figure 7B.
Figure 7.
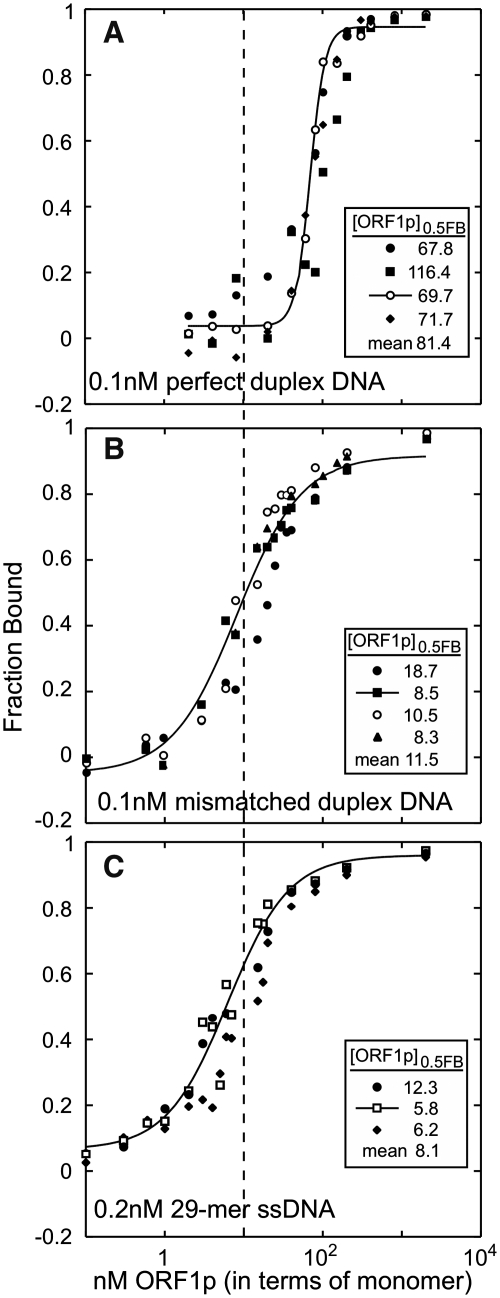
Comparison of single-stranded and double-stranded DNA binding by ORF1p. Each panel shows independent filter binding assays carried out at 37°C as described in the ‘Materials and Methods’ section. The insert shows the [ORF1p]0.5FB for each assay along with the mean. (A) The perfect duplex, d29:[32P]-d29_c (Table 1). (B) The mismatched duplex, d29:[32P]-d29_cmm. (C) The single-stranded 29-mer, d29_c, the same data presented in Figure 5E. The vertical dashed line marks the [ORF1p]0.5FB for ORF1p binding to the mismatched duplex (B).
The foregoing experiments showed that interaction of ORF1p with a molar excess of single-stranded DNA (with respect to the trimer content of ORF1p polymers) generated mainly cross-linked trimers. To determine whether nucleic acid binding and polymer formation were mutually exclusive properties of ORF1p, we determined if cross-linkable multimers of ORF1p trimers could assemble on sub-stoichiometric amounts of single-stranded DNA.
To examine this issue, we first determined the minimal length of DNA that prevented polymerization of ORF1p when the oligonucleotide was present in molar excess of, or stoichiometric with, the protein. Figure 2C shows that this is a 20-mer as judged by the recovery of cross-linked protein relative to that cross-linked in 0.5 M NaCl. We then determined if sub-stoichiometric amounts of oligonucleotides that were integer multiples of the 20-mer could prevent the polymerization of ORF1p trimers into aggregates too large to be recovered by gel electrophoresis.
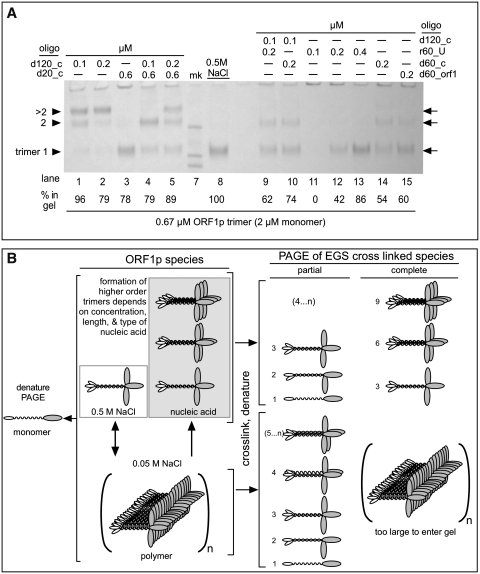
Figure 3A shows that this can be the case. We obtained similar recoveries of cross-linked protein by gel electrophoresis from reactions wherein 0.67 µM ORF1p been incubated with 0.1 μM 120-mer (lane 1), 0.6 μM 20-mer (lane 3) or 0.5 M NaCl (lane 8). Additionally, higher order cross-linked multimers of ORF1p trimers prevail on the 120-mer (lanes 1 and 2), even in the presence of a molar excess of the 20-mer (lanes 4 and 5). These multimers correspond to dimers of trimers (trimer2) and larger trimer complexes (>trimer2, which we presume is a trimer of the ORF1p trimer; see legend to Figure 3A and B).
Figure 3.
ORF1p polymers—effect of oligonucleotide length and concentration. (A) Cross-linking with 1 mM EGS was carried out as described in the ‘Materials and Methods’ section and all of the oligonucleotide-containing reactions were at 0.05 M NaCl. The arrows indicate the position of the trimer, and higher multiples thereof, i.e. trimer2, >timer2, the latter of which we presume is a trimer of the ORF1p trimer. While the mobility of the trimer band corresponds to its expected molecular (127 kDa), the higher trimer multiples correspond to molecular weights of 223 and 294 kDa, respectively, less than expected and likely the result of extrapolation error (see legend to Figure 3A). The marker bands are 200, 116 and 97 kDa. We determined the relative recovery of protein in each lane by densitometry using ImageJ (50) on a tiff file of the gel image captured by a Qimaging® Micropublisher 5.0 RTV camera. The ‘% in gel’ indicates the percent recovery of total protein in the various bands normalized to the amount recovered as trimer cross linked in 0.5 M NaCl, which we set to 100%. As noted in the text, the distribution of total protein among the various bands depends on the length and concentration of the oligonucleotide. (B) Schematic representation of possible ORF1p species and their cross-linked products generated in the experiments shown in Figures 2 and 3A. The numbers to the left of the cartoons of the cross-linked species indicate their monomer content.
Thus, binding to nucleic acid does not preclude the formation of cross-linkable multimers by the ORF1p trimers. Also, when maximally packed on the 120-mer, each trimer occupies ∼40 nt. Binding of ORF1p to nucleic acids that permit trimer–trimer interactions is apparently favored over those that do not (lanes 3–5). These results suggest that trimer–trimer contacts enhance the stability of nucleic acid binding. However, the recovery of ORF1p polymers on sub-stoichiometric amounts of DNA can be variable (see Supplementary Figure S4 and Supplementary Data), which suggests that the geometry that favors maximal packing is not always attained.
Even though the foregoing results suggest that a 60-mer DNA would only have ‘room’ for a single trimer, Figure 3A (lanes 14 and 15) shows that higher order cross-linkable trimer complexes can also assemble on 60-mer DNAs. Thus, the propensity for ORF1p to polymerize can force tighter packing of ORF1p trimers on the 60-mers than observed on the 120-mer DNA. Our finding that this phenomenon occurred in a similar fashion on distinct 60-mer DNAs (lanes 14 and 15, see Table 1) indicates that it is not sequence specific.
Fewer and smaller higher order trimer complexes assemble on 60-mer than 120-mer DNA. This would explain why less ORF1p was recovered on the gels using sub-stoichiometric amount of the 60-mer than the 120-mer DNA (cf. protein recovery on lanes 14 and 15 with lanes 1 and 2). The addition of 0.1 µM 120-mer to 0.2 µM 60-mer (d60_c) DNA (lane 10) increased the overall recovery of ORF1p to about that found with the 120-mer supplemented with the 20-mer (lanes 4). However, as Figure 3A reveals (and densitometry confirmed; data not shown), the relative abundance of trimer and trimer2 in lanes 4 and 10 are reversed. Thus the 60-mer more efficiently competes for trimer than the 20-mer, perhaps because it can make more contacts with the protein than the 20-mer.
In contrast to the 60-mer DNA, few if any higher order ORF1p trimer complexes assembled on sub-stoichiometric concentrations of the 60-mer RNA (Figure 3A, lanes 11–13). This difference could result from the RNA assuming a more compact structure than DNA upon binding to the protein. In contrast to DNA, RNAs can form highly folded structures via non-canonical base pairing, including U:U pairs, often abetted by protein contacts (e.g. ref. 42). Whatever the explanation, our results with the RNA agrees with the findings that one mouse ORF1p trimer was bound per 50 nt of RNA (29). However, our results with the DNA oligonucleotides show that as few as 20 nt are sufficient for productive nucleic acid–ORF1p interactions.
Figure 3B shows a schematic representation of ORF1p trimers and their cross-linked products consistent with the gel patterns in Figures 2 and 3. As the trimer is very stable (Supplementary Data; Furano,A.V., unpublished data and ref. 26), it is unlikely that the coiled-coil domain dissociates and reforms as four-stranded or higher order coiled-coils.
However, sequences in the C-terminal half of the protein could mediate polymerization as it contains an RNA recognition motif (RRM, Figure 1, Supplementary Figure S1; and ref. 25). Although the ORF1p RRM is non-canonical (25), some RRMs mediate protein–protein interactions in addition to nucleic acid–protein interactions (43,44). Additionally, studies on fusion proteins of both the human and mouse ORF1p synthesized in E. coli indicated that this region of the protein could form aggregates (23,36).
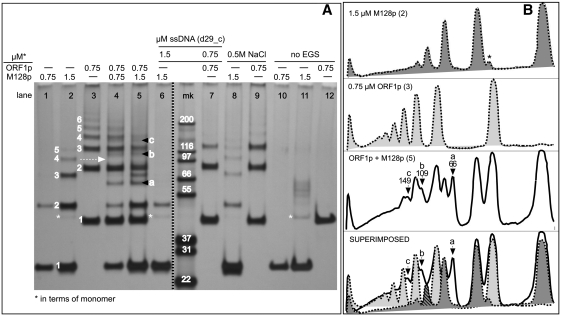
Therefore, we determined if the M128p deletion mutant, which contains the entire C-terminal half of ORF1p (Figures 1 and Supplementary Figure S1) and is largely a monomer at >20°C (see previous section) could form polymers. Figure 4 (lanes 1 and 2) shows that incomplete cross-linking generates protein bands that migrate as approximate integer multiples of the 24 kDa M128p monomers.
Figure 4.
Polymer formation by the M128p monomer. (A) Effect of oligonucleotide (in 0.05 M NaCl) or of 0.5 M NaCl on cross-linked products generated by reaction with 0.05 mM EGS with M128p (lanes 1 and 2), ORF1p (lane 3), or mixtures thereof (lanes 4 and 5). The protein was visualized using silver stain. The numbers indicate the estimated kDa of the indicated band as integer multiples of the kDa of the respective monomer band (#1) for M128p (between lanes 1 and 2) and ORF1p, (between lanes 2 and 3). For M128p, the respective values for bands 1–5 are: 27, 48, 75, 101 and 123 kDa. For ORF1p, the respective values bands for 1–6 are: 40, 86, 123, 155, 186, and 221 kDa. The dashed arrow indicates the band (#4) in the M128p pattern (lane 2), which is missing in the mixed cross-linking experiment displayed in lane 5 [cross-hatched in the superimposed traces shown at the bottom of (B)]. The letters, a, b and c indicate the novel bands that appear in the mixed cross-linking experiment, bottom two traces of panel B. Band #1 in lane 8 is a doublet (more obvious on the original gel) with an additional faster migrating band which likely represents internal cross-links of the 24-kDa monomer. An extra marker lane was removed from the gel image between lanes 6 and mk indicated by the broken dark line. The minor band at about 40 kDa in lane 11 (asterisks) is undigested Trx-HIS-TEV-M128p (see ‘Materials and Methods’ section). The other bands are unknown contaminants. (B) ImageJ densitometry traces of the indicated lanes. The numbers under letters a, b and c are the estimated kDa of the indicated bands. Note that the signal produced by silver staining, though quite sensitive and reproducible, not only is notoriously non-linear with respect to the amount of protein but also has a limited dynamic range. For example, the relative intensity of the bands in lanes 1 and 2 do not reflect the 2-fold difference in the amount of protein loaded in these lanes. Additionally, the expected increase in monomer intensity in instances where it would be expected because of a decrease in the amounts of the higher molecular weight multimers (e.g. lanes 6 versus 2, lanes 9 versus 3) is not obvious. However, the near-perfect superimposition of the density traces of the major protein species in lanes 2, 3 and 5 (B), indicates that little of these species were consumed by the formation of hybrid M128p / ORF1p products.
The polymerization is almost completely prevented by oligonucleotide (cf. lanes 2 and 6), but markedly less so by 0.5 M NaCl (cf. lanes 2 and 8). Note the relatively equal amounts of band 2 in lanes 6 and 8, and the absence of higher order bands in lane 6 (+ nucleic acid) but their presence in lane 8 (+0.5 M NaCl; also see legend to Figure 4). In contrast, 0.5 M NaCl almost completely inhibits polymerization of the ORF1p trimer (cf. lanes 3 and 9). Thus, subjecting the N-terminal half of the protein to high ionic strength inhibits cross-linkable interactions between the C-terminal regions of different trimers.
Although ORF1p trimers readily polymerize with each other (Figure 4, lane 3, also Figure 2A, lane 1) they co-polymerize poorly with the free M128p monomers. The banding pattern of cross-linked mixtures of M128p and ORF1p (lane 5, panels A and B) does reveal some novel bands (a, b and c). Their sizes are consistent with that of cross-linked hybrid products between a 24-kDa M128p monomer and respectively a monomer (40 kDa), dimer, and trimer (expected partially cross-linked products of ORF1p polymers, Figure 2A).
However, as the bottom trace of panel B illustrates, hybrid products account for only a minority of the cross-linked species generated when mixtures of the two proteins are cross-linked (lane 5). The relative concentrations of the M128p and ORF1p monomers and their major cross-linked products (panel B, ORF1p + M128p) are essentially the same as when the proteins were cross-linked separately. This is indicated by the nearly perfect superimposition of the densitometric traces of lane 2 (M128p alone) and lane 3 (ORF1p alone) on the densitometric trace of the mixture (lane 5, bottom panel, Figure 4B).
Binding of single-stranded nucleic acids by ORF1p
Work on mouse ORF1p showed that this protein exhibits both nucleic acid binding and nucleic acid chaperone activities. However, the activity of the full-length human protein had not been examined in this regard. Therefore, we felt it important to examine these properties of human ORF1p, particularly in the context of ORF1p polymer formation.
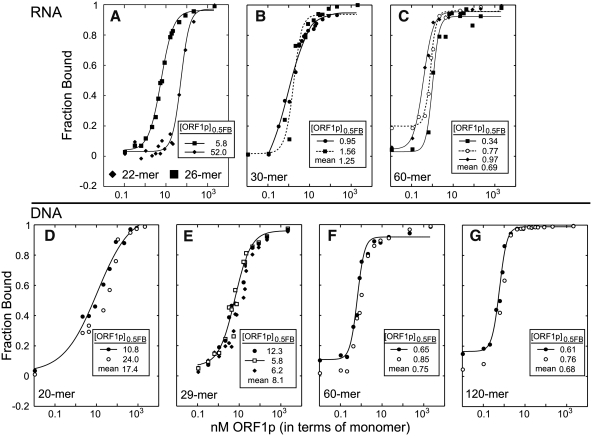
Figure 5 shows the results of independent filter-binding assays of various nucleic acids to a molar excess of ORF1p in 0.05 M NaCl. As the foregoing experiments showed, under these conditions the protein would be polymeric. Thus, the concentration of the species that is binding the nucleic acid is not known–it could be free trimer (or short oligomers thereof) in equilibrium with polymer, or fully polymer-bound trimers. Accordingly, we cannot determine Kds from these data and instead compare the relative affinities in terms of the concentration of total ORF1p that binds half the total nucleic acid ([ORF1p]0.5FB).
Figure 5.
Binding of single-stranded nucleic acids by ORF1p. Each panel shows the results of independent filter binding assays carried out for 1 h at 37°C as described in the ‘Materials and Methods’ section. The insert on each plot shows the [ORF1p]0.5FB (the concentration of protein at which half of the nucleic acid is bound) for each reaction along with the mean. The sequences of the oligonucleotides are given in Table 1. (A) 22-mer and 26-mer RNAs are, respectively, r22_3′ UTR and r26_3′ UTR. The 30-mer and 60-mer RNAs are respectively (B) r30_3′ UTR-a, and (C) r60_U. The single-stranded DNAs are respectively (D–G) d20_c, d29_c, d60_c and d120_c.
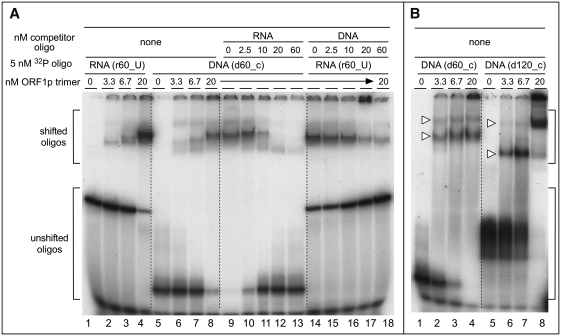
The affinities of ORF1p for RNA and DNA are length dependent, but the length that produces maximal affinity (∼1 nM) differs: a 30-mer for RNA but a 60-mer for DNA. In addition, Figure 3A had shown that higher order trimer complexes assemble on 60-mer DNA but not on 60-mer RNA. Although differences between the extent of compaction of the protein-bound RNA and DNA could explain these differences, they might also reflect distinct binding sites for the two nucleic acids. We examined this possibility by performing ORF1p-mobility shifts of [32P]-RNA or [32P]-DNA in the absence and presence of respectively, non-radioactive DNA or RNA.
Lanes 1–4 and 5–8 of Figure 6A, respectively, show the binding of [32P]-RNA and [32P]-DNA in the absence of non-radioactive competitor. Lanes 9–13 and 14–18 show that the non-radioactive competitors displaced their respective counter parts. Mixing the non-radioactive competitors and radioactive nucleic acids prior to adding ORF1p produced the same results, indicating rapid equilibrium between all of the reactants (data not shown). Thus, the binding sites for RNA and DNA either are the same or overlap.
Figure 6.
Electrophoresis of ORF1p–nucleic acid complexes. We carried out these electromobility shift assays of RNA or DNA by ORF1p as described in the ‘Materials and Methods’ section. The sequences of oligonucleotides are given in Table 1. Brackets indicate the shifted and un-shifted oligonucleotides. (A) ORF1p electromobility shifts in the absence (lanes 1–8) and presence of non-radiolabeled competitor (lanes 9–18). (B) Comparison of ORF1p electromobility shifts with 60-mer (d60_c, lanes 1–4) and 120-mer (d120_c, lanes 5–8) oligonucleotides. The white triangles indicate the slower and faster migrating bands mentioned in the text.
Figure 6B shows the effect of different molar ratios of ORF1p/DNA on mobility shifts of 60-mer and 120-mer DNA. In both cases, two shifted bands were seen (white arrowheads). A 4-fold molar excess of ORF1p hardly changed the relative amounts of the shifted bands with the 60-mer. In contrast, excess ORF1p dramatically increased the amount of the slower migrating band with the 120-mer (Figure 6B, lanes 7 and 8). Thus, the 120-mer can accommodate more of the higher order trimer complexes than the 60-mer. These results are consistent with those in Figure 3A, even though the results shown in Figure 6B were produced in the absence of cross-linker and at ∼40-fold lower concentrations of nucleic acid and protein.
Although the M128p monomer contains all of the residues implicated in nucleic acid binding (13,20,25,28–30), it does not bind nucleic acids with the same affinity as the ORF1p trimer. Supplementary Figure S5 (Supplementary Data) shows that the monomer bound 30-mer RNA with ∼50-fold lower affinity than the trimer and either inconsistently bound DNA or produced complexes that were not stable enough to survive either the filter-binding or gel shift assays (Supplementary Figure S5A and S5B, respectively). Nonetheless, as the next section shows, the M128p monomer and ORF1p were equally effective in the reactions required of a nucleic acid chaperone: annealing and melting nucleic acids.
Biphasic effect of ORF1p on mismatched duplex DNA
Mismatched double-stranded DNA is a proxy substrate for ORF1p chaperone activity, an activity likely required for one or more steps of the retrotransposition process (12,45). Figure 7A shows that while ORF1p has an ∼8-fold lower affinity for a perfectly matched duplex than a mismatched duplex (Figure 7B), it binds the latter with about same affinity as single-stranded DNA (cf. Figure 7B and C). These results are quite reproducible—each assay in Figure 7 represents an independent binding reaction.
Mouse ORF1p can lower the melting temperature of mismatched duplexes (45). Therefore, we initially thought that the similar affinities of the human protein for single-stranded and mismatched duplex DNA reflected dissociation of the duplex and subsequent binding to the resultant single strands. To examine this possibility we determined the fate of the mismatched double-stranded DNA for each of the binding reactions shown in Figure 7B.
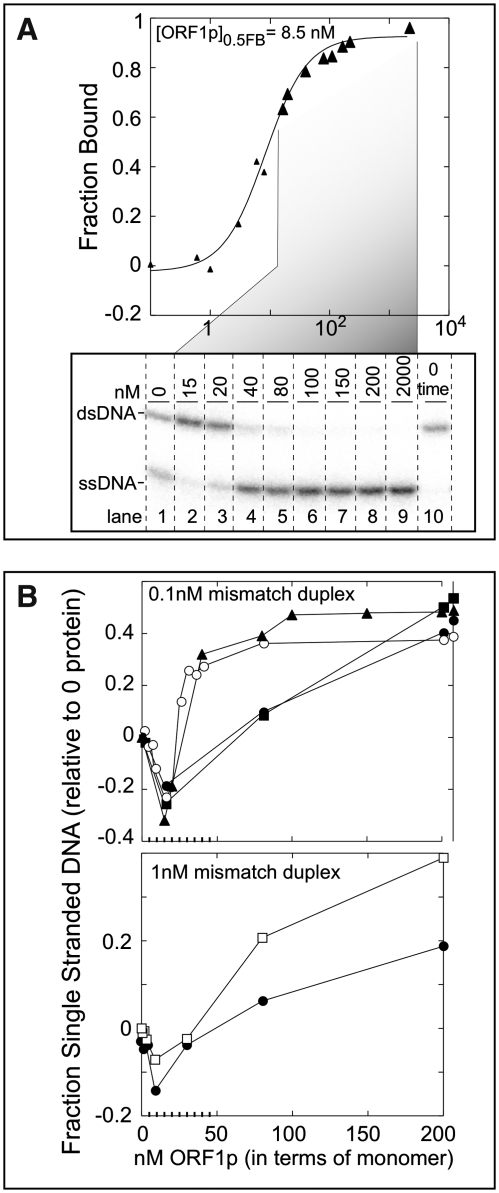
Figure 8A illustrates one such determination. Samples at different protein concentrations (enlarged triangles) from a binding assay (Figure 7B, triangles) were denatured in SDS and held on ice until electrophoresis on non-denaturing polyacrylamide gels at 4°C. The lower part of Figure 8A shows the nature of the mismatched duplex at the selected points. Lane 10 shows that all of the nucleic acid is double-stranded at the outset, and lane 1 shows the extent of melting without protein after 1 h at 37°C.
Figure 8.
Fate of the mismatched duplex. (A) Binding curve of ORF1p with the mismatched duplex, d29:[32P]-d29_cmm, from the experiment shown in Figure 7B (plot with filled triangles), which was also plotted here as filled triangles. Large triangles indicate the samples that were examined by PAGE in the lower part of the panel. The image of the dried gel shows the [32P]-d29_cmm products: double-stranded (ds) and single-stranded (ss) DNA, at the indicated ORF1p concentrations. Lane 10 shows the sample at 0 time in the absence of protein. (B) (Upper plot) The fraction single-stranded DNA was plotted after correcting for the amount of melting that occurred without protein after incubation for 1 h at 37°C. This plot shows the results from the four independent binding experiments shown in Figure 7B using the same symbols in both cases. As shown in lanes 6–9 in the lower part of (A), all of the duplex eventually melted. However, the fraction of single-stranded DNA is <1 due to subtraction of the fraction single-stranded DNA that occurred in the absence of protein. (Lower plot) This panel shows the results from two independent binding assays using 1 nM of the mismatched duplex, d29:[32P]-d29_cmm. These reactions were incubated at 37°C for just 5 instead of 60 min for the other binding assays.
In contrast to our expectations, lanes 2 and 3 show there is less single-stranded DNA in the presence of the protein than in its absence. Thus, rather than melting the mismatched duplex, the protein protected it from dissociating. Whatever the state of the protein-bound duplex, we presume that the two strands are ‘sensed’ as single strands, yet are recovered as a duplex when the protein is denatured. For want of a better term, the two strands are ‘caged’. As the protein concentration increases, the duplex is recovered as separate strands upon denaturation of the protein. At these molar ratios of protein to nucleic acid, ORF1p would be largely polymeric.
The densitometric analysis of the fraction single-stranded DNA from this experiment is plotted in the upper panel of Figure 8B (triangles) after correcting for the amount dissociated without protein. Doing so produced negative values at concentrations of the protein that caged the duplex. However, we could thereby readily compare the results of each of the other binding reactions shown in Figure 7B which we also present in the upper panel of Figure 8B. In some cases, we determined the fate of the mismatched duplex over the entire range of protein concentrations (e.g. empty circles). We note that the ‘window’ in the binding curve at which caging occurs is rather narrow and can be missed unless sufficient samples are examined.
Caging is not due to the 50-fold molar ratio of ORF1p to the mismatch duplex shown in the upper part of panel B, for it also occurred using 1 nM mismatched duplex and near stoichiometric amounts of ORF1p (lower half of Figure 8B). However, under these conditions too, dissociation of the duplex only occurred at high molar excesses of protein to nucleic acids. That caging is seemingly independent of ORF1p concentration supports the idea that it may be an intra-trimer phenomenon.
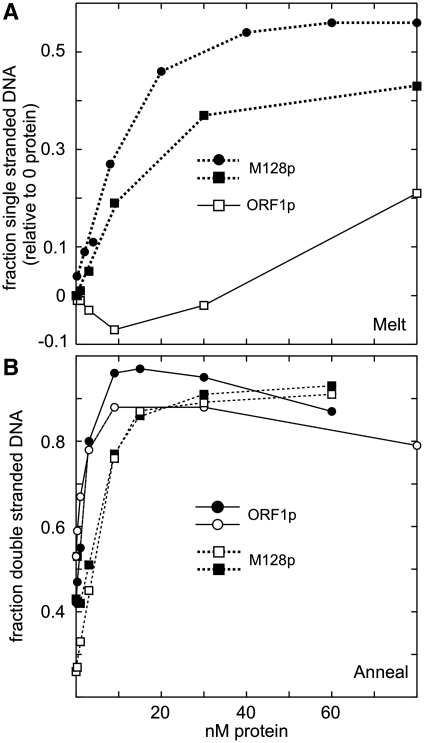
This contention is consistent with the dramatic difference between the melting of the mismatched duplex by ORF1p and the M128p monomer (Figure 9A). Despite the relatively low affinity of the M128p monomer for the mismatched duplex (Supplementary Figure S5 and Supplementary Data), it readily melts it, but does not cage it. (Neither protein melted the perfect duplex; data not shown.) Additionally, despite its relatively low affinity for single-stranded DNA (Supplementary Figure S5 and Supplementary Data), M128p is just as effective as ORF1p in promoting the annealing of DNA (Figure 9B). Therefore, the M128p monomer exhibits both the annealing and melting activities of a nucleic acid chaperone.
Figure 9.
DNA melting and annealing by ORF1p and M128p. These experiments were carried out as described in the ‘Materials and Methods’ section. The indicated amounts of protein (in terms of monomer) were incubated with the appropriate substrates for 5 min at 37°C, whereupon the fractions of single-stranded or double-stranded DNA were determined by gel electrophoresis as described for the experiments in Figure 8. (A) The ORF1p data are taken from the lower plot of Figure 8B (open squares in both cases). The mismatched duplex DNA (1 nM) used for M128p was (d29:[32P]-d29_cmm). The differences in the ultimate fraction of single-stranded DNA produced reflect the different values for the fraction of single-stranded DNA generated at 37°C in the absence of protein, which were subtracted from these values (see text). (B) Annealing assays with 2 nM d29 and 1.8 nM [32P]-d29_c (filled circles) or 1.8 nM [32P]-d29 and 2 nM d29_c (the other symbols). All of the results in (A) and (B) were generated in independent reactions.
These results suggest that caging is more than a function of the relative kinetics of annealing and melting, but rather a property of the trimeric arrangement of the C-terminal halves of the protein. The same is true for high-affinity binding for nucleic acids by ORF1p, which is only exhibited by the trimer (Figures 5 and 6), and which is essential for retrotransposition (12,25,28,31,32).
DISCUSSION
Here, we reveal properties of the purified human L1 ORF1p trimer that have important implications for its functions in retrotransposition as both a nucleic acid binding protein and a nucleic acid chaperone:
First, in the absence of nucleic acid, ORF1p exists as a polymer under the very conditions (0.05 M NaCl) that are optimal for high (∼1 nM)-affinity nucleic acid binding. In this state ORF1p can be cross-linked into large aggregates of indeterminate size (Figures 2A and 3B). However, the polymeric form of ORF1p is an active conformer of the protein for it is immediately resolved to trimers, or multimers thereof, by nucleic acids (Figures 2C, 3A and 6B).
Multiple mouse ORF1p trimers were also found to bind nucleic acids of sufficient length (see Figure 3 in ref. 29). But our finding that the trimer multimers can be cross-linked indicates that nucleic acid binding and the protein–protein interactions that mediate polymerization are not mutually exclusive (Figure 3A). This finding may be related to our second novel finding; i.e. the biphasic effect of the protein on mismatched duplex DNA, a proxy substrate for nucleic acid chaperone activity.
ORF1p not only can protect the duplex from dissociation (melting) at 37°C but also eventually melts the duplex when present at sufficient molar excess and thus largely polymeric (Figure 8). As the protein binds the mismatched duplex and single-stranded DNA with the same affinity (Figure 7), it presumably ‘senses’ the duplex as single strands. However, the complementary strands must be held in such close approximation (‘caged’) that they are recovered as a duplex upon denaturation of the protein.
As caging is relatively insensitive to the molar ratio of protein to nucleic acid (Figure 8), we presume that it is an intra-trimer phenomenon; e.g. the two nucleic acid strands could be bound to the juxtaposed C-terminal halves of a given trimer. That caging is solely a property of the trimer supports this contention. The M128p monomer does not cage the mismatched duplex even though it is as equally proficient as the trimer in promoting the annealing and melting of nucleic acids (Figure 9), the two activities required of a nucleic acid chaperone.
Mismatched double-stranded DNA is a proxy for a chaperone substrate that ORF1p could encounter during retrotransposition. Thus, understanding the biochemical mechanism of the hand off between the two states of the ORF1p-bound mismatched duplex could not only reveal how the ORF1p nucleic acid chaperone functions in retrotransposition, but also provide a biochemical rationale for its uniquely trimeric structure. We currently do not understand the biochemical features of ORF1p–nucleic acid complexes or trimer–trimer interactions in sufficient detail to usefully speculate on how these processes might be related or to the role of ORF1p in retrotransposition.
However, the results in Figure 4 do provide an important starting point for understanding trimer–trimer interactions as well as the effect of the coiled-coil domain on the structure of ORF1p. These results showed that polymerization of full-length ORF1p trimers is apparently mediated by interactions between the C-terminal halves of different trimers. Furthermore, inhibition of polymerization by 0.5 M NaCl is likely the result of alterations in the orientation of the C-terminal halves so as to mask the surfaces that mediate inter-trimer interactions.
Given the dependence of coiled-coil structures on inter-helical salt bridges (e.g. ref. 46), we suggest that this change in orientation is due to a salt-induced conformational change in the coiled-coil domain. The possibility that the configuration of the C-terminal halves of the trimer is sensitive to structural change in the coiled-coil domain is consistent with the findings that showed that minimal amino acid changes in the coiled-coil, which have no discernible effect on trimerization, can profoundly affect the activity of ORF1p in retrotransposition assays (Callahan, K., Perez-Gonzalez, C., Furano, A.V., unpublished data, 32).
Despite the sensitivity that ORF1p activity can exhibit in response to minor amino acid changes in the coiled-coil domain, this domain, paradoxically, is the most variable region of the L1 encoded proteins (23,24,33–35) and a strongly conserved feature of ORF1p evolution (Walser, J.-C. and Furano, A.V., unpublished observations, 24,33). Thus, even though preservation of trimer formation is essential for the survival of L1, the coiled-coil domain is continually being remodeled perhaps as an adaptive response to changes elsewhere in the L1 element or the host environment.
Although the role of ORF1p polymerization in retrotransposition awaits clarification, it could explain several phenomena related to L1 biology. For one, it could account for the accumulation of the protein in stress granules and other intra-cellular foci in cells transfected with L1-expression vectors (e.g. refs 47 and 48).
More provocatively, that polymeric ORF1p is an active form of the protein could provide a mechanism for cis preference—the assembly of ORF1p on the transcript from which it was translated (33,37). Cis preference is essential to the survival of L1 elements, for without it, ORF1p, which interacts non-specifically with nucleic acids, would be consumed by non-productive interactions with non-L1 nucleic acids. However, to date no mechanism to explain cis preference has been forthcoming.
We suggest that polymerization of newly synthesized ORF1p would prevent it from diffusing into the cytoplasm. Thus, it would likely accumulate near the translational apparatus. When the L1 transcript is released from the ribosomes, perhaps even mediated by ORF1p polymers, it will be incorporated into the L1 ribonucleoprotein particle, the putative retrotransposition intermediate. Kroutter et al. (49) have also suggested that ORF1p could compete with the translational machinery for the L1 transcript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Intramural Program of the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK)/National Institutes of Health (NIH). Funding for open access charge: NIDDK/NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate the skill and efficiency of the Protein Expression Laboratory, Advanced Technology Program, SAIC-Frederick in preparing baculovirus and infected insect cells. We also thank Dr John Moran for the JCC8 clone that contained the L1.3 element.
REFERENCES
- 1.Eickbush TH, Malik HS. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington DC: ASM Press; 2002. pp. 1111–1144. [Google Scholar]
- 2.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 3.IHGS-Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 5.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl Acad. Sci. USA. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing AD, Kazazian HH. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1272. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang CRL, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7:67–72. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 14.Mathias SL, Scott AF, Kazazian HHJ, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 15.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 16.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 17.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retransposable element. Mol. Cell. Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bibillo A, Eickbush TH. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 2002;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- 19.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 21.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 22.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 24.Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J. Biomed. Biotechnol. 2006;2006 doi: 10.1155/JBB/2006/45621. ID#45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl Acad. Sci. USA. 2009;106:731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl Acad. Sci. USA. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 28.Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. LINE-1 Retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Basame S, Wai-lun Li P, Howard G, Branciforte D, Keller D, Martin SL. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J. Mol. Biol. 2006;357:351–357. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 30.Januszyk K, Li PW-l, Villareal V, Branciforte D, Wu H, Xie Y, Feigon J, Loo JA, Martin SL, Clubb RT. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J. Biol. Chem. 2007;282:24893–24904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- 31.Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J. Biol. Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 32.Martin SL, Bushman D, Wang F, Li PWL, Walker A, Cummiskey J, Branciforte D, Williams MC. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic Acids Res. 2008;36:5845–5854. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acids Res. Mol. Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- 34.Demers GW, Matunis MJ, Hardison RC. The L1 family of long interspersed repetitive DNA in rabbits: sequence, copy number, conserved open reading frames, and similarity to keratin. J. Mol. Evol. 1989;29:3–19. doi: 10.1007/BF02106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boissinot S, Furano AV. The recent evolution of human L1 retrotransposons. Cytogenet. Genome Res. 2005;110:402–406. doi: 10.1159/000084972. [DOI] [PubMed] [Google Scholar]
- 36.Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl Acad. Sci. USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 39.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein–nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLean A, Munson P, Rodbard D. Simultaneous analysis of families of sigmoidal curves: application to bioassay, radioligand assay, and physiological dose-response curves. Am. J. Physiol.– Endocrinol. Metab. 1978;235:E97–E102. doi: 10.1152/ajpendo.1978.235.2.E97. [DOI] [PubMed] [Google Scholar]
- 41.Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two-and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heus H, Hilbers C. Structures of non-canonical tanemd base pairs in RNA helices: review. Nucleosides Nucleotides Nucleic Acids. 2003;22:559–571. doi: 10.1081/NCN-120021955. [DOI] [PubMed] [Google Scholar]
- 43.Cléry A, Blatter M, Allain FHT. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Kielkopf CL, Lücke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkhard P, Ivaninskii S, Lustig A. Improving coiled-coil stability by optimizing ionic interactions. J. Mol. Biol. 2002;318:901–910. doi: 10.1016/S0022-2836(02)00114-6. [DOI] [PubMed] [Google Scholar]
- 47.Doucet AlJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.