Abstract
MiR-145 can regulate cell apoptosis, proliferation, neural development and stem cell differentiation. Previous studies indicate that miR-145 is downregulated in human colon cancer cells. However, the molecular mechanisms of miR-145 used to regulate colon carcinogenesis and angiogenesis remain to be clarified. Here, we show that the expression of miR-145 is downregulated in colon and ovarian cancer tissues and cell lines. MiR-145 inhibits p70S6K1 post-transcriptional expression by binding to its 3′-UTR. The angiogenic factors hypoxia-inducible factor 1 (HIF-1) and vascular endothelial growth factor (VEGF), which are downstream molecules of p70S6K1, are decreased by miR-145 overexpression. P70S6K1 rescues miR-145-suppressed HIF-1 and VEGF levels, tumorigenesis and tumor angiogenesis. Furthermore, the miR-145 level is inversely correlated with the amount of p70S6K1 protein in colon cancer tissues. Taken together, these studies suggest that miR-145 serves as a tumor suppressor which downregulates HIF-1 and VEGF expression by targeting p70S6K1, leading to the inhibition of tumor growth and angiogenesis. The miR-145 rescue could be a rationale for therapeutic applications in colon cancer in the future.
INTRODUCTION
MicroRNAs are a class of small non-coding RNAs that have been identified as a new kind of gene expression regulators through targeting mRNAs for translational repression or degradation (1–3). More and more evidence has shown the important roles of miRNAs in regulating various cellular functions such as cell apoptosis, cell proliferation, neural development and stem cell differentiation (4–6). Recently, miRNAs are found to play pivotal roles in tumorigenesis, cancer invasion and metastasis (7,8). MiR-145 was first found to be reduced in colorectal neoplasia. Then, the deregulation of this miRNA was demonstrated in breast, ovarian, lung, nasopharyngeal, bladder, gastric and prostate cancers (9–16). The well-known central tumor suppressor p53 can enhance the post-transcriptional maturation of several miRNAs, including miR-145, in response to DNA damage, showing the vital growth-suppressive function of miR-145 (17). In addition to its part in cancer development, miR-145 plays an important role in regulating cell fate and plasticity of smooth muscle cells (18).
Angiogenesis is a process by which new microvessels sprout from existing vessels. It is vital for tumorigenesis and tumor development, because tumors cannot grow without angiogenesis after their diameters reach 1–2 mm (19). Among all the angiogenic factors, the most potent one is vascular endothelial growth factor (VEGF), which is responsible for the growth and permeability of vascular endothelial cells, vasculature and tumor angiogenesis (20,21). Hypoxia-inducible factor 1 (HIF-1) is a heterodimeric transcription factor composed of HIF-1α and HIF-1β subunits, and is a major regulator of VEGF, in response to hypoxia, by binding to the promoter of VEGF (22,23). However, there is no information pertaining to whether or not miR-145 regulates tumor growth and angiogenesis through mediating the expression of HIF-1α, and VEGF.
The mammalian target of rapamycin (mTOR)/p70S6K1 is an important signaling pathway in regulating cellular functions. The activation of p70S6K1 by mTOR enhances the translation of mRNAs that bear a 5′ terminal oligopyrimidine tract, which encodes proteins related with the translational apparatus like ribosomal proteins, elongation factors eEF1A, eEF2 and the poly A-binding protein (24). MTOR/p70S6K1 can be activated by the dysfunction or genetic alterations of its upstream PI3K/PTEN/AKT signaling pathway or by the overexpression or amplification of p70S6K1 (24,25). Given the important roles of these proteins in cancerous characteristics such as the cell cycle, cell apoptosis, cell growth and proliferation, mTOR/p70S6K1 is proven to be one of the most important targets in cancer therapy, and its inhibitors exhibit encouraging effects in animal experiments and clinical trials (24,26). Evidences by our lab and others demonstrate that the overexpression of p70S6K1 in both cancer cells and vascular endothelial cells induces tumor angiogenesis (27–29). Nevertheless, there is no miRNA that has been reported to target p70S6K1 up to this point in time.
Colorectal cancers are the third most commonly diagnosed types of cancer and the third leading cause of cancer-related death in both men and women (30). In most studies, miR-145 has been shown to act as a vital tumor suppressor in colorectal cancers, and its deregulation is correlated with clinicopathologic features and prognostic potential in colorectal cancers (12,31,32). On the contrary, Arndt et al. (33) reported that SW620 stably expressing miR-145 cells (SW620/miR-145) showed increased cell proliferation/metabolic activity and its mesenchymal-like cell morphology, which indicated the oncogenic potential of miR-145. The demonstrated miR-145 targets include the Rho-effector rhotekin (RTKN) (34), the metastasis gene mucin 1 (MUC1) (35), the Ets transcription factor Friend leukemia virus integration 1 (Fli1) (36), Clathrin Interactor 1 (CLINT1), core-binding factor b subunit (CBFB), the protein phosphatase 3 catalytic subunit a isoform (PPP3CA) (37), the well-known pluripotency factors OCT4, SOX2 and KLF4 (4), the epidermal growth factor receptor (EGFR), the nucleoside diphosphate linked moiety X-type motif 1 (NUDT1) (38), the fascin homologue 1 (FSCN1) (39), c-Myc (40) and the junctional adhesion molecule A and fascin (41). A recent study showed that miR-145 targets the Src family member YES, which plays a role in cell growth and transformation, and the transcription factor STAT1, which is related to cell apoptosis and survival, in colon cancer (42). Moreover, miR-145 is involved in controlling cell apoptosis by targeting DNA fragmentation factor-45 (DFF45) (43), and it inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene (FLI1) (44). However, the role and the underlying mechanisms of miR-145 in colorectal cancers still remain unclear.
In this study, we demonstrate that: (i) p70S6K1 is the direct target of miR-145; (ii) the level of miR-145 is decreased in colon cancers; (iii) miR-145 suppresses tumor growth and angiogenesis by decreasing HIF-1α and VEGF expression through targeting p70S6K1 in vitro and in vivo; (iv) the expression of miR-145 is negatively correlated with the p70S6K1 protein levels in colon cancer tissues. These results demonstrate for the first time that p70S6K1 is a direct target of miR-145 in the regulation of tumor growth and angiogenesis.
MATERIALS AND METHODS
Cell culture, tissue samples and reagents
The human colon cancer cells SW1116 and SW480 (American Type Culture Collection, Manassas, VA, USA) were cultured in a RPMI 1640 medium that contained a 10% heat-inactivated fetal bovine serum, 100 units/ml, penicillin G and 100 mg/ml streptomycin in a 5% CO2 incubator at 37°C. NCM356 cells were purchased from INCELL (Innovative Life Science Solution, TX, USA). Cells were maintained in INCELL's enriched M3:10 medium, which is M3 medium plus supplements and 10% fetal bovine serum and antibiotics, and cultured in 37°C, 5% CO2 humidified environment. Twenty pairs of human colon cancer samples, including some primary colon cancer tissues and paired adjacent normal colon tissues, were obtained from Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai, China. Nine malignant ovarian cancer tissues and nine normal ovarian tissues were obtained from the Nanjing Maternal and Child Health Hospital, Nanjing, China. In all the cases, the diagnoses and grading were confirmed by two experienced pathologists, which were done in accordance to the principles laid down in the latest World Health Organization Classification. Informed consent was also obtained from all patients, and the study was approved by Nanjing Medical University ethics committee. Tissues specimens that fit into the diagnostic categories of interest will be identified by computerized search of the clinical files (CoPath) using SNOMED as standardized terminology. Since the analysis will be carried out without access or notation of any information regulated by HIPAA, this approach qualifies for the status of NIH Exemption #4. Protocol for the study has been approved by the Thomas Jefferson University and Nanjing Medical University Human Assurances Committees.
P70S6K1, p-p70S6K, PDCD4 and VEGF antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The antibody against HIF-1α and the growth factor-reduced phenol red-free Matrigel were obtained from BD Biosciences (Franklin Lakes, NJ, USA). The CD31 antibody was from Dako (Carpinteria, CA, USA). The antibody against β-actin was from Sigma (St Louis, MO, USA). Moloney murine leukemia virus (M-MLV) reverse transcriptase, DNA Marker, Taq polymerase and oligo(dT)18 were from TaKaRa (Dalian, China). Lipofectamine 2000 was from Invitrogen (Carlsbad, CA, USA). A lentivirus-based human miRNA library was purchased from Open Biosystems (Huntsville, AL, USA). Anti-miR-145 and the negative control of miRNA inhibitors were from Ambion (Applied Biosystems, Carlsbad, CA, USA). TaqMan qRT-PCR Kit for miR-145, High Capacity RNA-to cDNA Kit and Power SYBR Green PCR Master Mix were from Applied Biosystems. The Luciferase (Luc) assay system was from Promega (San Luis Obispo, CA, USA). Horseradish peroxidase conjugated anti-mouse and anti-rabbit antibodies and visualized with enhanced chemiluminescence reagent, which were from Perkin-Elmer Life Sciences (Boston, MA, USA).
Northern blotting
Total RNAs were extracted using Trizol (Invitrogen) according to the manufacturer's instructions. Aliquots of RNA samples (25 μg/sample) were electrophorized on 15% acrylamide and 8 M urea denature gels and then transferred onto Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ, USA). The membranes were agitated at 80°C for 2 h and then hybridized with oligonucleotide probes corresponding to the complementary sequences of mature miR-145 and U6. The probes were labeled at the 5-end using the polynucleotide kinase in the presence of [γ-32P] ATP (GE Healthcare, Piscataway, NJ, USA). Hybridization was carried out at 39°C in ULTRAhyb™-Oligo Hybridization Buffer (Ambion) for 16 h. Membranes were washed at 42°C twice with SSC (2×) and 0.1% SDS. The membranes were stripped off by putting them in 1% SDS at 65°C for 30 min, and rehybridized. Images were obtained using the scanner Storm 860 (Molecular Dynamics, Sunnyvale, CA, USA).
Construction of p70S6K1 3′-UTR luciferase plasmids and reporter assays
A length of 510 bp PCR product of p70S6K1 (nt724-1233) containing its 3′-UTR was amplified by the following primers: 5′-CGGGGATCCGGGTGGACCTGGGGTTTATTT-3′ (sense) and 5′-CCCCTCGAGTTCATCAAAAGGCCATCAAAT-3′ (antisense). The PCR product was then cloned into a pcDNA6.2 vector. After identifying the orientation, the correct 3′-UTR of p70S6K1 (WT) was cut from pcDNA6.2 and cloned into pMIR-REPORTER (ambion, TX, USA). Site-directed mutagenesis of the miR-145 seed sequence in p70S6K1-3′-UTR (Mut) was performed by using a Quick change-mutagenesis kit (Stratagene, CA, USA), with pcDNA6.2-p70S6K1 WT as the template. The primers for site-directed mutagenesis were as follows: 5′-GCAGTACTGCTATGTGCTAAGCTTAACTCCAAGCCTTGGAATGGG-3′ (sense), 5′-CCCATTCCAAGGCTTGGAGTTAAGCTTAGCACATAGTACTGC-3′ (antisense). The mutant p70S6K1-3′-UTR was then cloned into pMIR-REPORTER. For reporter assays, human 293 cells were transiently transfected with WT or Mut reporter plasmid, pCMV β-galactosidase plasmid, and miRNA-145 plasmid (Open Biosystems) using lipofectamine 2000. Luciferase (Luc) activity was measured 24 h after transfection by using the Luc assay system.
Quantitative real-time PCR
A quantitative real-time RT–PCR was used to determine the expression levels of miR-145 and VEGF in cells and tumor tissues. To measure the miR-145 levels, total RNAs were extracted from cultured cells and tumor tissues using Trizol (Invitrogen, Carlsbad, CA, USA). The quantitative real-time RT-PCR analysis of miR-145 levels was performed using the TaqMan Reverse Transcription Kit and TaqMan MicroRNA Assays Kit (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturers’ instructions. The expression levels of miR-145 were calculated using the delta–delta Ct method with U6 as an internal control.
Real-time PCR was performed to detect the VEGF mRNA levels using SYBR Green. Reverse transcription reactions were performed using High Capacity RNA-to-cDNA Kit according to the manufacturer's instructions. The 100 ng of RT product was used for the PCR reaction using Power SYBR Green PCR Master Mix. The reaction contained the following: 10 µl of 2× PCR Master Mix, 1 µl of forward primer, 1 µl of reverse primer, 1 µl of cDNA template, and 7 µl of H2O. The reaction program was set at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 30 s, and the melt curve was included. The primers of VEGF are the forward primer, 5′-CGAGGGCCTGGAGTGTG-3′ and the reverse primer, 5′-CCGCATAATCTGCATGGTGAT-3′. The primers of GAPDH are the forward primer, 5′-ATGGGTGTGAACCATGAGAAGTATG-3′ and the reverse primer, 5′-GGTGCAGGAGGCATTGCT-3′.
Lentivirus transduction and establishment of stable colon cancer cell lines expressing miR-145
In order to make miR-145 stably expressed in colon cancer cells, we used a pLe-miR-145 plasmid from a lentivirus-based human miRNA library (Open Biosystems, USA), which can be used in order to overexpress human pre-miRNA, for packaging the lentivirus and the transduced colon cancer cell lines SW480 and SW1116. In short, pLe-miRNA-145 plasmid DNA and the transfection complex DNAs were transfected into 293T cells through the use of Arrest-In regent according to the company instruction. The validated, non-silencing, scrambled control was used as a negative control. Lentiviruses in the supernatant were collected and used to transduce SW1116 and SW480 cells. Stable cell lines were selected using puromycin.
Adenovirus transduction
Recombinant adenoviruses were made using the AdEasy system (25). In short, the p70S6K1 cDNA was subcloned into the vector pAdTrack-CMV and then transferred to AdEasy-1 plasmid through homologous recombination, as previously described (26). The viral vectors were then transfected into Ad-293 cells in order to generate viruses. A control virus carrying the green fluorescent protein (Ad-GFP) was derived from the same vector system. The viruses were titered and 20 multiplicity of infection (MOI) of Ad-p70S6K1 and Ad-GFP were used to infect cells.
Western blotting
The cells were homogenized in a chilled lysis buffer containing 10 mM Tris–HCl (pH 7.4), 1% NP-40, 0.1% deoxycholic acid, 0.1% SDS, 150 mM NaCl, 1 mM EDTA and 1% protease inhibitors. Total proteins were collected by centrifugation. For tissue samples, tissues were ground in liquid nitrogen in a radioimmunoprecipitation assay buffer, and the total tissue proteins were extracted as previously described. Twenty micrograms of lysate proteins were separated by SDS–PAGE and subsequently transferred to a nitrocellulose membrane. Membranes were blocked with 5% non-fat dry milk for 2 h and incubated with primary antibodies. Protein bands were detected by incubation with horseradish peroxidase-conjugated antibodies and visualized with an enhanced chemiluminescence reagent.
Cell proliferation assay
To determine the effects of miR-145 and its target p70S6K1 on colon cell proliferation, the parental SW1116 cells (mock), SW1116 cells stably expressing pLe-SC and pLe-miR-145 were transduced by Ad-GFP or Ad-p70 at 20 MOI for 24 h and subsequently typsinized and resuspended. Cells at 1 × 105 were seeded in a 6-well plate and cultured overnight. The cells were then trypsinized and counted each day using a hemocytometer for 7 days of being plated.
In vivo tumor growth assay
The 6-week-old nude mice [BALB/cA-nu (nu/nu)] were purchased from Shanghai Experimental Animal Center (Chinese Academy of Sciences, China), maintained in pathogen-free conditions, and sustained with standard diets. All procedures involving animals were approved by the Institutional Committee on Animal care, Nanjing Medical University. Twenty mice were randomly divided into four groups with five mice per group. Aliquots of SW1116 cells (1.0 × 106 cells in 50 µl) including stable miR-145-expressing cells and cells infected by Ad-p70S6K1 or Ad-GFP adenovirus for 24 h were mixed with 50 µl growth factor-reduced phenol red-free Matrigel and injected subcutaneously into both flanks of nude mice. The xenografts were removed from the mice 30 days after the implantation and trimmed of the surrounding tissues. Then, the xenografts were weighed. Part of the tissue samples was snap frozen in liquid nitrogen and stored at −80°C for analysis.
Semi-quantitative RT-PCR for HIF-1α mRNA levels
Total cellular RNAs were isolated using Trizol reagent (Invitrogen) according to the manufacturer's protocol. RNA at 1 μg was used for cDNA synthesis using oligo(dT)18 and M-MLV reverse transcriptase. The primers for HIF-1α and GAPDH were as follows: HIF-1α sense primer: 5′-TCGGGCCTCCGAAACCATGA-3′; anti-sense primer: 5′-CCTGGTGAGAGATCTGGTTC-3′. GAPDH sense primer: 5′-AATGCATCCTGCACCACCAACTGC-3′; anti-sense primer: 5′-GGAGGCCATGTAGGCCATGAGGTC-3′. HIF-1α and GAPDH mRNA levels were detected as described previously (45), and quantified using UVP VisionworksLS Software.
Immunohistochemical analysis
Tumor samples were fixed with Bouin's solution (saturated picric acid 300 ml, formaldehyde 100 ml, glacial acetic acid 20 ml) for 24 h, washed with 70% ethanol, and processed by the paraffin-embedded method. The tissues sections (5 μm thick) were then heat-immobilized or pepsin-immobilized according to the manufacturer's instructions. Antibodies against p70S6K1, HIF-1α, VEGF and CD31 were used for the immunostaining and detected through the Dako Envision two-step method of immunohistochemistry (Carpinteria, CA, USA). The relative angiogenesis levels were calculated by microvessel density (MVD). In short, slides were first scanned under low power (×40) in order to determine three areas with the maximum number of microvessels that were consequently evaluated at ×200 magnification. The data are presented as mean ± SE from 10 slides for each group.
Statistical analysis
The data represent mean ± SE except where indicated. Statistical analysis was performed based on a Student's t-test at the significance level of P < 0.05. Spearman's non-parametric correlation test was performed to test the correlation between the expression levels of miR-145 and p70S6K1 by SPSS.
RESULTS
MiR-145 is downregulated in human colon cancer tissues
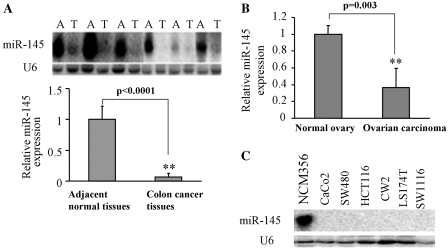
To assess whether or not miR-145 is dysregulated in colon cancer tissues, miR-145 expression levels from colon cancer tissues and their adjacent normal tissues are analyzed by northern blotting. As shown in Figure 1A, the expression levels of miR-145 in human colon cancer tissues are much lower than those in the adjacent normal tissues. To determine whether or not the miR-145 levels were also downregulated in other types of human cancer, the expression of the miR-145 level was tested in ovarian carcinoma tissues and normal tissues by the TaqMan real-time RT-PCR. When compared to normal ovarian tissues, the miR-145 level in ovarian carcinoma tissues decreased to 40%, indicating that the downregulation of miR-145 is found in both colon and ovarian cancers (Figure 1B). Further experiments were performed by using normal epithelial cell line (NCM356) and several different colon cancer cell lines to show that miR-145 expression was undetectable or very low in colon cancer cell lines including CaCo2, SW480, HCT116, CW2, LS174T and SW1116 cells in contrast with normal cells (Figure 1C). These results indicate that miR-145 is downregulated in both colon cancer tissues and cancer cell lines.
Figure 1.
The expression of miR-145 in colon cancer tissues and cell lines. (A) The expression levels of miR-145 were analyzed in colon cancer tissues (T) and its adjacent normal tissues (A) by Northern blotting. SnRNA U6 levels were used as an internal control. (B) Tissue samples from normal ovary and ovarian carcinoma were used to extract total RNAs. The levels of miR-145 were measured by qPCR. U6 was used as an internal control and the ratio of miR-145/U6 in tumor samples was normalized to that in normal ovary tissues. (C) The expression levels of miR-145 were analyzed by Northern blotting in normal colon epithelial cell line (NCM356), colon cancer cell lines including CaCo2, SW480, HCT116, CW2, LS174T and SW1116. The double asterisks indicates significant difference when compared to control at P < 0.01.
The p70S6K1 3′-UTR is a target of miR-145
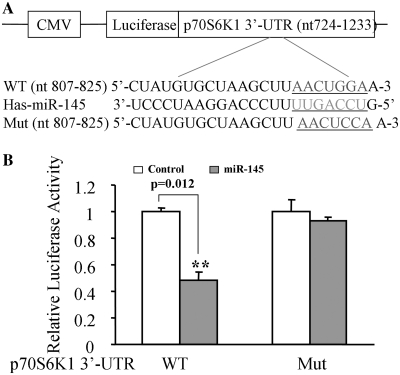
To search for the potential targets of miR-145, we found a putative miR-145 binding site located in the 3′-UTR of p70S6K1. This putative binding site is fully conserved in 13 species including human in the region complementary with the seed sequence (Table 1). To test whether or not miR-145 targets the 3′-UTR region of p70S6K1, p70S6K1 3′-UTR containing this seed sequence was cloned into the pMIR-REPORT miRNA reporter vector (WT). The forced expression of miR-145 markedly suppressed the luciferase activity of the wild-type reporter by 50%, suggesting that miR-145 inhibits its 3′-UTR function. We also mutated the miR-145 binding site in the reporter construct (Mut). Cells were co-transfected with the pLe-miR-145 or scrambled control plasmid with the wild-type or mutant p70S6K1 3′-UTR reporter and β-gal plasmids, then the luciferase activities were assayed. P70S6K1 wild-type but not mutant reporter activity was affected by the transfection of pLe-miR-145, suggesting the point mutation of this seed sequence abolished the effect of miR-145 (Figure 2B).
Table 1.
Comparison of nucleotides between the miR-145 seed-sequence and its target in different species
| Species | p70S6Kl 3′-UTR (nt 807-826) |
|---|---|
| Chimpanzee | UGUGCUAAGCUUAACUGGAA |
| Rhesus | UGUGCUAAGCUUAACUGGAA |
| Mouse | UGUGCUAAGCUUAACUGGAA |
| Rat | UGUGCUAAGCUUAACUGGAA |
| Rabbit | UGUGCUAAGCUUAACUGGAA |
| Shrew | UGUGCUAAGCUUAACUGGAA |
| Hedgehog | UGUGCUAAGCUUAACUGGAA |
| Dog | UGUGCUAAGCUUAACUGGAA |
| Cat | UGUGCUAAGCUUAACUGGAA |
| Horse | UGUGCUAAGCUUAACUGGAA |
| Cow | UGUGCUAAGCUUAACUGGAA |
| Chicken | UGUGCUAAGCUUAACUGGAA |
| Human | UGUGCUAAGCUUAACUGGAA |
| hsa-miR-145 | CUAAGGACCCUUUUGACCUG |
Figure 2.
MiR-145 binding resides at p70S6K1 3′-UTR region. (A) The schematic representation of p70S6K1 3′-UTR luciferase constructs. The p70S6K1 3′-UTR (nt724-1233) was cloned into pMIR-REPORTER vector (WT), and the mutant construct was obtained by site-directed mutagenesis by changing two base pairs of miR-145 seed-sequence (Mut) and inserted into the vector. (B) 293 cells were transfected with 0.2 μg of WT or Mut reporter, 0.3 μg of pLE-miR-145, and 0.1 μg of β-gal plasmids. The relative Luc activity was assayed and calculated by the ratio of luc/β-gal activity, which was normalized to that of the control. Results are presented as mean ± SE from three duplicates. Double asterisks indicate significant difference when compared to the control (P < 0.01).
P70S6K1 protein levels correlate inversely with miR-145 levels in human colon cancer cell lines
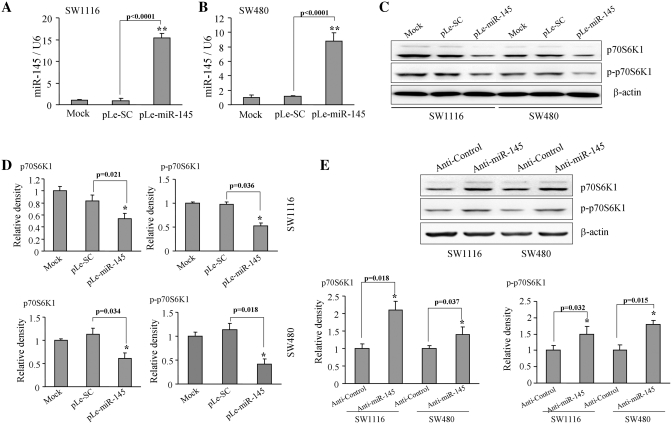
Next, to further ensure the validity of these results in human colon cancer cells, human colon cancer cell lines SW1116 and SW480 were infected with the lentivirus carrying miR-145 (pLe-miR-145) or the scrambled control (pLe-SC) and selected by puromycin. The expression levels of miR-145 and p70S6K1 were analyzed in these stable cell lines SW480-miR-145, SW480-SC cells, SW1116-miR-145 and SW1116-SC using TaqMan real-time RT-PCR and western blotting. As shown in Figure 3A and B, the expression levels of miR-145 were similar in mock cells and miR-SC cells, while increased by 15- and 9-fold in pLe-miR-145-expressing SW1116 and SW480 cells, respectively (Figure 3A and B), demonstrating that these stable cells were successfully expressing miR-145. The cells with miR-145 overexpression showed low levels of p70S6K1 and p-p70S6K1 proteins by 40–45% decrease compared to the scrambled control cells (Figure 3C and D). Furthermore, transfection of anti-miR-145 inhibitor in SW1116 and SW480 cells increased p70S6K1 and p-p70S6K1 expression (Figure 3D and F). These initial experiments indicate that miR-145 negatively regulated p70S6K1 expression.
Figure 3.
Overexpression of miR-145 decreased p70S6K1 protein expression. SW1116 (A) and SW480 (B) cells stably expressing scrambled control (SC) and miR-145 were subjected to TaqMan real-time RT-PCR analysis for miR-145 and U6 expression. Results are presented as mean ± SE from three duplicate experiments. Double asterisks indicate significant difference when compared to the control (P < 0.01). (C) The p70S6K1 and p-p70S6K1 protein levels were analyzed by western blotting in mock cells and cells expressing negative control SC and miR-145 precursor constructs. (D) Relative densities were quantified using ImageJ software. Results are presented as mean ± SE from three duplicates. Asterisk indicates significant difference when compared to the control (P < 0.05). (E) Cells were transfected with 25 nM of anti-miR-145 inhibitor or negative control inhibitor. The expression levels of p70S6K1, p-p70S6K1 and β-actin were determined by immunoblotting. Relative density was quantified using ImageJ software. Asterisk indicates significant difference when compared to the control (P < 0.05).
VEGF and HIF-1α expression is inhibited by miR-145
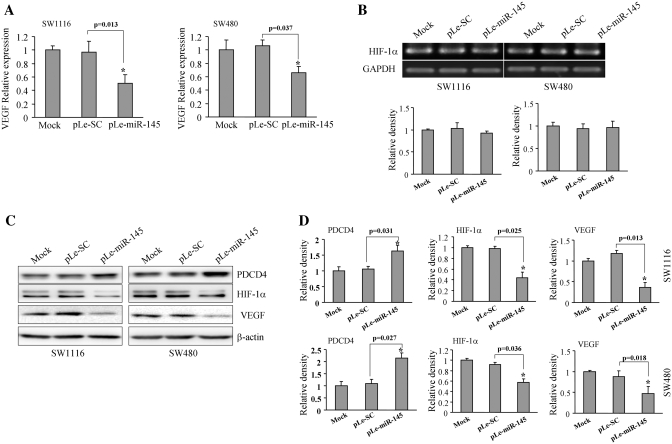
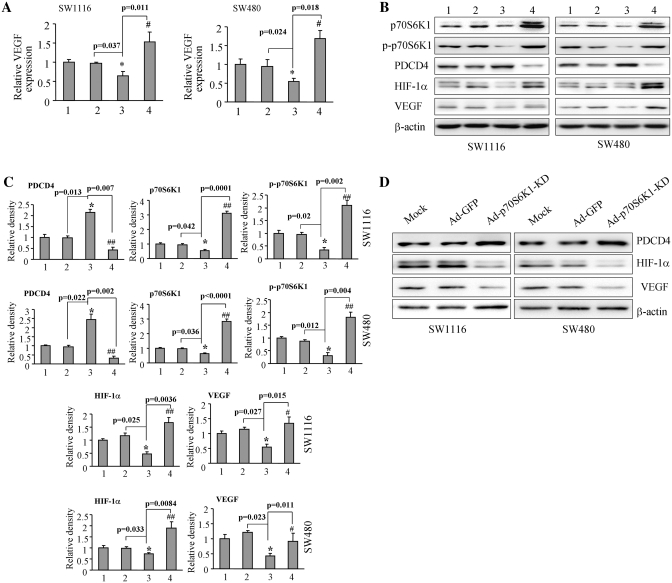
Programmed cell death 4 (PDCD4) is a known downstream target of p70S6K1 (46,47). We tested the PDCD4 levels in these cells. The results showed an increase in the levels of PDCD4 in the miR-145-overexpressing cells, which were consistent with our previous results, suggesting that miR-145 downregulated p70S6K1 for increasing PDCD4 expression (Figure 4C). VEGF is a crucial angiogenic factor for controlling vasculature and angiogenesis. HIF-1α is one of VEGF regulators for inducing its transcriptional expression by binding to its promoter. It is well known that p70S6K1 is a key molecule in regulation process of protein synthesis and eukaryotic translation. Our previous studies have shown that the mTOR/p70S6K1 signaling pathway mediates tumor angiogenesis and tumor growth by changing the levels of HIF-1α and VEGF (45,48,49). Furthermore, to test whether or not the overexpression of miR-145 in colon cells will have any effect on VEGF and HIF-1α expression, SW1116-miR-145, SW1116-SC, SW480-miR-145 and SW480-SC cells were used to test the levels of VEGF and HIF-1α expression. As shown in Figure 4A, when compared to the scrambled control, the VEGF expression in the miR-145-overexpressing cells was inhibited by about 55 and 40% by TaqMan RT-PCR in SW1116 and SW480 cells, respectively; while HIF-1α expression did not change at mRNA level (Figure 4B). Consistent with this result, the protein levels of VEGF were suppressed by >50% in both SW1116-miR-145 and SW480-miR-145 cells (Figure 4C and D). As we anticipated, the protein level of HIF-1α also showed a similar trend in the miR-145-overexpressing cells, demonstrating that miR-145 downregulates the expression of VEGF and HIF-1α (Figure 4C and D).
Figure 4.
Overexpression of miR-145 inhibited VEGF and HIF-1α expression. (A) SW1116 and SW480 cells overexpressing miR-145 were analyzed for VEGF mRNA levels by TaqMan real-time PCR. Results are presented as mean ± SE of VEGF mRNA levels from three duplicates. Asterisk indicates significant difference when compared to the control (P < 0.05). (B) Mock cells, cells transfected with pLe-SC and pLe-miR-145 were used to test HIF-1α mRNA levels by semi-quantitative RT-PCR. (C) Total cellular proteins were collected and subjected to western blotting. Forced expression of miR-145 increased PDCD4 level and inhibited HIF-1α and VEGF protein expression. (D) The protein levels of above study from three independent experiments are quantified and presented as mean ± SE. Asterisk indicates significant difference when compared to the control (P < 0.05).
MiR-145 inhibits VEGF and HIF-1α expression via p70S6K1
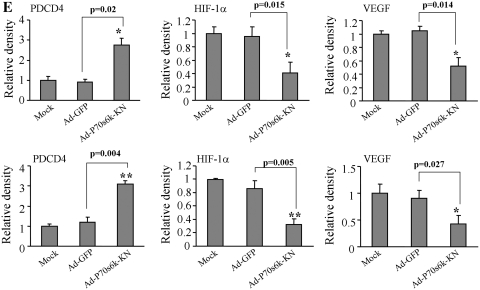
To assess whether or not miR-145 suppressed VEGF and HIF-1α expression by targeting p70S6K1, we restored p70S6K1 expression levels in the SW480-miR-145 and SW1116-miR-145 cells using adenoviruses carrying p70S6K1 (Ad-p70S6K1). Cells transduced by the adenovirus carrying GFP (Ad-GFP) were used as the control. The expression levels of PDCD4 were increased by miR-145 overexpression and attenuated by p70S6K1 overexpression, showing the functional effect of miR-145 (Figure 5B and C). Then, the expression levels of VEGF were tested by real-time RT-PCR and western blotting, and p70S6K1, p-p70S6K1 and HIF-1α levels were determined by western blotting. The overexpression of p70S6K1 in SW1116-miR-145 and SW480-miR-145 cells restored the miR-145-inhibited VEGF mRNA level (Figure 5A). Similarly, the miR-145 overexpression inhibited p70S6K1, HIF-1α and VEGF at the protein level by 40–50%, while the infection with Ad-p70S6K1 markedly recovered the p70S6K1 protein level and restored miR-145-inhibited p70S6K1, p-p70S6K1, HIF-1α and VEGF expression (Figure 5B and C). In addition, transduction of adenovirus carrying p70S6K1 dominant negative mimicked miR-145 overexpression, which showed increased PDCD4 levels, and decreased HIF-1α and VEGF expression (Figure 5D and 5E). These results suggest miR-145 inhibits the expression of HIF-1α and VEGF by targeting p70S6K1.
Figure 5.
Forced expression of p70S6K1 restored VEGF and HIF-1α levels inhibited by miR-145. (A) SW1116 and SW480 cells (mock), and cells stably expressing SC and miR-145 were infected with adenoviruses carrying GFP (Ad-GFP) or p70S6K1 (Ad-p70) at 20 MOI for 48 h. VEGF and GAPDH mRNA levels were determined by real-time RT-PCR. Lanes 1–4 represent cells expressing Ad-GFP alone, SC +Ad-GFP, miR-145 + Ad-GFP, and cells expressing miR-145 + Ad-p70S6K1, respectively. Asterisk indicates significant difference when compared to the control, hash indicates significant difference when compared to miR-145+Ad-GFP versus Ad-p70S6K1 groups. (B) The total PDCD4, p70S6K1, p-p70S6K1, HIF-1α and VEGF protein levels from the above experiment are analyzed by western blotting. Forced expression of p70S6K1 inhibited PDCD4 expression and restored p70S6K1, HIF-1α and VEGF protein levels in miR-145 stably expressed cells. (C) The signals of PDCD4, p70S6K1, p-p70S6K1, HIF-1α and VEGF protein levels were obtained and quantified from three independent experiments. Results are presented as mean ± SE. Asterisk indicates significant difference when the control vs miR-145 treatment. Hash and double hashes indicate significant difference when compared the miR-145-overexpresing cells treated with or without p70S6K1 overexpression at P < 0.05 or at P < 0.01, respectively. Groups 1–4: mock+Ad-GFP, pLe-SC+Ad-GFP, pLe-miR-145+Ad-GFP, pLe-miR-145+Ad-p70S6K1, respectively. (D) Mock cells, cells transduced by adenoviruses carrying GFP (Ad-GFP) and p70S6K1 dominant negative (Ad-p70S6K1-KD) were used to test expression of PDCD4, HIF-1α and VEGF by immunoblotting. (E) Quantification of signal density by Image J software. Asterisk and double asterisks indicate significant difference when compared to mock or Ad-GFP group at P < 0.05 or at P < 0.01, respectively.
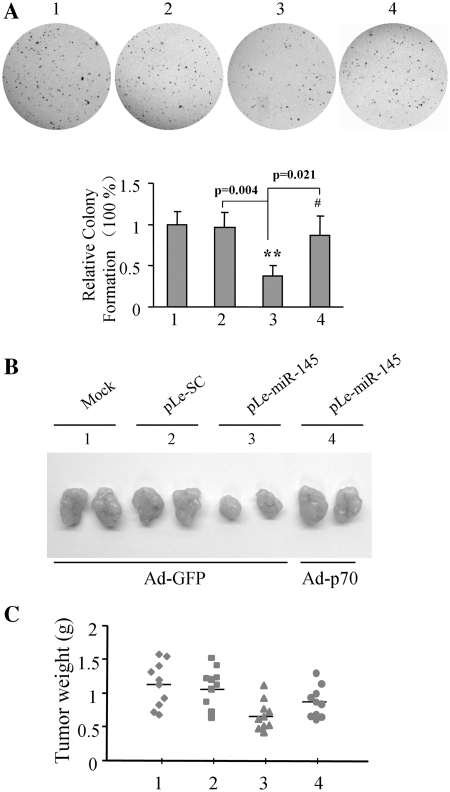
MiR-145 inhibits cell proliferation and tumor growth by downregulating VEGF and HIF-1α expression via targeting p70S6K1
Next, the cell proliferation assay showed miR-145 overexpression suppressed cell growth, while forced expression of p70S6K1 in miR-145-overexpressing cells restored the cell growth rate (Supplementary Figure S1), suggesting that miR-145 inhibits cancer cell proliferation through p70S6K1 expression. Then, we studied the effects of miR-145 and p70S6K1 on anchorage-independent growth. As shown in Figure 6A, compared to the mock (Group1) and scrambled control (pLe-SC, Group 2), the overexpression of miR-145 decreased the number of colonies to <50% (Group 3), while the presence of p70S6K1 in miR-145-overexpressing cells restored the number of colonies to 75% (Group 4). These results indicate that overexpression of p70S6K1 restores miR-145-inhibited tumor growth in vitro. Given the important roles of HIF-1α, and VEGF in inducing angiogenesis and tumor growth, we tested whether or not miR-145 inhibited tumor angiogenesis and tumor growth through the reduction of HIF-1α, and VEGF expression by targeting p70S6K1. Parental SW1116 cells (mock), SW1116 cells stably expressing pLe-SC and pLe-miR-145, were transduced by Ad-GFP or Ad-p70 at 20 MOI for 24 h, and the cells were subsequently typsinized and resuspended. The cells were mixed with growth factor reduced phenol-free Matrigel and injected subcutaneously into both flanks of the nude mice. Xenografts were collected after 30 days of injection. As shown in Figure 6B, the sizes of xenographs were similar from mock cells and SW1116-pLe-SC infected cells, and were much bigger than those from the SW1116-pLe-miR145 cells. The result indicates that stably expressed miR-145 in colon cancer cells reduces tumor growth. When transduced with Ad-p70S6K1, the SW1116-pLe-miR145 cells formed bigger tumors, showing that the overexpression of p70S6K1 in miR-145 overexpression cancer cells restored the rate of tumor growth. Consistent with these results, the overexpression of miR-145 decreased the tumor weight by 40%, while the infection of Ad-p70S6K1 caused restoration by 20% when compared to the miR-145 treated with Ad-GFP group (Figure 6C).
Figure 6.
Role of miR-145 in colony formation and tumor growth. (A) SW1116 mock cells, and cells stably expressing SC and miR-145 were infected with Ad-GFP or Ad-p70S6K1 at 20 MOI for 24 h, then cells were typsinized, and counted. Cells at 5 × 103 were plated in each well on 6-well plates with 0.05% soft agar layer. After the incubation of the cells for 14 days, the number of cell colonies was counted under the microscope, and the cells were fixed with 100% methanol and stained with 0.5% crystal violet dye. (B) SW1116 mock cells, and cells stably expressing SC and miR-145 were infected with Ad-GFP or Ad-p70S6K1 at 20 MOI for 24 h. Cells were implanted into nude mice. Xenografts were taken out after implantation for 30 days, and represented tumors are photographed. (C) Tumor weight was obtained and presented.
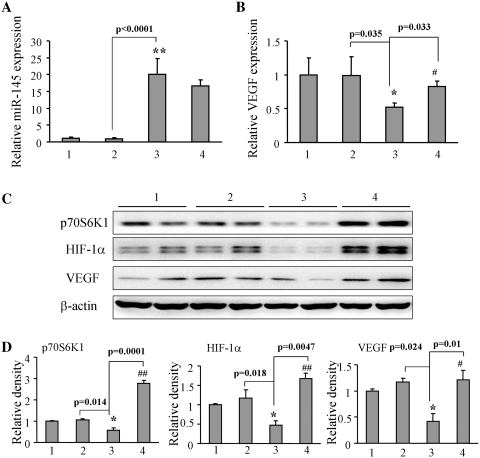
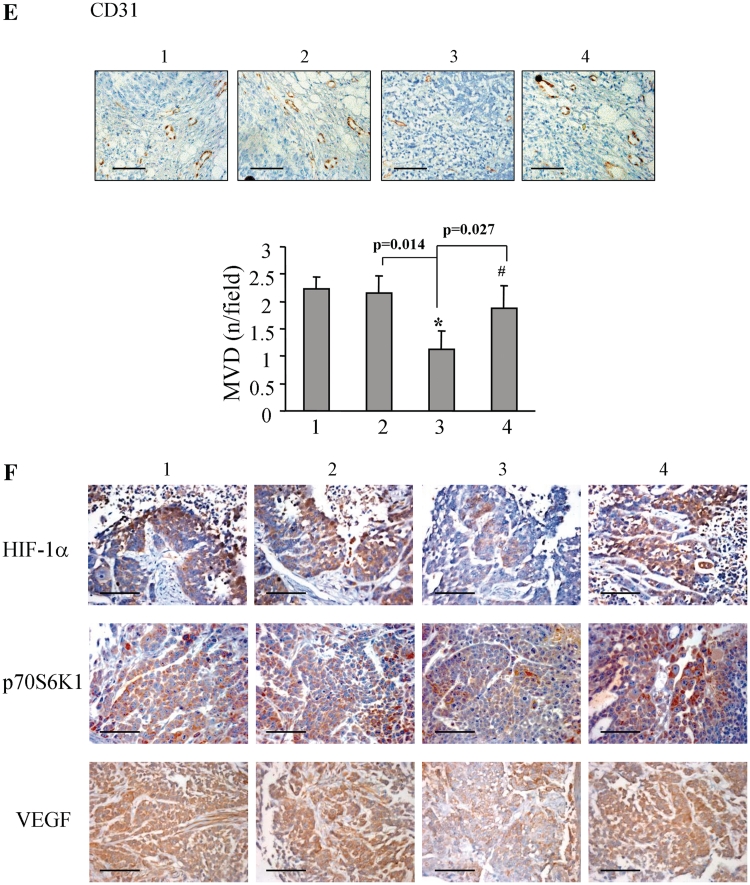
To further test the expression levels of miR-145 in tumor tissues from the nude mice, we randomly chose tumor tissues from each group and analyzed miR-145 level using the TaqMan real-time RT-PCR. The higher levels of miR-145 expression in the tumors from SW1116-pLe-miR-145 cells were inversely correlated with the lower expression levels of VEGF mRNA. P70S6K1 overexpression restored miR-145-inhibiting VEGF expression level (Figure 7A and B). Similarly overexpression of miR-145 decreased p70S6K1, HIF-1α and VEGF protein levels, while the forced expression of p70S6K1 restored miR-145-inhibited protein expression of p70S6K1, HIF-1α and VEGF (Figure 7C and D), indicating that p70S6K1 is upstream regulator of HIF-1α and VEGF. Immunohistochemistry assays showed that the overexpression of miR-145 decreased the number of CD-31 positive microvessels to 50%, while the forced expression of p70S6K1 restored the angiogenesis responses (Figure 7E). Similar trends were obtained with the expression levels of HIF-1α, p70S6K1 and VEGF in tumor tissues, which showed that miR-145 inhibited these protein levels. The p70S6K1 overexpression increased miR-145-inhibited HIF-1α, p70S6K1 and VEGF levels (Figure 7F). Taken together, these results given above, confirmed that miR-145 decreased HIF-1α and VEGF expression by targeting p70S6K1.
Figure 7.
MiR-145 inhibited HIF-1α and VEGF expression through p70S6K1 in vivo. (A and B) Total mRNAs were obtained from tumor tissues of mock+Ad-GFP, SC+Ad-GFP, miR-145+Ad-GFP, miR-145+Ad-p70S6K1 (Lanes 1–4, respectively). MiR-145 and VEGF mRNA levels were analyzed by TaqMan real-time RT-PCR using RNAs from 10 different tumors per treatment, and presented as mean ± SE. Asterisk and double asterisks indicate significant difference when compared control versus miR-145 treatment at P < 0.05 and P < 0.01, respectively. Hash indicates significant difference when compared miR-145 treatment with versus without p70S6K1 overexpression (P < 0.05). (C) The protein levels of p70S6K1, HIF-1α and VEGF were analyzed in the tumor tissues by western blotting. (D) The signals of p70S6K1, HIF-1α and VEGF protein expression were obtained from three replicate experiments, and presented as mean ± SE. Asterisk indicates significant difference when the control verus miR-145 treatment (P < 0.05). Hash and double hashes indicate significant difference when miR-145 treatment with versus without p70S6K1 overexpression at P < 0.05 and P < 0.01, respectively. (E) The blood vessels were stained using CD31 antibodies, and positive-stained blood vessels were counted in five areas with maximum number of microvessels under the microscope for each slide with 10 slides per experiment. The results are presented as mean ± SE (n = 10). Asterisk and hash indicate significant difference when compared the control versus miR-145, and miR-145 treatment with versus without p70S6K1 overexpression, respectively (P < 0.05). Bar, 100 µm. (F) The expression of HIF-1α, p70S6K1 and VEGF were determined in tumor tissues by immunohistochemistry with representative images showed. Bar, 50 µm.
MiR-145 expression is negatively correlated with the p70S6K1 level in human colon cancer tissues
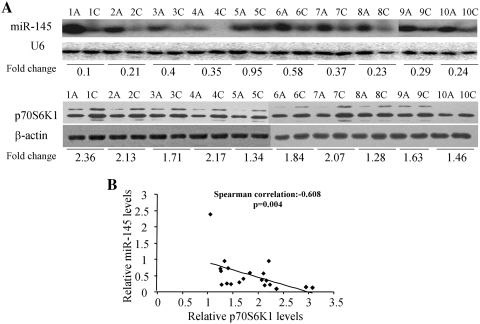
Twenty pairs of primary colon cancer tissues and adjacent normal colon tissues were tested for levels of miR-145 expression and U6 mRNA by northern blotting and for levels of p70S6K1 and β-actin protein expression by western blotting (Figure 8A and Supplementary Figure S2). The non-parametric correlation between miR-145 and p70S6K1 levels was assayed by SPSS software. Spearman's correlation test showed a significant inverse correlation between the miR-145 and p70S6K1 expression levels exist in colon cancer tissue with Spearman's correlation = −0.608, P = 0.004, confirming that higher expression levels of miR-145 were significantly associated with lower levels of p70S6K1 protein expression (Figure 8B).
Figure 8.
The p70S6K1 protein levels were negatively correlated with miR-145 levels in colon cancer tissues. (A) The expression levels of miR-145 were determined by northern blotting and normalized to U6 levels. The fold changes were obtained by the ratio of miR-145 level in cancer tissues (C) to miR-145 level in adjacent normal tissues. The p70S6K1 protein levels were assayed by immunoblotting and normalized to β-actin. The fold changes were obtained by the ratio of p70S6K1 to β-actin level. (B) Non-parametric correlation Spearman's correlation test was used to test the correlation between the levels of miR-145 and p70S6K1 from 20 pairs of colon cancer and adjacent normal tissues by SPSS software. The protein levels of p70S6K1 were negatively correlated with miR-145 levels in 20 colon cancer tissues (P < 0.05).
DISCUSSION
Growing lines of studies have demonstrated that miRNAs are important regulators of protein encoding genes. In addition to regulating cell reprogramming, neural development, cytoskeletal dynamics and smooth muscle cell fate and plasticity (4,6,18,50,51), miR-145 has been reported to be frequently downregulated in various kinds of cancers including colon cancer (9,11,12,14,31,32). Recently, an important discovery indicated that miR-145 was involved in the death-promoting regulatory loop of p53 (17,52,53), showing the crucial role of miR-145 in carcinogenesis and cancer development. Our results show that compared with adjacent normal colon tissues, the miR-145 levels in colon cancer from clinical samples is downregulated. These results suggest that miR-145 plays an important role in colon cancer.
P70S6K1, one of the downstream targets of mTOR, functions as a key regulator in protein synthesis, thus controlling various cellular functions such as cell proliferation, cell cycle and cell apoptosis. The activation and/or overexpression of p70S6K1 is a hallmark of cancer. This is the first study to show that p70S6K1 is negatively regulated by miR-145 at the post-transcriptional level by binding a specific target site within the 3′-UTR (Table 1 and Figure 2). Overexpression of miR-145 in human colon cancer cell lines SW480 and SW1116 inhibits p70S6K1 at protein level, further confirming that miR-145 targets p70S6K1 protein. Some clinical studies have shown that miR-145 is downregulated in colon cancers (12,31,32,54) and miR-145 can inhibit tumor growth (31,32). However, there was opposite conclusion in a different study showing that miR-145 has oncogenic potential in metastatic colorectal cancer (55). In this study, we showed that higher levels of miR-145 are closely correlated with lower expressions of p70S6K1 in human colon carcinoma tissues, indicating that miR-145 may inhibit p70S6K1 expression in clinical cancer tissues.
HIF-1α and VEGF are important regulators in tumor angiogenesis, which is required for tumorigenesis and cancer development (20,56). HIF-1α regulates VEGF at the transcriptional level by binding to its promoter (23). Our previous studies demonstrated that p70S6K1 is the upstream regulator of HIF-1 and VEGF in controlling tumor growth and angiogenesis in ovarian, lung and prostate cancers (45,48,49). Consistent with the inhibitory effect on p70S6K1, the overexpression of miR-145 also suppresses HIF-1α and VEGF expression in this study. Furthermore, p70S6K1 overexpression in colon cancer cells expressing pLe-miR-145 restored miR-145-inhibiting HIF-1α and VEGF levels, suggesting that miR-145 controls HIF-1α and VEGF expression by targeting p70S6K1. In vivo studies showed that miR-145 inhibited colon cancer growth, while forced expression of p70S6K1 restored tumor growth. The expression levels of p70S6K1 in patient colon cancer samples were also reversely related with miR-145 levels. Consistent with our prediction, miR-145 expression inhibited angiogenesis by decreasing p70S6K1, HIF-1α and VEGF levels in tumor tissues, while p70S6K1 overexpression rescued miR-145-inhibited p70S6K1, HIF-1α and VEGF expression, and tumor angiogenesis. Taken together, these in vitro and in vivo results suggest that miR-145 is a tumor suppressor, which inhibits tumor growth and angiogenesis through the inhibition of HIF-1α and VEGF expression by targeting p70S6K1.
In summary, our present study suggests that miR-145 negatively regulates p70S6K1 expression at the post-transcriptional level via a specific target motif at nt 807–825 of the p70S6K1-3′-UTR, miR-145 suppresses HIF-1α and VEGF expression by targeting p70S6K1, and inhibits tumor growth and angiogenesis. We also demonstrate that levels of miR-145 expression in resected patient colon tumor tissues are much lower than adjacent normal tissues, and are inversely correlated with the expression levels of p70S6K1 protein in the tumors. These results implicate that our results are clinically relevant, and miR-145 overexpression and the inhibitory strategies against p70S6K1/HIF-1/VEGF signaling pathway may be a rationale for therapeutic applications in colon cancer in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Basic Research Program of China (2007CB947002, 2007CB947001, in part); National Natural Science Foundation of China (30570962 and 30871296, in part); Key Basic Research Program of the Jiangsu Higher Education Institutions of China (09KJA310001, in part); National Cancer Institute, NIH (R01CA109460, in part). Funding for open access charge: National Key Basic Research Program of China (2007CB947002).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 4.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 5.le SC, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–948. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Iorio MV, Visone R, Di LG, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, Ma Y, Fiering S, Memoli V, Li H, et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin. Cancer Res. 2009;15:1177–1183. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 13.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int. J. Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 14.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 15.Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, Chang YS, Chen SJ. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br. J. Cancer. 2009;100:1002–1011. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int. J. Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 18.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folkman J. Tumor angiogenesis: therapeutic implications. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 20.Plate KH, Breier G, Weich HA, Risau W. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature. 1992;359:845–848. doi: 10.1038/359845a0. [DOI] [PubMed] [Google Scholar]
- 21.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 22.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat. Rev. Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 25.Jiang BH, Liu LZ. Role of mTOR in anticancer drug resistance: perspectives for improved drug treatment. Drug Resist. Updat. 2008;11:63–76. doi: 10.1016/j.drup.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldo P, Cecco S, Giacomin E, Lazzarini R, Ros B, Marastoni S. mTOR pathway and mTOR inhibitors as agents for cancer therapy. Curr. Cancer Drug Targets. 2008;8:647–665. doi: 10.2174/156800908786733513. [DOI] [PubMed] [Google Scholar]
- 27.Liu LZ, Zheng JZ, Wang XR, Jiang BH. Endothelial p70 S6 kinase 1 in regulating tumor angiogenesis. Cancer Res. 2008;68:8183–8188. doi: 10.1158/0008-5472.CAN-08-0819. [DOI] [PubMed] [Google Scholar]
- 28.Skinner HD, Zheng JZ, Fang J, Agani F, Jiang BH. Vascular endothelial growth factor transcriptional activation is mediated by hypoxia-inducible factor 1alpha, HDM2, and p70S6K1 in response to phosphatidylinositol 3-kinase/AKT signaling. J. Biol. Chem. 2004;279:45643–45651. doi: 10.1074/jbc.M404097200. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Tan D, Zhang Z, Liang JJ, Brown RE. Activation of Akt-mTOR-p70S6K pathway in angiogenesis in hepatocellular carcinoma. Oncol. Rep. 2008;20:713–719. [PubMed] [Google Scholar]
- 30.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 31.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 32.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 33.Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, Zhou H, Zhao RC. miR-145 inhibits breast cancer cell growth through RTKN. Int. J. Oncol. 2009;34:1461–1466. [PubMed] [Google Scholar]
- 35.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70:378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsson E, Fredlund FP, Heldin J, Barkefors I, Bondjers C, Genove G, Arrondel C, Gerwins P, Kurschat C, Schermer B, et al. Discovery of microvascular miRNAs using public gene expression data: miR-145 is expressed in pericytes and is a regulator of Fli1. Genome Med. 2009;1:108. doi: 10.1186/gm108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostenfeld MS, Bramsen JB, Lamy P, Villadsen SB, Fristrup N, Sorensen KD, Ulhoi B, Borre M, Kjems J, Dyrskjot L, et al. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene. 2010;29:1073–1084. doi: 10.1038/onc.2009.395. [DOI] [PubMed] [Google Scholar]
- 38.Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA. Biol. 2011;8:125–131. doi: 10.4161/rna.8.1.14259. [DOI] [PubMed] [Google Scholar]
- 39.Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br. J. Cancer. 2010;102:883–891. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, Hu J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J. Exp. Clin. Cancer Res. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotte M, Mohr C, Koo CY, Stock C, Vaske AK, Viola M, Ibrahim SA, Peddibhotla S, Teng YH, Low JY, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene. 2010;29:6569–6580. doi: 10.1038/onc.2010.386. [DOI] [PubMed] [Google Scholar]
- 42.Gregersen LH, Jacobsen AB, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS ONE. 2010;5:e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Guo H, Qian G, Ge S, Ji H, Hu X, Chen W. MiR-145, a new regulator of the DNA fragmentation factor-45 (DFF45)-mediated apoptotic network. Mol. Cancer. 2010;9:211. doi: 10.1186/1476-4598-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Guo H, Zhang H, Wang H, Qian G, Fan X, Hoffman AR, Hu JF, Ge S. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu LZ, Hu XW, Xia C, He J, Zhou Q, Shi X, Fang J, Jiang BH. Reactive oxygen species regulate epidermal growth factor-induced vascular endothelial growth factor and hypoxia-inducible factor-1alpha expression through activation of AKT and P70S6K1 in human ovarian cancer cells. Free Radic. Biol. Med. 2006;41:1521–1533. doi: 10.1016/j.freeradbiomed.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 47.Carayol N, Katsoulidis E, Sassano A, Altman JK, Druker BJ, Platanias LC. Suppression of programmed cell death 4 (PDCD4) protein expression by BCR-ABL-regulated engagement of the mTOR/p70 S6 kinase pathway. J. Biol. Chem. 2008;283:8601–8610. doi: 10.1074/jbc.M707934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Q, Liu LZ, Fu B, Hu X, Shi X, Fang J, Jiang BH. Reactive oxygen species regulate insulin-induced VEGF and HIF-1alpha expression through the activation of p70S6K1 in human prostate cancer cells. Carcinogenesis. 2007;28:28–37. doi: 10.1093/carcin/bgl085. [DOI] [PubMed] [Google Scholar]
- 49.Liu LZ, Fang J, Zhou Q, Hu X, Shi X, Jiang BH. Apigenin inhibits expression of vascular endothelial growth factor and angiogenesis in human lung cancer cells: implication of chemoprevention of lung cancer. Mol. Pharmacol. 2005;68:635–643. doi: 10.1124/mol.105.011254. [DOI] [PubMed] [Google Scholar]
- 50.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng L, Carter AD, Childs SJ. miR-145 directs intestinal maturation in zebrafish. Proc. Natl Acad. Sci. USA. 2009;106:17793–17798. doi: 10.1073/pnas.0903693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl Acad. Sci. USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death. Differ. 2009;17:246–254. doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC. Cancer. 2009;9:374. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.