Abstract
Although microRNAs (miRNAs) are important regulators of gene expression, the transcriptional regulation of miRNAs themselves is not well understood. We employed an integrative computational pipeline to dissect the transcription factors (TFs) responsible for altered miRNA expression in ovarian carcinoma. Using experimental data and computational predictions to define miRNA promoters across the human genome, we identified TFs with binding sites significantly overrepresented among miRNA genes overexpressed in ovarian carcinoma. This pipeline nominated TFs of the p53/p63/p73 family as candidate drivers of miRNA overexpression. Analysis of data from an independent set of 253 ovarian carcinomas in The Cancer Genome Atlas showed that p73 and p63 expression is significantly correlated with expression of miRNAs whose promoters contain p53/p63/p73 family binding sites. In experimental validation of specific miRNAs predicted by the analysis to be regulated by p73 and p63, we found that p53/p63/p73 family binding sites modulate promoter activity of miRNAs of the miR-200 family, which are known regulators of cancer stem cells and epithelial–mesenchymal transitions. Furthermore, in chromatin immunoprecipitation studies both p73 and p63 directly associated with the miR-200b/a/429 promoter. This study delineates an integrative approach that can be applied to discover transcriptional regulatory mechanisms in other biological settings where analogous genomic data are available.
INTRODUCTION
Regulation of gene expression at the post-transcriptional level is governed in part by microRNAs (miRNAs), which are approximately 22 nucleotide non-protein-encoding RNAs that modulate the stability and/or translation of messenger RNAs (mRNAs) via partially complementary base-pairing interactions (1). Most microRNAs are transcribed by RNA polymerase II (2), and miRNA expression can be regulated by transcription factor (TF) binding sites present in their promoters (3–7). However, for the majority of miRNAs, promoters have not been defined and the TF binding sites upstream of these miRNA loci have not been experimentally tested.
Dysregulation of miRNA expression is common in human disease and contributes to pathology, since miRNAs regulate significant disease-relevant processes such as cell division, differentiation, and apoptosis (8,9). In addition, in certain cancer contexts, the pattern of miRNA expression captures important features of the developmental origin of cancers (10) and may predict the course of disease (11). However, the mechanisms underlying miRNA dysregulation are not clear, in part because the transcriptional regulation of most miRNAs is not well characterized.
In this study, we implemented an integrative computational approach to dissect the transcriptional regulation of miRNAs. We focused on the dysregulation of miRNAs in ovarian carcinoma of the serous histologic sub-type, which has a high mortality and accounts for approximately two-thirds of ovarian carcinomas. Although a subset of miRNAs dysregulated in ovarian carcinomas is associated with changes in genomic copy number and epigenetic modifications, for many miRNAs additional, unknown mechanisms appear to contribute to the reprogramming of miRNA expression (12,13). We therefore sought to discover the TFs that may drive the dysregulation of miRNAs in ovarian carcinoma. We implemented a computational pipeline to annotate miRNA transcription start sites (TSS) and putative promoter regions, and then to identify the TFs with binding sites enriched in the promoters of overexpressed miRNAs in ovarian carcinoma. This approach yields putative regulatory interactions between TFs and miRNA promoters for subsequent experimental validation.
We report here that the best candidate driver of miRNA overexpression in ovarian carcinoma is the p53/p63/p73 family of TFs. Although p53 has been shown to transactivate several miRNAs, including the miR-34 family (14–17), the transcriptional regulation of miRNA genes by p73 and p63 has not been well-described. Further analysis using data from The Cancer Genome Atlas (TCGA) suggested that, in ovarian carcinoma, p73 and p63 are primarily responsible for the altered expression of miRNAs with p53 family binding sites. We experimentally validated our approach by confirming that p73 and p63 directly regulate transcription of the miR-200 family, a novel target predicted by our analysis that is an important regulator of epithelial–mesenchymal transitions (EMTs) and of the cancer stem cell phenotype (18–22).
This study illustrates how an integrative computational analysis can identify new regulatory interactions between TFs and miRNAs. We also provide a resource by defining putative miRNA promoters and associating TF binding sites with these miRNA promoters on a genome-wide scale, and we discuss how our approach is broadly applicable to dissect TF–miRNA regulatory networks in other systems where miRNA expression data from distinct physiologic states are available.
MATERIALS AND METHODS
Clinical materials
Normal primary human ovarian surface epithelial (HOSE) cell and serous ovarian carcinomas from which RNA was analyzed in this study have been previously described (23), and are described in Supplementary Data. HOSE specimens were obtained under a protocol that was approved by the Research Review Committee and Internal Review Board at the Fox Chase Cancer Center. All ovarian carcinoma tissue specimens were collected by the Pacific Ovarian Cancer Research Consortium Repository under a protocol approved by the Fred Hutchinson Cancer Research Center Institutional Review Board.
Analysis of miRNA microarray data generated in the current study
We analyzed miRNA microarray data generated on a locked nucleic acid (LNA) probe-based platform designed to profile 480 human miRNAs (Exiqon, Inc.) (23). Array quality control and data normalization was performed as previously described (23). We defined expressed miRNAs as the miRNAs expressed in at least two of the four normal or at least four of the 16 serous ovarian carcinomas. Non-specific filtering was performed as previously described (23), and genes that showed low variability across samples were removed from the analysis, which yielded 181 genes for downstream analyses. Differentially expressed miRNAs were identified by considering miRNAs to be over- or underexpressed in carcinoma compared to normal if log2 fold change was greater than 1.0 and adjusted P < 0.01.
Identification of TFs commonly expressed in serous ovarian carcinoma
We analyzed data from Hendrix et al. (24), where Affymetrix GeneChip Human Genome U133 Array Set HG-U133A microarrays were used to profile gene expression across a panel of 4 normal ovaries and 41 serous ovarian carcinoma tissues. The carcinoma tissues comprised the following stages: Stage IA (n = 1), Stage IC (n = 3), Stage II (n = 2), Stage IIC (n = 2), Stage III (n = 2), Stage IIIB (n = 1), IIIC (n = 25), IIID (n = 1) and IV (n = 5) and the following grades: well-differentiated (n = 2), moderately differentiated (n = 5), poorly differentiated (n = 18), grade information not available (n = 16). All samples were selected to contain at least 70% malignant epithelial cells. We considered genes to be expressed if they showed expression [as determined by Affymetrix PMA (Present/Marginal/Absent) calls] in at least 2 out of 4 normal samples or 8 out of 41 carcinoma samples. We identified 83 TFs expressed in normal ovary and/or serous ovarian carcinoma tissue specimens by intersecting expressed genes with all TFs in tfbsConsFactors files downloaded from the UCSC genome browser (25,26). Out of these 83 TFs, 79 corresponded to binding sites found upstream of miRNAs overexpressed in ovarian carcinoma.
Annotation of putative miRNA promoters and their TF binding sites
miRNA coordinates were downloaded from miRBase (version 10) (27) and used to map miRNAs to their genomic locations as described in Supplementary Data. To assign TSS for each miRNA locus, we used RefSeq, AceView, expressed sequence tags (ESTs) and Eponine predictions downloaded from the UCSC genome browser (hg 18 version of the genome assembly) (26). If both 5′ and 3′ ESTs were available from the same clone and formed a transcript containing the miRNA, the miRNA was considered expressed by this transcript and its TSS was the 5′-end of the EST.
We examined the putative promoters of each miRNA/miRNA locus for conserved TF binding sites defined in the UCSC files tfbsConsSites and tfbsConsFactors (26), as described in Supplementary Data. The conservation of binding sites is based on human/mouse/rat multiple alignments and the score is computed using the Transfac Matrix Database (v7.0) (28).
Enrichment of TF binding sites in promoters of overexpressed miRNA loci
We performed a Fisher's exact test for each of the 79 expressed TFs to compare the number of TF binding sites in the promoters of overexpressed miRNA loci versus control miRNA loci. To assess the false discovery rate, predicted Q-values were estimated from P-values (derived from Fisher's exact test) by using the ‘Q-value’ package in R (29).
Correlations between expression of TFs and expression of miRNAs in TCGA data set
We analyzed data from 253 serous ovarian carcinoma specimens from TCGA. Serous ovarian carcinoma tissue specimens in TCGA are generally from patients with advanced stage and high-grade disease. Clinical data on stage and grade was available on 217 of the 253 specimens we studied, and confirmed that the vast majority of these were high-grade (202 were poorly differentiated and 13 were moderately differentiated) and advanced stage [Stage IIB (n = 2), Stage IIC (n = 4), Stage IIIA (n = 2), Stage IIIB (n = 9), Stage IIIC (n = 161) and Stage IV (n = 39)]. Data on percentage malignant epithelium was available for 202 of the 253 specimens we studied: >75% malignant epithelial cells (n = 150 specimens), ≥50% and <75% (n = 43 specimens), <50% (n = 9 specimens). To derive correlations between TF and miRNA expression, we retrieved Agilent gene expression microarray and miRNA microarray data from the TCGA for 253 serous ovarian carcinomas, for which both gene expression (run on AgilentG4502A_07_3) and miRNA expression profiling data were available (http://cancergenome.nih.gov/index.asp). Agilent Level 1 microarray expression data were used for both miRNA and mRNA analyses. We used 447 miRNAs for calculating correlations; all of these miRNAs had at least one TF binding site associated with their promoters as determined by our method. All TCGA Level 1 files used to calculate correlations are listed in Supplementary Table S1. Correlation between the expression of the miRNAs and TFs was assessed by calculating Pearson correlation coefficients. Bioconductor packages ‘limma’ and ‘ltm’ were used to process the raw signal data and to calculate P-values for correlation between the expression of miRNAs and TFs, respectively (30,31).
Sequencing of TP53
All coding and flanking regulatory regions for exons 4-10 of TP53 were sequenced as previously described (32).
Cell culture
The 2008 ovarian carcinoma cell line was maintained in RPMI (Invitrogen) with 10% fetal bovine serum (Atlanta Biologicals) under 5% CO2. All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed on 2008 ovarian carcinoma cells as previously described (33). Sheared chromatin was incubated with 12 µg of antibody overnight at 4°C and then incubated with 40 µl prewashed Protein G agarose beads (Millipore). Antibodies used for immunoprecipitation included 4A4 (Santa Cruz) for p63, 259A (Imgenex) for p73, and normal mouse IgG (Santa Cruz) to control for non-specific binding. Quantitative PCR (qPCR) was performed as described in Supplementary Data using the primers listed in Supplementary Table S2. Triplicate immunoprecipitations were performed using each antibody, using independent batches of chromatin.
Promoter assays
Firefly luciferase promoter activity reporter plasmids were generated by PCR amplification of the region of interest from human genomic DNA (BioLine) followed by subcloning into the MluI and BglII sites of the pGL3-Enhancer vector (Promega), as described in Supplementary Data. Primers used are in Supplementary Table S3. To assay promoter activity, 2008 cells were plated in 96-well plates at 8 × 103 cells/well 24 h prior to transfection. The cells were co-transfected with 100 ng of each firefly luciferase promoter reporter vector, or pGL3-Enhancer as a control, and 25 ng of pRL-TK (Promega). Twenty-four hours after transfection, firefly and Renilla luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega). Promoter activity was calculated as the ratio of firefly to Renilla luciferase signal.
RESULTS
Overview of computational pipeline for the identification of TFs regulating miRNA expression
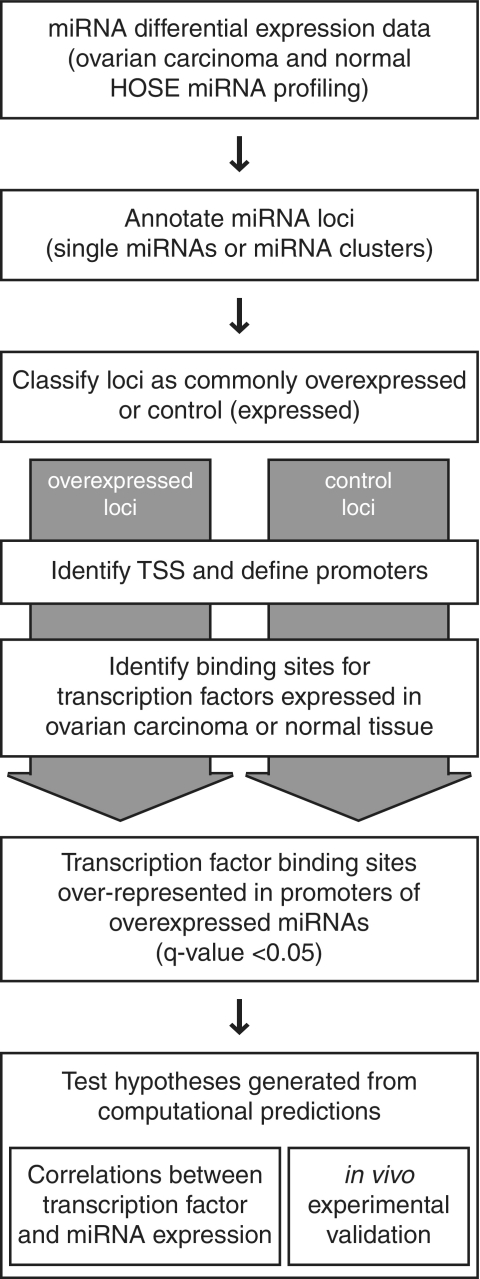
We implemented a computational pipeline to study the transcriptional regulation of miRNA expression (Figure 1). Briefly, the pipeline starts with miRNA differential expression data as input and classifies miRNAs into two categories: miRNAs that are overexpressed in the samples of interest and control miRNAs that are expressed but not overexpressed. Putative promoters of both the overexpressed and control miRNAs are then defined via genomic mapping and annotation of TSS using a series of criteria described below. After identifying the TFs expressed in the samples based on mRNA expression data, the number of predicted binding sites for each expressed TF in the promoters of both the overexpressed miRNA and control miRNA loci is determined. The output of the pipeline is TFs with binding sites overrepresented in the promoters of the overexpressed miRNAs, and these regulatory predictions can then be experimentally validated.
Figure 1.
Analysis pipeline for identifying candidate TFs responsible for miRNA overexpression. MicroRNA and mRNA expression profiling data were used to identify TF binding sites overrepresented in the promoters of miRNAs overexpressed in ovarian carcinoma. TF binding sites were based upon Transfac position weight matrices. (TSS, transcriptional start site; HOSE, human ovarian surface epithelial cells).
Identification of a set of miRNAs commonly differentially expressed in serous ovarian carcinoma
As input to our pipeline, we defined miRNAs that are differentially expressed in serous ovarian carcinomas compared to normal specimens. To avoid bias, we used multiple studies to identify miRNAs that are commonly altered in ovarian carcinoma. We previously profiled miRNA expression in primary serous ovarian carcinomas from 16 individuals and in cultured normal human ovarian surface epithelial (HOSE) cells from 4 other individuals using miRNA microarrays (23). We analyzed that data set here to identify miRNAs significantly dysregulated in serous ovarian carcinoma as compared to normal. We found 43 miRNAs to be overexpressed and 20 to be underexpressed in ovarian carcinoma (log2 fold change >1.0 and adjusted P < 0.01) (Supplementary Table S4). We then compared the results of this analysis and six other ovarian carcinoma miRNA profiling studies (13,34–38). We found 30 miRNAs to be significantly overexpressed in two or more of the entire set of seven studies (Supplementary Table S5), which should provide a more reliable list of miRNAs commonly overexpressed in serous ovarian carcinomas. We also identified a set of 18 commonly underexpressed miRNAs (Supplementary Table S6), although these did not yield statistically significant overrepresentation of any specific TF binding sites in subsequent analysis and are therefore not discussed further.
In order to define a set of control miRNAs for comparison, we used data from our microarray analysis of ovarian carcinoma and normal specimens to identify 238 expressed miRNAs, which includes the 30 commonly overexpressed miRNAs defined above. We then converted this list of expressed miRNAs to a list of miRNA loci, where each locus corresponds to a single promoter, which may drive expression of multiple miRNAs. Specifically, we mapped each miRNA to its genomic position, and miRNAs that are encoded at multiple locations were assigned to each locus separately. Because many miRNAs are found in clusters in single polycistronic transcripts (39,40), we annotated all miRNAs within a genomic cluster as a single locus. After accounting for the expansion of the number of miRNA locations as a result of miRNAs encoded at multiple loci, and contraction of this number because of miRNAs in clusters with shared promoters, the 238 miRNAs corresponded to 151 loci representing a miRNA or miRNA cluster that is expressed in serous ovarian carcinomas and/or normal samples (Supplementary Table S7). We divided the 151 loci into two classes: those that encode overexpressed miRNAs and those that do not. If a single miRNA within a cluster was overexpressed, the entire miRNA locus was classified into the overexpressed class for the purpose of this analysis. From this approach, we annotated 30 loci (miRNAs or miRNA clusters) that were overexpressed in serous ovarian carcinomas, and we used the remaining 121 loci as the control set in all further analyses (Supplementary Table S8).
Annotation of miRNA TSSs and identification of candidate TFs controlling miRNA expression
To define putative promoter regions for subsequent analysis, we first annotated the TSS of each overexpressed or control miRNA locus (Supplementary Table S7). We found 43% of miRNAs to be located within and in the same orientation as a RefSeq gene (41). The TSS for these miRNAs was assumed to be the same as for the host gene, as it has been shown that miRNAs within host genes are generally co-transcribed from a shared promoter (42,43). For miRNA genes that did not match to RefSeq, we used AceView, which provides comprehensive transcriptional evidence from full length cDNAs and ESTs (44), to identify the TSS of 38% of the miRNAs. We next used predictions by Eponine (45) to define the TSS of 7% of the miRNAs and EST clones to locate the TSS of 3% of the miRNAs. For the remaining 9% of miRNAs whose TSS could not be found by the above methods, the position 500 bp upstream of the miRNA was taken as the TSS. In the case of miRNAs that lie in genomic clusters, the TSS of the most 5′ miRNA was assigned to all miRNAs in the cluster, because such miRNAs are expressed as a single primary transcript from a shared promoter (40).
To ensure our analysis would involve only TFs commonly expressed in serous carcinoma and/or normal ovarian tissue, we used mRNA microarray data from serous ovarian carcinomas and from normal ovarian tissues available in the Gene Expression Omnibus (GSE6008) (24) to identify 79 TFs expressed in normal ovary and/or serous ovarian carcinomas. For each of the 151 miRNA (or miRNA cluster) loci identified above, we examined a region extending from 5000 bp upstream to 500 bp downstream of the TSS for predicted TF binding sites (Supplementary Table S8). If a miRNA was less than 500 bp downstream from the TSS, we instead used the distance between the miRNA and TSS for the downstream boundary of the region of interest. We only included binding sites conserved between human, mouse and rat, as annotated by the UCSC genome browser (26), for the 79 expressed TFs.
To determine which TFs are likely to drive miRNA overexpression in ovarian carcinoma, we performed a Fisher's exact test for each of the 79 TFs to compare the number of binding sites in the putative promoters of the 30 overexpressed miRNA loci with the number of sites for the 121 control miRNA loci (Supplementary Table S9). We found an overrepresentation of binding sites for the tumor protein 53 (p53) family, early growth response 2 (EGR2), and SP1 in the promoters of miRNAs overexpressed in serous ovarian carcinoma [Q-value < 0.05, (29); Table 1]. The most significant Q-value, 0.005, was found for the p53 family, which consists of three proteins (i.e. p53, p63 and p73) that recognize the same consensus binding site (46,47). We identified 15 p53 family binding sites among the 30 overexpressed miRNA promoter loci versus 17 such sites in the 121 control miRNA promoter loci (Table 1). Thus, this analysis suggests that the three members of the p53 family, as well as EGR2 and SP1, may be drivers of miRNA dysregulation in serous ovarian carcinoma.
Table 1.
TF binding sites enriched in promoters of miRNAs overexpressed in serous ovarian carcinoma
| TF binding site | Promoters of overexpressed miRNA |
Promoters of expressed miRNAs |
P-value | Q-value | ||
|---|---|---|---|---|---|---|
| With site | Without site | With site | Without site | |||
| p53/p63/p73 | 15 | 15 | 17 | 104 | 6.83E-05 | 0.0054 |
| EGR2 | 10 | 20 | 9 | 112 | 0.0006 | 0.0246 |
| SP1 | 10 | 20 | 10 | 111 | 0.0011 | 0.0283 |
| NFKB1 | 10 | 20 | 16 | 105 | 0.0145 | 0.2856 |
| RELA | 9 | 21 | 14 | 107 | 0.0206 | 0.2954 |
| NR2F1 | 6 | 24 | 7 | 114 | 0.0233 | 0.2954 |
| EGR3 | 8 | 22 | 11 | 110 | 0.0262 | 0.2954 |
| PAX4 | 14 | 16 | 31 | 90 | 0.0429 | 0.4036 |
| XBP1 | 7 | 23 | 10 | 111 | 0.0460 | 0.4036 |
MicroRNAs were classified into 30 loci overexpressed in ovarian carcinoma and a control group of 121 loci expressed in carcinoma and/or normal HOSE samples. The number of promoters with binding sites for a given TF was compared between the two classes. A Fisher's exact test was performed for each of the 79 TFs that are expressed in carcinoma and/or normal samples. P-values computed by Fisher's exact test were converted to Q-values. TFs with P < 0.05 are shown, while those with Q < 0.05 are in bold.
Correlation between expression of TFs and expression of miRNAs in TCGA data
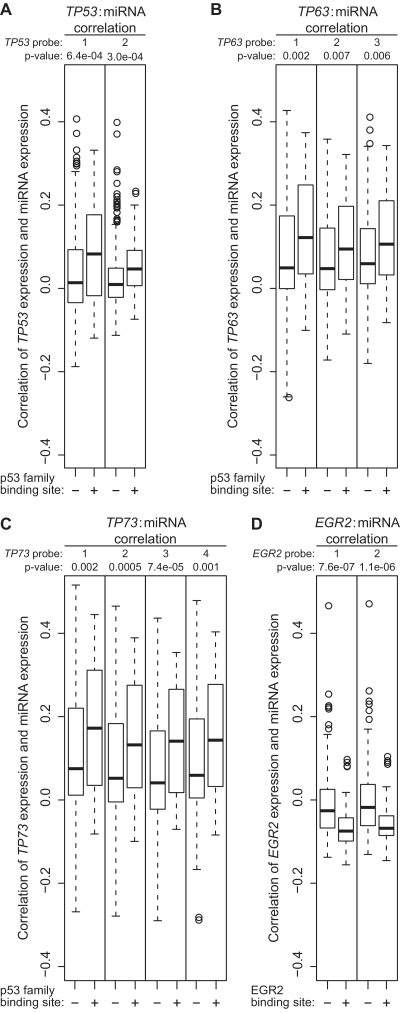
To determine if expression of the candidate miRNA TFs we identified correlates with expression of miRNAs with binding sites for those TFs, next we employed a larger, independent data set from TCGA, which provides comprehensive genomic profiling data from high-grade serous ovarian carcinomas. We selected samples from TCGA for which both miRNA and mRNA data were available for the same carcinoma tissue specimen. There were 253 such samples available. We calculated Pearson correlations between the expression of each of the five TFs identified above and the expression of each of 447 miRNAs in the 253 TCGA samples. We then compared correlation coefficients between the miRNAs whose promoters contain binding sites for a given TF and the remaining miRNAs lacking binding sites. We found no significant difference in the correlation of SP1 and miRNAs with or without SP1 binding sites (data not shown). However, the remaining four TFs (i.e., TP53, TP63, TP73, and EGR2) showed significantly higher expression correlations with miRNAs bearing their respective binding sites versus those without (P < 0.01, t-test) (Figure 2). This analysis suggests that our pipeline yields functional regulators of miRNA expression. Interestingly, we found significant negative correlations between the expression of both EGR2 probes and the expression of miRNAs with EGR2 binding sites, which suggests that EGR2 may serve as a transcriptional repressor (Figure 2D).
Figure 2.
Expression of miRNAs with TF binding sites is correlated with TF expression in TCGA ovarian carcinoma specimens. (A–D) Data from 253 TCGA serous ovarian carcinomas were used to calculate Pearson correlations between expression of the indicated TFs and expression of 447 miRNAs. Boxplots show the correlations using mRNA expression data from two TP53 probes (A), three TP63 probes (B), four TP73 probes (C), and two EGR2 probes (D). miRNAs were classified by the presence (+) or absence (−) of the indicated TF binding site in the miRNA promoter. For each probe, t-tests were used to compare mean correlation values of the two miRNA classes, and P-values computed from the t-tests are shown. Probes: TP53-1 (NM_000546_2_2048), TP53-2 (A_23_P26810), TP63-1 (A_23_P91979), TP63-2 (A_32_P114473), TP63-3 (A_32_P114475), TP73-1 (A_23_P74078), TP73-2 (A_23_P74081), TP73-3 (A_24_P413470), TP73-4 (NM_005427_1_1638), EGR2-1 (A_23_P46935) and EGR2-2 (A_23_P46936).
All three members of the p53 family featured probes that showed positive, significantly higher correlations with miRNAs bearing p53 family binding sites than those without (Figure 2A–C). Three out of seven TP63 probes and one out of three TP53 probes showed lower correlations that were not statistically significant (data not shown), which may be related to sequence-specific variation in probe performance. Of the three members of the p53 family, TP73 expression showed the strongest positive correlations with miRNAs bearing p53 family binding sites (Figure 2C). Taken together, these results suggest that members of the p53 family, and in particular p73, may drive the overexpression of a panel of miRNAs in ovarian carcinoma.
Overexpression of miRNAs with p53 binding sites in their promoters is not associated with the mutational status of TP53 in serous ovarian carcinomas
While expression of p53 mRNA appeared to correlate with miRNA expression, regulation of p53 occurs primarily at the protein level. Therefore, we sought to determine whether changes in the activity of the p53 protein may affect the expression of miRNAs bearing p53 family binding sites. Mutation is the leading cause of p53 dysfunction in serous ovarian carcinoma, as 60–70% of serous ovarian cancers are known to have TP53 mutations (48,49), which in general ablate the ability of p53 to transactivate its target genes. However, because mutations to TP53 can result in a dominant negative protein that alters the activity of the other p53 family members, we examined whether TP53 mutational status affects expression of miRNAs overexpressed in ovarian carcinoma. Somatic mutation data for ovarian carcinomas in TCGA were not available. We therefore determined the p53 mutation status of the 16 ovarian carcinoma specimens that we previously used for identifying differentially expressed miRNAs. We sequenced all coding and flanking regulatory regions for exons 4-10 of TP53, which include mutation hotspots and also coincide with the most highly conserved region of the gene (50). We found non-synonymous TP53 mutations accompanied by loss of the wild-type allele in 9 out of our 16 ovarian carcinoma specimens (Supplementary Table S10). Comparing the expression of miRNAs in the TP53 mutant versus wild-type specimens demonstrated no significant difference in the expression of the 17 overexpressed miRNAs that are encoded within the 15 overexpressed loci with p53 binding sites (Supplementary Figure S1). While these results do not exclude p53 as a possible driver of differential miRNA expression, this analysis suggests that p53 is not the primary regulator of these miRNAs in ovarian carcinoma.
p73 and p63 as putative positive regulators of miRNA expression in ovarian cancer
Our miRNA expression correlation analyses from TCGA data suggested that p73, and potentially p63, may drive the overexpression of miRNAs in ovarian carcinoma (Figures 2B and C). To identify specific miRNAs that may be regulated by p73 or p63 for further study, we used the P-values from the Pearson correlations to rank the miRNAs that were overexpressed in ovarian carcinoma and contained p53 family binding sites. We found multiple miRNAs with significant positive correlation with TP73 or TP63 expression (Table 2). A complete list of miRNAs showing positive correlations (Pearson correlation coefficient >0.25 and P < 0.05) with all p73 and p63 probes is provided in Supplementary Tables S11 and S12. To provide a biologically relevant reference for interpreting these correlation values, we also determined the correlations of 88 host gene-intronic miRNA pairs using the same data set of 253 TCGA samples. Intronic miRNAs (i.e., lying in host genes on the same strand) are generally co-transcribed with their host gene and therefore show strong correlations in expression with their host gene. Among miRNA-host gene pairs analyzed by the same criteria (correlation coefficients >0.25 and P < 0.05), host gene-intronic miRNA pairs showed an average correlation of 0.37 (±0.16). The correlations between the expression of p73 and p63 and miRNAs in Table 2 range between 0.25 and 0.39, and thus reflect a similar magnitude of correlation coefficients as observed with the host gene-intronic miRNA correlations.
Table 2.
Correlations between TP73 or TP63 and miRNA expression in TCGA ovarian carcinomas
| miRNA | Correlation | P-value |
|---|---|---|
| TP73 | ||
| miR-107 | 0.389 | 1.39E-10 |
| miR-200a | 0.389 | 1.47E-10 |
| miR-29c | 0.383 | 2.77E-10 |
| miR-23a | 0.373 | 8.72E-10 |
| miR-24 | 0.371 | 1.17E-09 |
| miR-200c | 0.364 | 2.36E-09 |
| miR-141 | 0.355 | 6.12E-09 |
| miR-30c | 0.348 | 1.30E-08 |
| miR-29b | 0.346 | 1.57E-08 |
| miR-429 | 0.337 | 4.03E-08 |
| miR-103 | 0.332 | 6.62E-08 |
| miR-200b | 0.318 | 2.31E-07 |
| miR-28-5p | 0.303 | 9.31E-07 |
| miR-30e | 0.303 | 9.41E-07 |
| miR-212 | 0.302 | 9.77E-07 |
| miR-93 | 0.301 | 1.06E-06 |
| miR-7 | 0.295 | 1.85E-06 |
| miR-106b | 0.291 | 2.46E-06 |
| let-7c | 0.289 | 2.88E-06 |
| miR-20a | 0.284 | 4.62E-06 |
| miR-130a | 0.278 | 7.05E-06 |
| miR-34a | 0.276 | 8.11E-06 |
| miR-27a | 0.274 | 9.62E-06 |
| miR-135b | 0.271 | 1.25E-05 |
| miR-219-5p | 0.266 | 1.77E-05 |
| miR-19a | 0.259 | 3.02E-05 |
| TP63 | ||
| miR-200a | 0.322 | 1.64E-07 |
| miR-29c | 0.320 | 1.91E-07 |
| miR-30e | 0.316 | 2.94E-07 |
| miR-30c | 0.311 | 4.50E-07 |
| miR-107 | 0.310 | 5.06E-07 |
| miR-429 | 0.294 | 1.99E-06 |
| miR-141 | 0.293 | 2.09E-06 |
| miR-24 | 0.287 | 3.50E-06 |
| miR-103 | 0.283 | 4.66E-06 |
| miR-29b | 0.278 | 7.10E-06 |
| miR-200c | 0.275 | 9.19E-06 |
| miR-200b | 0.263 | 2.20E-05 |
| miR-93 | 0.257 | 3.57E-05 |
Correlation coefficients for expression of miRNAs with TP73 expression and TP63 expression across 253 TCGA ovarian carcinomas were calculated. MicroRNAs listed showed positive correlation coefficients >0.25, P < 0.05 and contain p53/p63/p73 binding sites in their promoters. MicroRNAs are ranked in order of increasing P-value (unadjusted). The miR-200 family miRNAs, indicated in bold, had significant positive correlations with TP73 and TP63 expression. Correlation coefficients shown were calculated using representative probes for TP73 (A_23_P74081) and TP63 (A_32_P114473).
p73 and p63 directly regulate miR-200 transcription
To experimentally validate our computational predictions, we examined the transcriptional regulation of the miR-200 family of miRNAs. Expression of all five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) was positively correlated with p73 and p63 (all with P < 2.2 × 10−5), with correlation coefficients ranging from +0.318 to +0.389 for p73, and from +0.263 to +0.322 for p63 (Table 2). Our previous work has implicated the miR-200 family in ovarian carcinoma pathogenesis (51), and the miR-200 family has a well-established role in governing EMTs and repressing tumor-initiating cells (i.e. cancer stem cells) (18–20,22) in a variety of other cancer contexts.
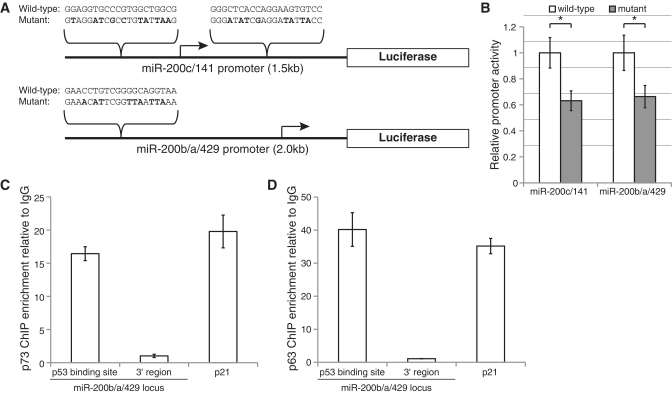
The miR-200 family miRNAs are transcribed from two chromosomal clusters: miR-200b/a/429 from chromosome 1, and miR-200c/141 from chromosome 12. The promoters of both miR-200 clusters contain p53 family binding sites. We first tested whether the predicted p53 family binding sites in the miRNA promoters modulate miR-200 transcription by employing a luciferase reporter system. We cloned a 2-kb fragment of the miR-200b/a/429 promoter, which contains one predicted p53 family binding site, upstream of the luciferase gene in the pGL3-Enhancer vector. Similarly, we also produced a miR-200c/141 promoter construct by cloning a 1.5-kb region of the miR-200c/141 promoter containing two predicted p53 binding sites into the vector. We then generated mutant versions of each promoter construct by mutating the consensus nucleotides in each predicted p53 family binding site, with the miR-200c/141 mutant promoter featuring mutations at both of the two predicted binding sites (Figure 3A). We transfected the wild-type and mutated constructs into 2008 ovarian carcinoma cells. Transfection with the wild-type promoter constructs led to increased luciferase activity as compared to the empty vector lacking a promoter, indicating that the minimal miR-200 promoters were functional (data not shown). To determine if the p53 family binding sites contribute to promoter activity, we compared the luciferase activity of the wild-type versus mutated promoters (Figure 3B). We found that mutation of the p53 binding sites of the miR-200b/a/429 and miR-200c/141 promoters each resulted in an ∼40% decrease in luciferase activity (P < 0.003, t-test), consistent with the notion that members of the p53 family directly regulate miR-200 transcription.
Figure 3.
p73 and p63 directly regulate transcription of the miR-200 miRNA families. (A) Schematic of the miR-200c/141 and miR-200b/a/429 promoter fragments cloned upstream of the luciferase gene in the pGL3-Enhancer vector. To generate a mutant version of each promoter, the p53 family consensus binding sites were mutated at the nucleotides shown in bold. (B) Activity of the indicated mutant and wild-type miR–200 promoter constructs transfected into 2008 ovarian carcinoma cells. Promoter activity is expressed relative to the wild-type construct for each promoter, and bars represent the mean relative activity ± SD of four replicates. Asterisk indicates P < 0.003, t-test. (C and D) Chromatin immunoprecipitation of p73 (C) and p63 (D) from 2008 cells. qPCR was used to assess the enrichment of the indicated regions relative to an IgG control chromatin immunoprecipitation. Enrichment of the p53 family binding sites and negative control regions 1 kb 3′ to the miRNAs at the miR–200b/a/429 loci are shown. Enrichment of the p21 promoter served as a positive control. Bars represent mean enrichment ± SD of technical replicates from a representative experiment that was repeated three times.
We next sought to determine which p53 family members are physically associated with the miR-200 promoters in ovarian carcinoma cells. We performed chromatin immunoprecipitations on the 2008 cell line using antibodies directed against p53, p63 or p73, as well as normal IgG as a control for non-specific binding. We did not find enrichment of p53 at several known targets, including p21 and MDM2, nor at the miR-200 promoter regions, which is consistent with western blots revealing that 2008 cells do not express stable p53 protein (data not shown). In the p73 and p63 immunoprecipitations, however, we found a strong enrichment of the miR-200b/a/429 p53 family binding site relative to the IgG control (Figure 3C and D). The levels of enrichment were similar to those seen at the p21 promoter, a known transcriptional target of the entire p53 family (46,52). As a further negative control for non-specific enrichment, we assayed the immunoprecipitates for a region 1-kb downstream of the 3′-end of miR-429, the last miRNA of the miR-200b/a/429 cluster. No enrichment was found at this control region, indicating that we were specifically enriching the promoter of the miR-200b/a/429 locus. When the locus on chromosome 12 corresponding to the miR-200c/141 promoter was analyzed in the p73 and p63 chromatin immunoprecipitates, no enrichment was observed compared to the IgG control (data not shown). Although such negative results are difficult to interpret, this observation may be a false negative due to the limitations of the sensitivity of chromatin immunoprecipitation, such as the limited set of promoter regions assayed by qPCR as well as potentially limited antibody accessibility related to chromatin structure or interference from transcriptional co-factors. As an additional negative control, we assayed the promoter of miR-29a/b-1, which is overexpressed in ovarian carcinomas yet in our computational analysis was not found to have p53 family TF binding sites. Our chromatin immunoprecipitation analysis found no enrichment of p63 or p73 at the miR-29a/b-1 promoter (Supplementary Figure S2). Taken together, our results demonstrate that both p73 and p63 associate with the miR-200b/a/429 promoter and may also contribute to the expression of the miR-200c/141 locus in ovarian carcinoma.
DISCUSSION
Our study demonstrates how an integrative computational approach can take advantage of increasingly comprehensive genomic profiling data sets to understand the transcriptional regulation of miRNAs. Although miRNA transcriptional regulation mechanisms have been investigated for specific miRNAs in the past, genome-scale studies have been limited, partially due to the lack of well-annotated miRNA primary transcripts and their corresponding transcriptional start sites. MicroRNAs are excised through a series of processing steps from primary transcripts, which are highly variable in length and can be several kilobases long; identifying the TSS for miRNAs at a genome scale is therefore not trivial. Although experimental methods using chromatin signatures to identify miRNA promoters have been useful in identifying approximate locations of miRNA TSS in some cases (53,54), such approaches have their limitations because miRNA expression is highly tissue specific and therefore experimental data applies to a relatively limited subset of miRNAs in specific cell line or tissue contexts. Our genome-wide approach relies heavily on transcript information derived from ESTs, both from EST clusters (AceView) and from ESTs derived from a common clone, to provide transcript-based evidence for defining the TSS of intergenic miRNAs. AceView is one of the most comprehensive assemblies of human transcriptome data, drawing cDNA sequences from dbEST, Genbank and RefSeq and representing a broad panel of tissue and cell type contexts. There are currently more than 8 million ESTs in dbEST and notably approximately 2 million 5′ capped ESTs in Genbank submitted by Kimura et al. (55). An important aspect of our study, therefore, is the genome-wide definition of presumptive miRNA TSS and promoters using such data, which provides a resource for further investigations of miRNA transcriptional regulation.
The key biological finding of this work is the identification of the p63 and p73 TFs as significant activators of miRNA overexpression in ovarian carcinoma. We found conserved TF binding sites for the p53/p63/p73 family upstream of 17 miRNA loci overexpressed in serous ovarian carcinoma, suggesting p63 and/or p73 may coordinately regulate an ensemble of miRNAs that could significantly modify the cellular gene expression landscape via repression of a cohort of mRNA targets. We also observed an overrepresentation of conserved EGR2 binding sites upstream of miRNA loci overexpressed in ovarian carcinoma, with 10 overexpressed miRNA loci containing EGR2 binding sites. In the TCGA ovarian carcinoma data set, EGR2 expression was significantly negatively correlated with the expression of miRNAs containing EGR2 binding sites. EGR2 has been suggested to be a tumor suppressor, based on the fact that it is frequently underexpressed in human cancers (including ovarian) and cancer cell lines (56,57). We hypothesize that the 10 miRNA loci mentioned above are normally repressed by EGR2, but that decreased EGR2 expression in ovarian carcinoma leads to their overexpression.
It is important to note that both p63 and p73 can be expressed as multiple isoforms that were not distinguishable on the expression platforms involved in this study. The isoforms of p63 and p73 can be divided into two classes: transactivating TAp63/TAp73 isoforms and the inhibitory ΔNp63/ΔNp73 isoforms. In ovarian carcinoma, multiple inhibitory isoforms of p73 and p63 are overexpressed and can be associated with poor prognosis (58,59). In contrast, transactivating TAp73 increases the response rate to chemotherapy in BRCA-1-associated ovarian carcinoma (60). We hypothesize that p73 and p63 may drive the expression of multiple miRNAs that contribute to tumor aggressiveness and/or treatment response. For instance, low levels of miR-200 family miRNA expression are associated with poor treatment response in ovarian carcinoma (61); thus, TAp73-mediated activation of miR-200 could serve as a mechanism for increased sensitivity to chemotherapy. Future studies will be required to determine the role of specific isoforms of p73 and/or p63 in determining miRNA transcription in ovarian carcinoma.
In experimental validation of the pipeline predictions, we focused on the miR-200 family because of its important roles in tumor development and progression. Members of the miR-200 family inhibit EMT and suppress tumor invasion by directly repressing the TFs Zeb1 and Zeb2 (18–20). Additionally, the miR-200 family has been shown to repress Bmi1 and Suz12, two essential components of polycomb repressor complexes that are responsible for the maintenance of tumor-initiating cells (21,22). Previous work on miR-200 regulation has largely focused on mechanisms of repression, including transcriptional inhibition and epigenetic modifications (18,62,63); mechanisms of transcriptional activation of this miRNA family are not well-understood. Our results with p63 and p73 shed light onto positive transcriptional regulation of the miR-200 family miRNAs, which has potential therapeutic value in the development of approaches aimed at modifying miR-200 family miRNA expression for the treatment of diverse forms of human cancer.
Although our work focused on miR-200 transcriptional regulation by p73 and p63, recent studies have identified different mechanisms by which the p53 family can modulate miR-200 expression. p63 can serve as a transcriptional regulator of Dicer to modulate miRNA maturation, including processing of the miR-200 family (64). Given our observation of p63 binding to the miR-200b/a/429 promoter, these studies suggest that p63 can act both directly and indirectly to alter miRNA expression, although the mechanism may be cellular context-dependent. Additionally, a recent report found that in mammary epithelial cells, p53 serves as a transcriptional activator of miR-200c, but not of the miR-200b/a/429 cluster (17). In ovarian carcinomas, TP53 is commonly mutated and thus unlikely to drive the overexpression of miR-200 that we detected in our samples; however, TP53 mutations can frequently yield dominant negative proteins that interfere with the actions of p63 and p73 through promoter competition and through the formation of inactive heterotetrameric complexes (65). Although we did not find a relationship between the TP53 mutational status of ovarian carcinomas and their expression of miRNAs with p53 family binding sites, we cannot exclude the role of p53 in the transcriptional regulation of the miR-200 family. In contrast, our results indicate that p63 and p73 are involved in overexpression of this miRNA family in ovarian carcinoma. Taken together with the report of p53-driven miR-200c expression in mammary epithelial cells, our study suggests that cellular context determines the relative importance of different p53 family members on the transcriptional activation of the miR-200 family.
Through our integrative analysis, we identified transcriptional mechanisms that govern miRNA expression in ovarian carcinoma and also found novel activators of miRNAs implicated in tumorigenesis. Our results indicate that p73 and potentially also p63 contribute to broad dysregulation of miRNA expression in ovarian carcinoma. Although we have directed our analysis here toward ovarian cancer, our approach can be broadly applied to other cancer types (e.g., TCGA data sets are expected to expand to include nearly 25 cancer types), or any biological state(s) in which mRNA and differential miRNA expression data are available (e.g., expression profiles across stages of development or across different cell differentiation states). Because different cancer and tissue types show unique mRNA and miRNA expression patterns, utilizing our computational pipeline with other systems may reveal distinct TFs that regulate miRNA activity in varied biological contexts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (PHS NRSA T32 GM07270 from the National Institute of General Medical Sciences to E.C.K.); the National Cancer Institute (Interdisciplinary Training in Cancer Research Grant CA80416 to K.G., R01 CA140323 to A.K.G., Chromosome Metabolism and Cancer Training Grant NCI CA09657 to J.D.A.); the Pacific Ovarian Cancer Research Consortium Ovarian SPORE Award (P50CA083636 to N.D.U. and M.T.); the caBIG In Silico Center of Excellence (contract #HHSN261200800001E to M.T.); the American Cancer Society (Fellowship to J.D.A.); the Department of Defense (Ovarian Cancer Career Development Award OC080159 to M.T.); Jaconette L. Tietze Young Scientist Award (to M.T.). Funding for open access charge: Pacific Ovarian Cancer Research Consortium Ovarian SPORE Award (NIH P50CA083636); Department of Defense Ovarian Cancer Career Development Award (OC080159 to M.T.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
E.C.K., K.G. and J.D.A. wrote the manuscript. K.G. designed and implemented the computational pipeline and TCGA data analyses, D.S. performed the Affymetrix mRNA microarray analysis and R.G. and W.L.R. supervised parts of the computational analyses. E.C.K., J.D.A., Y.C. and R.K.P. performed the experimental validation. K.W. and E.M.S. sequenced TP53, and E.M.S. edited the manuscript. C.W.D., A.K.G., K.C.O. and N.D.U. helped procure and/or process specimens, and C.W.D. helped design the study. M.T. designed and supervised the study and helped to write the manuscript. All authors read and approved the final manuscript. The content of this publication is solely the responsibility of the authors and does not necessarily reflect the views or policies of the NIH or Department of Health and Human Services or other funders, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin. Cancer Res. 2006;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 5.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schanen BC, Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2011;97:1–6. doi: 10.1016/j.ygeno.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Latronico MV, Catalucci D, Condorelli G. MicroRNA and cardiac pathologies. Physiol. Genomics. 2008;34:239–242. doi: 10.1152/physiolgenomics.90254.2008. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl Acad. Sci. USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 15.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin. Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 17.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 20.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar D, Parkin R, Wyman S, Bendoraite A, Sather C, Delrow J, Godwin AK, Drescher C, Huber W, Gentleman R, et al. Quality assessment and data analysis for microRNA expression arrays. Nucleic Acids Res. 2009;37:e17. doi: 10.1093/nar/gkn932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth GK. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galic V, Willner J, Wollan M, Garg R, Garcia R, Goff BA, Gray HJ, Swisher EM. Common polymorphisms in TP53 and MDM2 and the relationship to TP53 mutations and clinical outcomes in women with ovarian and peritoneal carcinomas. Genes Chromosomes Canc. 2007;46:239–247. doi: 10.1002/gcc.20407. [DOI] [PubMed] [Google Scholar]
- 33.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 34.Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih Ie M, Zhang Y, Wood W, 3rd, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 36.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 37.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 39.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 40.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl Acad. Sci. USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Down TA, Hubbard TJ. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 47.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 48.Havrilesky L, Darcy M, Hamdan H, Priore RL, Leon J, Bell J, Berchuck A. Prognostic significance of p53 mutation and p53 overexpression in advanced epithelial ovarian cancer: a Gynecologic Oncology Group Study. J. Clin. Oncol. 2003;21:3814–3825. doi: 10.1200/JCO.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 49.Leitao MM, Soslow RA, Baergen RN, Olvera N, Arroyo C, Boyd J. Mutation and expression of the TP53 gene in early stage epithelial ovarian carcinoma. Gynecol Oncol. 2004;93:301–306. doi: 10.1016/j.ygyno.2004.01.043. [DOI] [PubMed] [Google Scholar]
- 50.Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 51.Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O’Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol. Oncol. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unoki M, Nakamura Y. Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene. 2001;20:4457–4465. doi: 10.1038/sj.onc.1204608. [DOI] [PubMed] [Google Scholar]
- 57.Unoki M, Nakamura Y. EGR2 induces apoptosis in various cancer cell lines by direct transactivation of BNIP3L and BAK. Oncogene. 2003;22:2172–2185. doi: 10.1038/sj.onc.1206222. [DOI] [PubMed] [Google Scholar]
- 58.Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, Daxenbichler G, Zeimet A, Zeillinger R, Marth C, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- 59.Marchini S, Marabese M, Marrazzo E, Mariani P, Cattaneo D, Fossati R, Compagnoni A, Fruscio R, Lissoni AA, Broggini M. DeltaNp63 expression is associated with poor survival in ovarian cancer. Ann. Oncol. 2008;19:501–507. doi: 10.1093/annonc/mdm519. [DOI] [PubMed] [Google Scholar]
- 60.Ibrahim N, He L, Leong CO, Xing D, Karlan BY, Swisher EM, Rueda BR, Orsulic S, Ellisen LW. BRCA1-associated epigenetic regulation of p73 mediates an effector pathway for chemosensitivity in ovarian carcinoma. Cancer Res. 2010;70:7155–7165. doi: 10.1158/0008-5472.CAN-10-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leskela S, Leandro-Garcia LJ, Mendiola M, Barriuso J, Inglada-Perez L, Munoz I, Martinez-Delgado B, Redondo A, de Santiago J, Robledo M, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr. Relat. Cancer. 2011;18:85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 62.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 64.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.