Abstract
TFIIH is a multisubunit factor essential for transcription initiation and promoter escape of RNA polymerase II and for the opening of damaged DNA double strands in nucleotide excision repair (NER). In this study, we have analyzed at which step of the transcription cycle TFIIH is essential for transcription by RNA polymerase I. We demonstrate that TFIIH associates with the rDNA promoter and gene-internal sequences and leaves the rDNA promoter in a complex with RNA polymerase I after start of transcription. Moreover, mutations in the TFIIH subunits XPB and XPD found in Cockayne syndrome impair the interaction of TFIIH with the rDNA, but do not influence initiation complex formation or promoter escape of RNA polymerase I, but preclude the productivity of the enzyme by reducing transcription elongation in vivo and in vitro. Our results implicate that reduced RNA polymerase I transcription elongation and ribosomal stress could be one factor contributing to the Cockayne syndrome phenotype.
INTRODUCTION
RNA polymerases are dependent on auxiliary factors to recognize their promoters and to initiate, elongate and terminate transcription. These transcription factors are specific for each class of RNA polymerase. TATA-binding protein (TBP) was the first transcription factor shown to be essential for all three classes of RNA polymerases (1,2). TFIIH, which was supposed to be primarily a general transcription factor of RNA polymerase II, was described to play an essential role in RNA polymerase I transcription (3–5). TFIIH can be isolated in a complex with RNA polymerase I, the basal initiation factor TIF-IB and with the DNA repair factors CSB and XPG. TFIIH is essential for rDNA transcription in vivo and in vitro and resides in the nucleolus where photobleaching experiments determined a residence time of 25 s in comparison to 6 s at a RNA polymerase II promoter indicating a differing function of TFIIH in Pol I than in Pol II transcription.
TFIIH is a basal or general transcription factor of RNA polymerase II and necessary for the transcription of every protein-coding gene. TFIIH is composed of 10 subunits with three enzymatic activities, the ATP-dependent helicases XPB and XPD and the CAK sub-complex with the kinase cdk7. The ATPase domain of the helicase XPB opens the DNA double strand at the promoter (6) and creates the transcription bubble. XPB plays a major role in promoter escape, a phase of instability and pausing of the early elongation phase until nucleotide 15, whereas XPD is a necessary structural component for this step (7,8). The cdk7 subunit of TFIIH phosphorylates the C-terminal domain (CTD) of the largest subunit of RNA polymerase II and thus initiates elongation. Thus TFIIH is involved in initiation, promoter clearance and elongation of RNA polymerase II.
Mutations in TFIIH subunits cause three distinct diseases: the cancer prone skin disease xeroderma pigmentosum (XP) and the premature aging diseases trichothiodystrophy (TTD) and Cockayne syndrome (CS) (9). XP is due to non-repaired DNA lesions. In nucleotide excision repair (NER), the XPB and XPD subunits of TFIIH serve an essential function in opening the DNA strand around helix distorting lesions and the accumulation of UV-induced DNA damage is highly mutagenic.
The pathomechanisms of the premature aging phenotypes of CS and TTD are less well defined. As a sub-pathway of NER is defective in these tumor-free syndromes, accumulating DNA damage could drive tumor suppression at the expense of premature aging (10). However, total NER deficiency by mutation of the central NER factor XPA is not followed by premature aging, thus indicating that the mutations causing premature aging might impair another common function of the involved genes. As TFIIH is a basal transcription factor, transcriptional deficiencies might be causal for premature aging (11–13).
In this study, we have investigated at which step of the transcription cycle TFIIH is involved in RNA polymerase I transcription. TFIIH binds to the rDNA promoter and gene-internal sequences and leaves the rDNA promoter with the polymerase and complexes with the polymerase during transcription. Mutations in the helicase subunits of TFIIH found in CS impair the interaction of the factor with the rDNA in vivo and in vitro and severely reduce Pol I transcription. Purified TFIIH stimulates the elongation activity of RNA polymerase I. TFIIH is not needed for efficient initiation complex formation and does not influence the stability of RNA polymerase I–template interaction after transcription start, but is essential for productive transcription. Our study revealed a novel role for TFIIH as an elongation factor of RNA polymerase I. Elongation of RNA polymerase I transcription might be a common function of CS-causing genes.
MATERIAL AND METHODS
Cell growth
HEK 293 and HeLa cells were grown in Dulbecco's MEM (PAA Laboratories GmbH) medium with additional 10% fetal calf serum (Biochrom AG) as well as l-glutamine (Biochrom AG), penicillin G and streptomycin (Pen-Strep, Biochrom AG).
XPA/XP cells from a XP patient complementation group A (GM02344), XPB/CS cells from patient XP11BE (GM02252,14) and XPD/CS cells from patient XP-CS2 (GM03249,15) were obtained from the Coriell Institute for Medical Research and grown in RPMI 1640 (PAA Laboratories GmbH) supplemented with15% fetal calf serum (Lonza Group Ltd.), l-glutamine (Biochrom AG) and penicillin G and streptomycin (Pen-Strep, Biochrom AG). The XPB/CS mutations from patient XP11BE (GM02252) result in a paternal allele with a stop codon in exon 6 (14) that is not detectable at the RNA level (16) and a maternal allele that contains a transversion at a splice acceptor site and an inactivating frameshift at the last 41 amino acids of the C-terminus (15). Accordingly the XPD/CS cells from patient XP-CS2 (GM03249) express XPD from a single allele mutated in the DNA-binding domain of helicase motiv V (G602D) (17). Transcriptionally active nuclear extracts were prepared from logarithmically growing mammalian cell culture as described (18).
Cell transfection
Using amaxa nucleofector I (Lonza) device, a transfection protocol was established with a plasmid encoding GFP. An amount of 4 µg pcDNA, pcDNA XPB and XPD were transfected in 4 × 106 cells by electroporation and RNA was harvested from 1 × 106 cells with RNeasy Plus Kit (Qiagen). For western blots cells were lysed by a buffer containing 1% Triton X-100.
RNA analysis
Exponentially growing cells of equal density were harvested by centrifugation. Total cellular RNA was prepared from XPA/XP-, XPB/CS- and XPD/CS- cells by using the peqGold TriFast reagent (Peqlab Biotechnologie GmbH). RNA (1 µg) was reverse transcribed using random primer p(dN)6 (F.Hoffmann-La Roche Ltd.) and 45S pre rRNA was quantified by real time PCR (LightCycler, F.Hoffmann-La Roche Ltd.) using primers that amplify a fragment from +307 to +442 of the human rDNA (hrDNA primer, Thermo Fischer Scientific GmbH: forward 5′-TGT CAG GCG TTC TCG TCT C-3′; reverse 5′-AGC ACG ACG TCA CCA TAT C-3′). Data were normalized to the level of GAPDH mRNA (GAPDH primer, Thermo Fischer Scientific GmbH: forward 5′-TGG ACC TGA CCT GCC GTC TA-3′; reverse 5′-CCC TGT TGC TGT AGC CAA ATT C-3′). Northern blot analysis of the 45S rRNA was performed as described (19).
Purification of TFIIH
A HeLa cell line stably expressing FLAG-tagged XPB (20) was used for the purification of TFIIH. An amount of 2.5 mg nuclear extract was incubated with anti-FLAG M2-agarose beads (Sigma-Aldrich Co.) in the presence of protease inhibitors on the rotating wheel at 4°C over night. After two washes with AM300 + 0.1% NP-40, bound TFIIH was eluted from the beads by incubation at 4°C for 2 h in AM300 + 0.1% NP-40 plus 0.25 µg/µl FLAG peptide and subsequently dialyzed against buffer AM100.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were done as described with minor modifications (21,22). After precipitation and washing, crosslinked proteins were digested by incubation with elution buffer (TE buffer; 1% SDS) and Proteinase K for 2 h at 68°C. DNA was extracted using the QIAquick Nucleotide Removal Kit (QIAGEN GmbH) and analyzed by PCR. The primers used for PCR have been published with the names: H0 (rDNA promoter); H1, H4, H8 and H13 (gene-internal) and H23/27 [intergenic spacer (IGS)] (23). Statistical analysis of qPCR was performed according to Tukey's multiple comparison test.
Template immunoprecipitation
An amount of 40 ng rDNA template (pHrP2 linearized with EcoRI) was incubated with 40 µg of the indicated nuclear extracts and 0.825 µmol guanine for 40 min at 30°C and 350 rpm. Addition of sarcosyl (final concentration: 0.025%) and salmon sperm DNA (final concentration: 400 ng) was used to provide single-round transcription. Transcription was started by adding ribonucleotides and stopped after the indicated time by crosslinking proteins to the template with formaldehyde (final concentration: 1%) for 10 min at 30°C. Crosslinking was stopped with glycine (final concentration 0.125 M) for 5 min at 30°C. Each sample was diluted with IP buffer and incubated with indicated antibodies over night at 4°C. Isolation of precipitated complexes, washing steps, digestion of proteins and DNA extraction as well as DNA amplification was done as described for ChIP analysis.
Preparation of immobilized template and isolation of pre-initiation complexes
The preparation of immobilized complexes has been described previously (22).
In vitro transcription
In vitro transcription assays were done as described (22).
Protein detection
Proteins were analyzed by immunoblotting. Proteins from eluates of immobilized complexes or nuclear extracts were separated by SDS–PAGE and transferred to nitrocellulose membrane (Whatman GmbH). Adequate secondary antibodies conjugated to horseradish peroxidase were used to detect immunocomplexes on the membrane by chemoluminescence according to the instructions of the manufacture (LumiGLO Reagent, Cell Signaling Technology).
Antibodies
α-actin, α-XPB, α-XPD, α-cdk7 and α-p62 were obtained from Santa Cruz (Santa Cruz Biotechnology, Inc.). α-Pol II CTD, α-XPB and α-XPD antibodies recognizing mutant subunits were purchased from Acris (Acris Antibodies GmbH). α-RPA116, α-TAF1110, α-TAF168, α-UBF and α-TIF IA were kindly provided by Ingrid Grummt. α-RPA135 was harvested from rabbits immunized with a mixture of two synthetic peptides (MDPGSRWRNLPSGPSC, LEKPPPSWSAMRNRKYNC; Peptide Specialty Laboratories GmbH). The α-p44 was affinity purified from rabbit serum immunized with one synthetic peptide (MDEEPERTKRWEGGYER; Peptide Specialty Laboratories GmbH).
RESULTS
The rDNA is decorated with TFIIH
Recently, it has been suggested that TFIIH plays an essential role in RNA polymerase I transcription. TFIIH can be identified in functional complexes with RNA polymerase I in the nucleolus and initiation factors and is essential for RNA polymerase I transcription in vitro and in vivo (3–5).
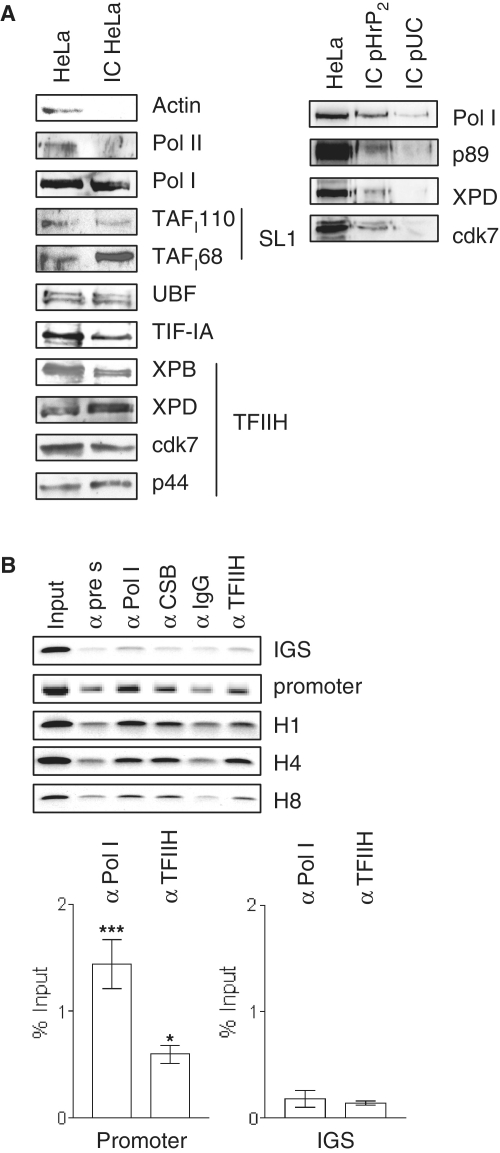
To elucidate the mechanisms of TFIIH action on rDNA transcription, we first asked if TFIIH is part of the initiation complex and binds to the rDNA promoter in vitro. Bead-bound immobilized rDNA template was used to isolate initiation complexes by incubation with HeLa nuclear extracts, followed by washing with buffer AM100 and elution of bound proteins with urea. Western blot analysis (Figure 1A) of these immobilized complexes (IC) revealed specific binding of RNA polymerase I and initiation factors SL1, UBF and TIF-IA to the rDNA promoter. TFIIH subunits could clearly be detected in these immobilized complexes thus indicating that TFIIH binds in the absence of active transcription to the rDNA in vitro. RNA polymerase I and TFIIH did also bind to immobilized pUC DNA. However, these complexes were not resistant to washing with buffer AM 100 in the presence of sarcosyl. To ascertain whether TFIIH directly binds to the rDNA promoter region in vivo, we performed ChIP analysis with HeLa cells expressing a flag-tagged XPB subunit of TFIIH (20). Antibodies against RNA polymerase I and CSB precipitated promoter and gene-internal regions of the rDNA (Figure 1B) (22). Flag-antibodies recognizing the tagged TFIIH subunit XPB specifically precipitated the rDNA promoter and all gene-internal sequences of the rDNA, but not the IGS. Real-time PCR analysis revealed significant binding of RNA polymerase I and TFIIH to the promoter, but not to the IGS. Hence, TFIIH directly binds to the rDNA promoter. Moreover, TFIIH is also bound to the coding rDNA sequences, implying a post-initiation role for TFIIH in RNA polymerase I transcription.
Figure 1.
TFIIH binds to the rDNA in vitro and in vivo. (A) One microgram of biotin labeled rDNA was bound to 20 µl of streptavidin magnetic beads and incubated with HeLa nuclear extract. After washing with buffer AM containing 100 mM KCl, bound proteins were eluted with 4 M urea and analyzed on western blots with the indicated antibodies. (Pol II-RNA polymerase II, Pol I-RNA polymerase I, TAFI110/TAFI68-subunits of selectivity factor 1, UBF-upstream binding factor, TIF-IA-transcription initiation factor IA, XPB/XPD/cdk7/p44-subunits of transcription factor of RNA polymerase II H). To control the specificity of binding, immobilized complexes formed on specific (pHrP2) and unspecific (pUC) DNA were washed with buffer AM100 containing 0.125% sarcosyl. Binding of RNA polymerase I and TFIIH was stronger to the human rDNA promoter than to unspecific DNA. (B) ChIPanalysis with chromatin from HeLa cells expressing flag-tagged XPB (20). The antibodies are indicated above (α-TFIIH = α-flag). For PCR analysis, primers amplifying the IGS or promoter and gene-internal regions (H1, H4, H8) were used as depicted. Primer position is described in detail in O'Sullivan et al. (23). Promoter region and IGS from three independent experiments was analyzed by qPCR and normalized to the respective negative controls (***P < 0.001; *P < 0.05). Pictures are representatives from at least three independent experiments.
TFIIH leaves the rDNA promoter in a complex with RNA polymerase I
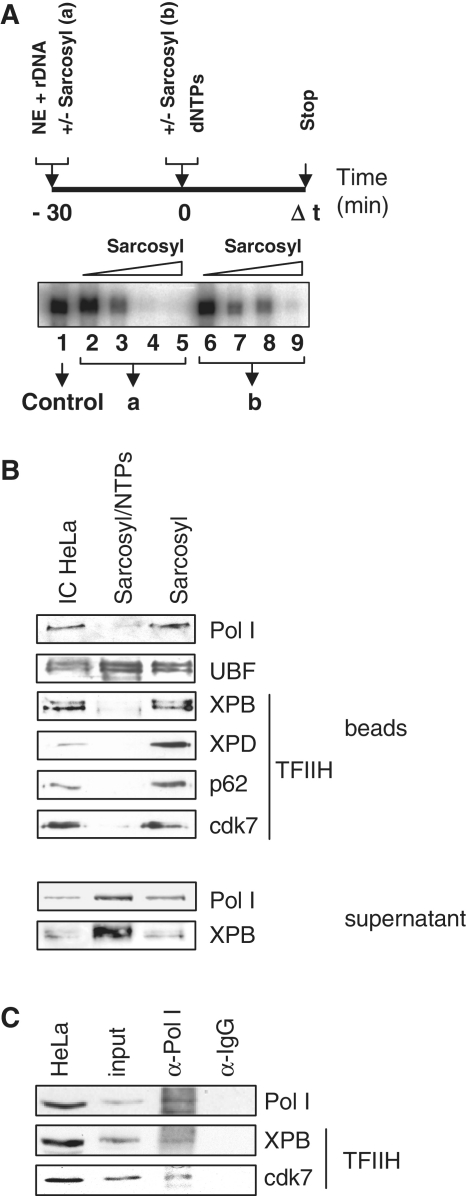
The 0.025% sarcosyl reduces transcription to a single round by precluding initiation complex formation, but allowing transcription from preformed initiation complexes (Figure 2A, compare lane 4 to lane 8). We asked whether TFIIH leaves the initiation complex after transcription initiation. Immobilized complexes were challenged by the addition of nucleotides (NTPs) and transcription restricted to a single round by sarcosyl. Proteins remaining on the template after single-round transcription and the supernatants were analyzed in western blots. Under these conditions RNA polymerase I left completely the template whereas UBF stayed bound to the rDNA (Figure 2B). Similar to RNA polymerase I, TFIIH is completely released from the rDNA template by the addition of nucleotides and can be detected in the supernatant.
Figure 2.
TFIIH leaves the rDNA promoter with RNA polymerase I. (A) In vitro transcription experiment with HeLa nuclear extract titrating the sarcosyl concentration for single-round transcription. (B) Bead-bound immobilized complexes (IC HeLa) were challenged by the addition of nucleotides (NTPs). Transcription was restricted to a single round by sarcosyl and the remaining proteins analyzed after urea elution by western blots. Sarcosyl/NTPs denominates the single-round transcription experiment and sarcosyl the control reaction without nucleotides. RNA polymerase I and the XPB subunit of TFIIH were also monitored in the supernatants. (C) Supernatants of single-round transcriptions with immobilized-complex isolated transcription factors were subject to immunoprecipitation with control antibodies (IgG) or antibodies directed against RNA polymerase I (Pol I) and visualized by western blots. Pictures are representatives from at least three independent experiments.
As TFIIH binding to the rDNA can be determined throughout the whole gene we next asked if TFIIH travels with the polymerase and can be detected in a complex after transcription or is released and able to re-initiate transcription. This was addressed by co-immunoprecipitation experiments with supernatants after one round of transcription (Figure 2C). Antibodies directed against RNA polymerase I clearly co-precipitated TFIIH after transcription. Thus, TFIIH associates with RNA polymerase I during transcription and does not leave the elongating enzyme.
Template-immunoprecipitation reveals binding of TFIIH to the rDNA throughout the whole transcription process
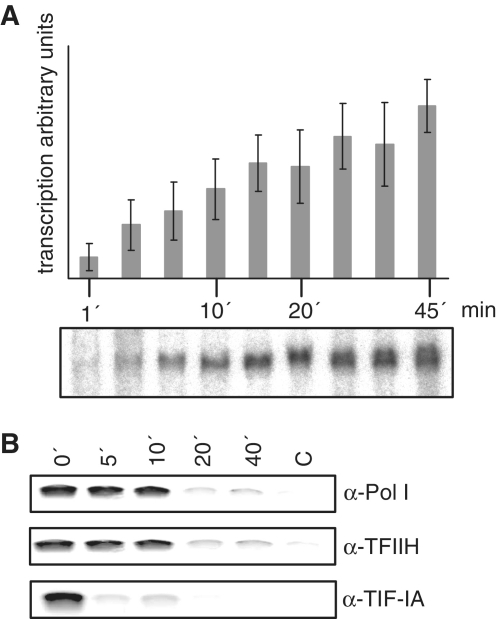
As the human rDNA promoter extends over the transcription start side, a construction of a g-less cassette to directly study elongation is not possible. To investigate the protein–DNA interactions after transcription start, we developed an assay combining in vitro transcription with formaldehyde crosslinking and immunoprecipitation followed by semi-quantitative PCR analysis. In this assay after synchronization of the reaction through formation of initiated complexes by pre-incubation of template with nuclear extracts in the presence of the first nucleotides, we determined single-round transcription kinetics by starting transcription reaction by addition of the missing nucleotides (Figure 3A). After linear increase of transcription in the first 15 min, a plateau phase is reached after transcription for 20 min.
Figure 3.
TFIIH associates with the rDNA throughout the transcription process. (A) Kinetics of single-round transcription with initiated complexes. Transcription was started after pre-incubation with the first 2 nt and restricted to a single round by addition of sarcosyl. Bars represent the mean value of three independent experiments. A typical autoradiograph of a single-round transcription from initiated complexes formed at the pHrP2 template is shown below. (B) TIP experiments showing the time-course of the association of RNA polymerase I, TFIIH and TIF-IA with the rDNA in single-round transcriptions. Nuclear extracts were pre-incubated with rDNA template in the presence of the first 2 nt and reaction was started by addition of the lacking NTPs. After the indicated time-points formaldehyde was added to stop the reaction and to crosslink proteins to the DNA. These complexes were subsequent used for immunoprecipitation with the indicated antibodies and template association was detected in PCR reactions with primers directed against the human rDNA promoter. Pictures are representatives from at least three independent experiments.
Using non-labeled nucleotides for transcription start we stopped transcription after different time points by crosslinking the proteins to the template with formaldehyde. The indicated proteins were immunoprecipitated and the crosslinked template isolated and quantified in semi-quantitative PCR. This allows a time course of the association of a particular protein with the rDNA template in vitro. RNA polymerase I could be quantitatively crosslinked to the rDNA until 10 min after start of transcription, whereas TIF-IA left the template directly after transcription start (Figure 3B). TFIIH was as long as the RNA polymerase I associated with the rDNA, indicating that TFIIH does not leave the enzyme. As the elongation of RNA polymerase I occurs with 30-nt/s (24), the used template is transcribed in 13 s. Our new method shows, that the polymerase and TFIIH are associated with the template for 10 min before entering the elongation mode and may undergo conformational changes after TIF-IA left the template.
In summary, template-immunoprecipitation (TIP) analysis revealed that TFIIH binds as long to the rDNA as RNA polymerase I. This new developed assay allows to study steps after transcription initiation e.g. promoter clearance of RNA polymerase I, that so far were difficult to address.
Mutations in TFIIH affect RNA polymerase I transcription
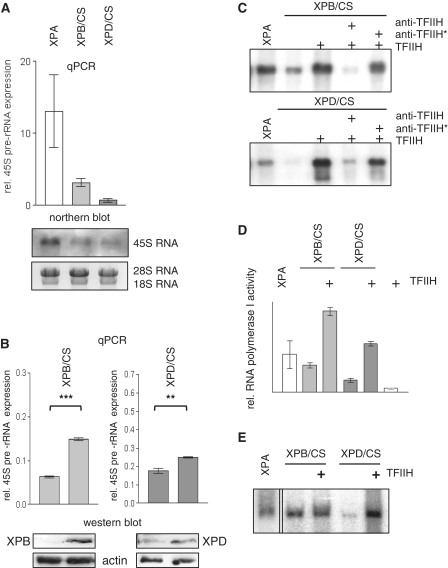
In recent reports it has been shown that mutations in the CSB protein found in CS patients impair RNA polymerase I elongation (4,25,22). To study the functional consequences of CS mutations in TFIIH for RNA polymerase I transcription, we isolated RNA from exponentially growing cells with mutations in XPA from a XP patient as a control, or XPB and XPD mutant cells from CS patients with monoallelic expression of mutant XPB or XPD subunits. Analysis of the 45S precursor rRNA allows a determination of ongoing RNA polymerase I transcription at the time-point of harvest and showed a considerable reduction of rRNA synthesis in cells from CS patients in real-time PCR analysis and in northern blots (Figure 4A).
Figure 4.
RNA polymerase I transcription is reduced in TFIIH/CS cells and can be rescued by wild-type TFIIH subunits in vivo and purified TFIIH in vitro. (A) Real-time PCR and northern blot analysis of 45S pre-rRNA expression of XPA cells as a wild-type control or CS cells harvested at equal density. Autoradiograph of the hybridized 45S pre-rRNA and ethidiumbromide stained 28S/18S rRNA as a loading control are shown below. (B) Real-time PCR analysis of pre-rRNA transcription and western blots of reconstituted lymphoblastoid cells with vector plasmid as a control, or wild-type XPB respective XPD. The XPB antibody recognizes only wild-type XPB, whereas the XPD antibody also detects mXPD. (C) Promoter-dependent RNA polymerase I in vitro transcription with the indicated nuclear extracts and purified TFIIH. Antibodies against the TFIIH subunit p44 inhibit the reconstitution (anti-TFIIH) and are inactivated by heat treatment (anti-TFIIH*). (D) Non specific RNA polymerase I transcription is stimulated by TFIIH. Sonified calf-thymus DNA was used as a template in the presence of α-amanitin with the indicated nuclear extracts and purified TFIIH. (E) The autoradiography shows end-to-end transcription of RNA polymerase I performed with a pUC template without rDNA promoter in the presence of α-amanitin. Pictures are representatives from at least three independent experiments.
In an attempt to correct the reduction in rRNA transcription, we transfected the wild-type TFIIH subunits into the cells with mutant TFIIH. Transfection of wt-XPB into the XPB/CS cell line resulted in a significant upregulation of rRNA transcription as detected by quantitative PCR. Moreover, wild-type XPD transfection into the XPD/CS lymphoblastoid cells clearly stimulated rRNA transcription (Figure 4B). Thus wild-type TFIIH subunits are able to restore the transcriptional defect of rDNA transcription in TFIIH/CS cells.
With nuclear extracts from the TFIIH mutant cells we asked the question whether mutation in TFIIH is followed by a reduced rDNA transcription activity in vitro and if so, if this defect can be overcome by the addition of purified TFIIH. In vitro transcription analysis uncovered a severely reduced transcriptional activity that could be rescued by the addition of affinity-purified TFIIH (Figure 4C). Moreover, the reconstitution of these extracts by purified TFIIH was abolished by pre-incubation of TFIIH with TFIIH antibody. Heat-inactivation of the antibody resulted in a clear stimulation of Pol I transcription by TFIIH.
TFIIH is necessary for RNA polymerase I transcription at a step subsequent to initiation (3) and TFIIH associates with the polymerase. We tested the effect of TFIIH on non-specific RNA polymerase I transcription. A−amanitin resistant transcription was reduced in nuclear extracts of TFIIH/CS cells and could be stimulated by the addition of affinity purified TFIIH thus indicating that TFIIH stimulates the elongation activity of the enzyme (Figure 4D).
A tailed template derived from the pUC plasmid was employed to assay RNA polymerase I transcription in the presence of α-amanitin. Addition of purified TFIIH resulted in a clear stimulation of RNA polymerase I activity (Figure 4E). As this end-to-end transcription assay reflects the elongation activity of the enzyme (26), TFIIH stimulates RNA polymerase I transcription by enhancing the elongation step of the transcription cycle. CS mutations in TFIIH impair RNA polymerase I transcription in vivo and in vitro and can be rescued by transfection of wild-type TFIIH subunits in vivo and by addition of purified TFIIH in vitro.
CS mutations in XPB and XPD reduce the affinity of TFIIH to the rDNA
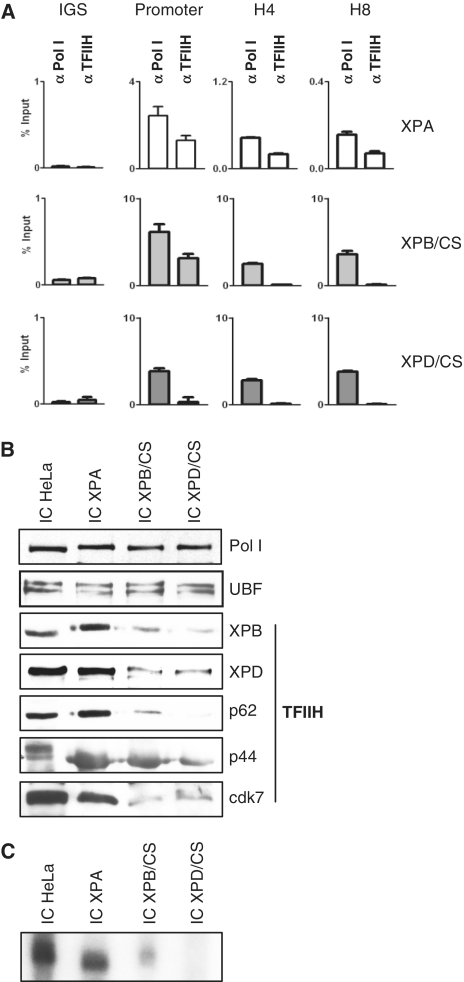
As mutations in TFIIH impair RNA polymerase I elongation, we next asked if mutations in TFIIH are accompanied by a decreased binding of TFIIH to the rDNA in vivo. ChIP analysis of chromatin isolated from lymphoblastoid cells of an XP-patient or donors with CS-mutations in XPB and XPD was used to address this question. TFIIH antibodies precipitated promoter and gene-internal sequences of the rDNA from chromatin of a XPA patient (Figure 5A). Mutation in XPB allows promoter-binding, but not binding of TFIIH to gene-internal sequences as primers generated against gene-internal sequences did not amplify products from the TFIIH immunoprecipitate. Moreover, mutation in XPD completely abolished binding of TFIIH to the rDNA in vivo as no rDNA sequences could be amplified. We next tested whether in vitro TFIIH presents reduced affinity to the rDNA. A human rDNA template was immobilized with magnetic beads and used to isolate initiation complexes from nuclear extracts from either HeLa cells, or cells from patients with XPA or XP/CS and bound proteins were analyzed on immunoblots. As demonstrated in Figure 5B, RNA polymerase I and UBF bound to the immobilized rDNA to the same extent independent of differences in the nuclear extracts. However, TFIIH mutations in XPB or XPD severely reduced the affinity of the complete TFIIH complex to the rDNA, indicating that the binding to the rDNA is mediated by the helicase subunits of TFIIH. Moreover, these results show that RNA polymerase I is efficiently recruited to the rDNA even in the absence of functional TFIIH. When these experiments were performed under in vitro transcription conditions, the lack of TFIIH resulted in an impaired RNA polymerase I single-round transcription (Figure 5C). There are no shorter transcripts or smears detectable in the gel, indicating that reduced TFIIH at the promoter does not result in the generation of partial transcripts (Supplementary Data). In summary CS mutations in TFIIH result in a reduced affinity of TFIIH to the rDNA in vivo and in vitro and impair RNA polymerase I transcription.
Figure 5.
Reduced binding of mutant TFIIH to the rDNA in vitro and in vivo. (A) Quantitative PCR analysis of ChIP of the indicated cell lines with antibodies against RNA polymerase I and TFIIH (anti-cdk7) as indicated above. Primer were directed against the IGS, promoter or gene-internal regions (H4, H8). (B) Immobilized complexes were formed with the indicated nuclear extracts, washed with buffer AM50 and analyzed after urea elution on immunoblots. (C) Immobilized complexes from nuclear extracts as indicated above were challenged by addition of nucleotides and radioactive UTP. Sarcosyl was added to limit transcription to a single round. Synthesized RNA was isolated after 60 min and subject to eletrophoresis and autoradiography. Pictures are representatives from at least three independent experiments.
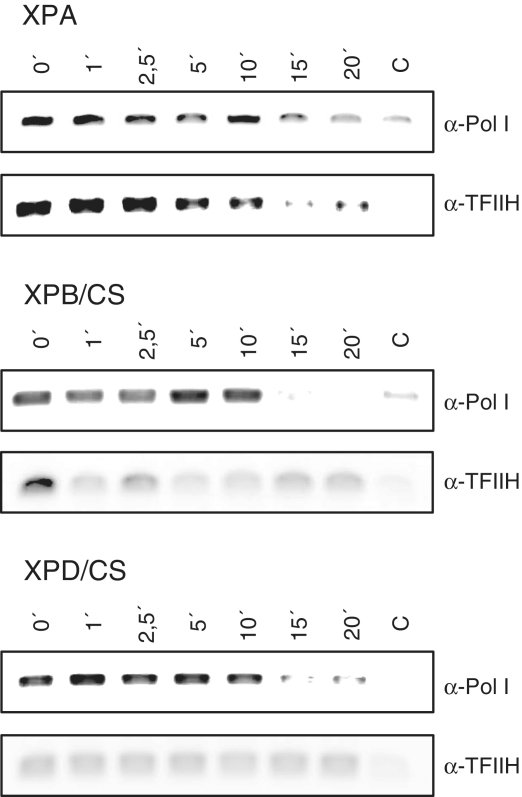
TFIIH does not influence the stability of the initiation complex of RNA polymerase I
CS mutations in TFIIH are accompanied by a reduced TFIIH binding to the rDNA promoter in vivo and in vitro. To investigate if this lack of TFIIH is followed by a reduced residence time of the polymerase, we performed template-immunoprecipitation (TIP) analysis with extracts from CS-cells. Experiments with nuclear extracts from cells of a XPA patient showed that TFIIH and RNA polymerase I display a comparable time course of rDNA binding (Figure 6). In nuclear extracts from XPB/CS and XPD/CS cells a rDNA binding of TFIIH was barely detectable, whereas RNA polymerase I showed similar binding kinetics to the rDNA as in the XPA control cells. The lack of TFIIH does not result in a prolonged residence time of the polymerase indicating that RNA polymerase I does not encounter promoter proximal stalling and that promoter clearance is not dependent on TFIIH. These results further support our finding that TFIIH does not influence the first steps of transcription like pre-initiation complex formation, formation of the first phosphodiester bonds and promoter clearance, but is essential at the subsequent step of elongation of RNA polymerase I transcription.
Figure 6.
Lack of TFIIH does not reduce residence time of the polymerase at the rDNA. TIP was performed by pre-incubation for 20 min of the indicated nuclear extracts with template and the first nucleotides to form initiated complexes. These complexes were challenged by the missing nucleotides and the reaction stopped after the indicated time points by addition of the crosslinking agent formaldehyde to 1%. Crosslinked proteins were immunoprecipitated with the indicated antibodies and associated template was amplified in semi-quantitative PCR. Pictures are representatives from at least three independent experiments.
DISCUSSION
TFIIH exemplifies the intimate structural link of transcription and DNA repair, and it exerts multiple different functions in both processes. In RNA polymerase II transcription TFIIH is necessary for promoter opening by the ATPase domain of XPB (27), promoter escape (7), phosphorylation of the CTD of RNA polymerase II (28) and transactivation of a variety of promoters (29). In NER, TFIIH opens the DNA double strand by the helicase subunits XPB and XPD (30). Mutations in TFIIH can result in three devastating diseases: the cancer prone skin disease XP and the premature aging disorders CS and TTD. Recently we and others could show, that TFIIH is essential for rDNA transcription by RNA polymerase I (3–5). However the precise molecular function of TFIIH in RNA polymerase I transcription remained undefined. Here, we identified the step of transcription of RNA polymerase I that is dependent on TFIIH. TFIIH is an elongation factor of RNA polymerase I and CS mutations in the helicase subunits of TFIIH reduce the affinity of TFIIH to the rDNA in vivo and in vitro and impair RNA polymerase I transcription.
We unraveled that TFIIH binds the whole rDNA gene in vivo. The ChIP experiments also demonstrate a binding of TFIIH to the rDNA promoter. TFIIH may be recruited to the rDNA promoter by its interaction with the enzyme or by interaction with the central initiation factor SL1/TIF-IB as shown in previous publications, but does not contribute to initiation (3,4). According to an alternative function to initiation of TFIIH in Pol I transcription, the ChIP experiments revealed that TFIIH binds to gene-internal regions. This indicates a possible function of TFIIH in a step after initiation as promoter clearance or elongation. The hypothesis that TFIIH is not a mere initiation factor in RNA polymerase I transcription is further supported by the experiments showing an association of TFIIH with the enzyme after transcription and the experiments with the newly developed method of TIP. They demonstrated that TFIIH is connected with the rDNA as long as the enzyme itself. These results are in accordance with previous studies by Hoogstraten presenting evidence that TFIIH stays for 2–10 s at a RNA polymerase II promoter, where it is the last factor to enter the initiation complex and the first to leave. The residence time at the rDNA was up to 25 s indicating a differing function of TFIIH in rRNA transcription than in transcription by RNA polymerase II (5).
We further investigated the consequences of CS mutations in TFIIH for rDNA transcription. The XPB/CS and XPD/CS cells were chosen for the following reasons: first: both cell lines exhibit mutations that result in one non-expressed null allele of either XPB or XPD and one expressed allele with a defined mutation (17,14); allowing easier interpretation of results than from two different mutant expressed alleles; second: a former publication described already a disturbance of the CSB-RNA polymerase I–TFIIH complex in these cells (4); third: as CS mutations in CSB affect RNA polymerase I transcription elongation (4,22) we asked whether RNA polymerase I transcription elongation might generally be defective in CS. RNA polymerase I transcription is severely affected in both cell lines in vivo and in vitro. Cellular rRNA synthesis can significantly be stimulated by transfection of the wild-type TFIIH subunit indicating that rRNA transcription in vivo is dependent on the integrity of the TFIIH complex. In vitro transcription can specifically be rescued by affinity-purified TFIIH and reconstitutes promoter-dependent and end-to-end non-specific transcriptional activity of RNA polymerase I that reflects the elongation activity of the enzyme (26). The western blots with nuclear extracts show that the mutant mono-allelic TFIIH subunits are reduced in comparison to controls (Supplementary Data); however, mutation in the C-terminus of XPB or mutation in the DNA-binding domain of XPD severely reduced the affinity of the complete TFIIH complex to the rDNA in vivo and in vitro as shown in ChIP experiments and experiments with immobilized complexes. The reduction of XPB or XPD subunits in nuclear extracts as detected in western blots (Supplementary Data) should result in a 50% reduced binding and activity of TFIIH in both cell lines. For XPD mutant cells and extracts there is no binding to the rDNA in vivo and a severely reduced binding in vitro detectable. This argues for a qualitative dysfunction of TFIIH in these mutant cells. The binding of TFIIH to the rDNA might be mediated by the helicase subunits of TFIIH although their activity is not needed in RNA polymerase I transcription (3). Our data imply that TFIIH does not influence the efficiency of promoter binding or the initiation rate of RNA polymerase I, but stimulates the elongation rate of the enzyme. Semi-quantitative (Supplementary Data) and quantitative PCR analysis (Figure 5A) of gene-internal regions do not show a comparable reduction in RNA polymerase I loading when TFIIH is severely reduced. As this reduction is accompanied by an impaired RNA synthesis, a reduced transcription elongation velocity might be the reason. As in vivo experiments of former publications imply an essential role of TFIIH in RNA polymerase I transcription (3,31), TFIIH might be essential for the processivity of the enzyme and low levels of TFIIH in the XPB/XPD cells at gene-internal regions of the rDNA might be below the detection limit of the assays. Alternatively, TFIIH might be indispensable for RNA polymerase I to read through damaged, oxidized templates resulting in reduced elongation. Such a failure of extracts from CS cells was shown in vitro for RNA polymerase II transcription with the same XPB/CS cell line as used in our study (32). Failure to read through oxidized template may be sensed by the cell as massive DNA damage and provoke cellular responses that result in premature aging. Moreover, TFIIH might be critical to resolve RNA/DNA intermediates during transcription elongation as the rDNA is densely packed in the nucleolus.
RNA polymerase I completely leaves the rDNA promoter even in the absence of TFIIH suggesting that RNA polymerase I does not encounter promoter proximal stalling in the absence of TFIIH as RNA polymerase II (7). However, in the absence of TFIIH RNA polymerase I does not enter a productive mode, indicating that TFIIH is essential for RNA polymerase I transcription elongation. Further support of our findings that TFIIH does not influence promoter proximal stability of RNA polymerase I is provided by TIP. We expected an instability of RNA polymerase I in the absence of TFIIH, but this assay shows that the enzyme–DNA interaction in TFIIH mutant extracts is as long stable as in TFIIH competent extracts. This assay does not investigate elongation, as the elongation phase is quite fast with 30 nt/s (24) but demonstrates, that RNA polymerase I does not leave the rDNA promoter directly after TIF-IA release but undergoes TFIIH-independent transitions until it enters the TFIIH-dependent stage of elongation.
Mutations in CSB, that accounts for 70% of CS cases, repress elongation by RNA polymerase I (22). Here we identified a common function of three genes that, when mutated cause CS. CSB and TFIIH with its helicase subunits XPB and XPD are involved in transcription elongation by RNA polymerase I. RNA polymerase I transcription accounts for up to 60% of total transcription in a growing cell and disturbances in ribosomal biogenesis (ribosomal stress) are followed by an imbalance of the rRNA components and ribosomal proteins. Ribosomal proteins like L5 and L11 target MDM2 and stabilize p53 (33,34). Ribosomal stress through inhibition of RNA polymerase I transcription is followed by p53 dependent apoptosis (35), a typical hallmark of CS (36). Impaired RNA polymerase I transcription elongation and ribosomal stress might contribute to the pathogenesis of CS by sensitizing cells to apoptosis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [Deutsche Forschungsgemeinschaft; IB83 2-2 (KFO142), IB 83 3-1]. Funding for open access charge: German Research foundation (DFG).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The Hela-flag XPB cells were a kind gift of M. Teichmann. pcDNA XPB was a kind gift from F. Coin and pcDNA XPD from A. Lehmann. A. Ushmorov helped to transfect the lymphoblastoid cells. The authors thank I. Grummt and R.Voit for continuous support and H. Geiger for critical reading of the manuscript. K. Scharffetter-Kochanek, M. Wlaschek and the other members of the lab helped in fruitful discussions. R. Assfalg and S. Koch are members of the International Graduate School in Molecular Medicine Ulm.
REFERENCES
- 1.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 2.Lobo SM, Tanaka M, Sullivan ML, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 3.Iben S, Tschochner H, Bier M, Hoogstraten D, Hozak P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 4.Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 5.Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, Vermeulen W, Houtsmuller AB. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 2002;10:1163–1174. doi: 10.1016/s1097-2765(02)00709-8. [DOI] [PubMed] [Google Scholar]
- 6.Douziech M, Coin F, Chipoulet JM, Arai Y, Ohkuma Y, Egly JM, Coulombe B. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol. Cell. Biol. 2000;20:8168–8177. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvir A. Promoter escape by RNA polymerase II. Biochim. Biophys. Acta. 2002;1577:208–223. doi: 10.1016/s0167-4781(02)00453-0. [DOI] [PubMed] [Google Scholar]
- 8.Bradsher J, Coin F, Egly JM. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J. Biol. Chem. 2000;275:2532–2538. doi: 10.1074/jbc.275.4.2532. [DOI] [PubMed] [Google Scholar]
- 9.Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 10.Garinis GA, Schumacher B. Transcription-blocking DNA damage in aging and longevity. Cell Cycle. 2009;8:2134–2135. [PubMed] [Google Scholar]
- 11.Coin F, Bergmann E, Tremeau-Bravard A, Egly JM. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer J, de Wit J, van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol. Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 13.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol. Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, Friedmann PS, Emmert S, Gratchev A, Lachlan K, et al. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum. Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- 15.Weeda G, van Ham RC, Vermeulen W, Bootsma D, van der Eb AJ, Hoeijmakers JH. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990;62:777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- 16.Hwang JR, Moncollin V, Vermeulen W, Seroz T, van Vuuren H, Hoeijmakers JH, Egly JM. A 3′ –> 5′ XPB helicase defect in repair/transcription factor TFIIH of xeroderma pigmentosum group B affects both DNA repair and transcription. J. Biol. Chem. 1996;271:15898–15904. doi: 10.1074/jbc.271.27.15898. [DOI] [PubMed] [Google Scholar]
- 17.Cleaver JE, Thompson LH, Richardson AS, States JC. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutomska A, Lebedev A, Scharffetter-Kochanek K, Iben S. The transcriptional response to distinct growth factors is impaired in Werner syndrome cells. Exp. Gerontol. 2008;43:820–826. doi: 10.1016/j.exger.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Li P, Roeder RG, Wang Z. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol. Cell. Biol. 2001;21:4614–4625. doi: 10.1128/MCB.21.14.4614-4625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebedev A, Scharffetter-Kochanek K, Iben S. Truncated Cockayne syndrome B protein represses elongation by RNA polymerase I. J. Mol. Biol. 2008;382:266–274. doi: 10.1016/j.jmb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 23.O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Panov KI, Friedrich JK, Russell J, Zomerdijk JC. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006;25:3310–3322. doi: 10.1038/sj.emboj.7601221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat. Struct. Mol. Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 28.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drane P, Compe E, Catez P, Chymkowitch P, Egly JM. Selective regulation of vitamin D receptor-responsive genes by TFIIH. Mol. Cell. 2004;16:187–197. doi: 10.1016/j.molcel.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Oksenych V, Coin F. The long unwinding road: XPB and XPD helicases in damaged DNA opening. Cell Cycle. 2010;9:90–96. doi: 10.4161/cc.9.1.10267. [DOI] [PubMed] [Google Scholar]
- 31.Guzder SN, Sung P, Bailly V, Prakash L, and Prakash S. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994;369:578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- 32.Dianov GL, Houle JF, Iyer N, Bohr VA, Friedberg EC. Reduced RNA polymerase II transcription in extracts of cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25:3636–3642. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 34.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 36.Laposa RR, Huang EJ, Cleaver JE. Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc. Natl Acad. Sci. USA. 2007;104:1389–1394. doi: 10.1073/pnas.0610619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.