Abstract
Localization of the P1 plasmid requires two proteins, ParA and ParB, which act on the plasmid partition site, parS. ParB is a site-specific DNA-binding protein and ParA is a Walker-type ATPase with non-specific DNA-binding activity. In vivo ParA binds the bacterial nucleoid and forms dynamic patterns that are governed by the ParB–parS partition complex on the plasmid. How these interactions drive plasmid movement and localization is not well understood. Here we have identified a large protein–DNA complex in vitro that requires ParA, ParB and ATP, and have characterized its assembly by sucrose gradient sedimentation and light scattering assays. ATP binding and hydrolysis mediated the assembly and disassembly of this complex, while ADP antagonized complex formation. The complex was not dependent on, but was stabilized by, parS. The properties indicate that ParA and ParB are binding and bridging multiple DNA molecules to create a large meshwork of protein–DNA molecules that involves both specific and non-specific DNA. We propose that this complex represents a dynamic adaptor complex between the plasmid and nucleoid, and further, that this interaction drives the redistribution of partition proteins and the plasmid over the nucleoid during partition.
INTRODUCTION
Stable inheritance of low-copy-number plasmids in bacteria requires plasmid-encoded partition systems, which actively position daughter copies in opposite cell halves prior to cell division. The most common partition systems in bacteria encode two proteins, a partition ATPase (ParA) and a site-specific DNA-binding protein (ParB) that act on a partition site, parS (1). The ParA ATPase is responsible for the movement and localization of the plasmid via interactions with the ParB protein when ParB is bound to parS. The P1 plasmid system in Escherichia coli is an excellent model to study DNA movement in bacteria due to extensive biochemical and genetic characterization of its components (2–7). The P1 system is a ‘Type I’ system, defined by the nature of the partition ATPase; Type I ATPases are specific variants of Walker-type ATPases (8). Recent evidence suggests that localization of Type I plasmids is a highly dynamic process that results in the uniform distribution and positioning of plasmids over the nucleoid regions of the cell (5,6,9–11). In vivo, in the presence of ParA, P1 plasmids co-localize with the bacterial nucleoid, and daughter plasmids are separated to either side of the nascent cell division site (12). In vitro, ParA in its ATP-bound form can bind DNA non-specifically (7) and interact with ParB bound to parS (13). The requirement for the ATP-dependent non-specific DNA (nsDNA) -binding activity of ParA in plasmid partition in P1 and several other Type I systems (7,14,15) has led to models in which plasmids use the nucleoid as a track or matrix upon which they move inside the cell.
Type II partition systems encode an actin-like ATPase, and the best studied example is that of the R1 plasmid (16,17). In the presence of ATP, the R1 ParM ATPase forms self-supporting filaments, which physically push plasmids to opposite cell poles through a mechanism of insertional polymerization between plasmids (18–20). The mechanism of Type I ATPases is less well defined. Several type I ATPases have been shown to form filament bundles in vitro (4,9,15,21–24), which led to the suggestion that Type I ATPases may resemble ParM in mechanism. However, Type I ATPases show no structural or sequence similarity to cytoskeletal ATPases such as ParM or actin. They do however show sequence and structural similarity to bacterial MinD ATPases (4,25–27), which are dynamic proteins necessary for the positioning of the cell division septum at mid-cell (28,29). MinD possesses an ATP-dependent membrane-binding activity, and MinD-GFP has been shown to oscillate over the entire cell length using the membrane as a matrix for movement. These patterns require stimulation of MinD ATPase activity by MinE on the membrane. The parallels between ParA and MinD in their biochemical properties and patterning activities suggest both systems are acting via similar mechanisms (7,27). We have shown recently that ParA undergoes a slow ATP-dependent conformational change that is necessary for it to bind DNA non-specifically (7). A time delay in MinD membrane-binding activity has also been considered as one of the critical requirements for its patterning behavior in vivo. We proposed a model where the time delay, combined with stimulation of ParA ATPase activity by the ParB–parS partition complex, generates an uneven distribution of ParA bound to the nucleoid, which provides a pulling force for plasmid movement. A pulling mechanism is also supported by examination of several fluorescent Type I ParAs, which were observed to dynamically associate with the nucleoid on the poleward side of segregating plasmids rather than in between them (6,10,11,30). Microscopy of P1 plasmids in live cells indicates that in between periods of motion, they spend some time attached to or stationary on the nucleoid (5,6).
Although our previous findings provide a basis for our model, the association between nucleoid-bound ParA and plasmid-bound ParB has yet to be defined. In this study we investigate ParA, ParB and DNA interactions in vitro. We used sucrose gradient sedimentation and light scattering assays to identify a large complex of ParA and ParB bound to DNA that is regulated by the ATP binding and hydrolysis activities of ParA. Complex assembly occurred on nsDNA but was significantly stabilized by the presence of parS. We believe that the nucleoprotein complex identified and characterized here represents the bridging interactions between the partition complex and the nucleoid that promote plasmid localization and movement.
MATERIALS AND METHODS
Protein and DNA
P1 ParA and ParB proteins were purified as previously described (13,31). The plasmids pBEF165 (containing parS) and pBEND5 (the vector) were the primary substrates for complex formation (32). Other DNA substrates were generated by PCR or by sonication of salmon sperm DNA (Supplementary Data).
Sucrose gradient sedimentation
Sucrose gradients (1.8 ml of 5–20% sucrose in Buffer F: 30 mM Tris–HCl pH 8.0, 100 mM NaCl, 2 mM DTT and 5 mM MgCl2) were prepared over a 0.2 ml 60% sucrose shelf. Standard reaction mixtures contained 3 µM ParA, 1.5 µM ParB and 18 µg/ml plasmid DNA in Buffer F with 0.5 mg/ml α-casein and, when present, 0.5 mM of ATP, ADP, or ATPγS. Mixtures were incubated for 5 min at room temperature, loaded on sucrose gradients, and then centrifuged in a Beckman TLS-55 rotor at 200 000 g for 45 min at 20°C. Two drop fractions (∼100 µl), were collected after manually inserting an 18 gauge needle into the 60% sucrose shelf. The sucrose below the needle was collected in a separate fraction. Samples were examined by agarose and/or polyacrylamide gel electrophoresis.
Light scattering
All light scattering experiments were performed at 23°C in Buffer A: 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2 and 0.1 mg of α-casein/ml. Measurements were performed in a Photon Technology International fluorescence system (Birmingham, NJ). The 60 µl sample was illuminated with 467 nm light, and scattered light was collected at a 90o angle. For steady-state data, samples were incubated for 30 min prior to measurement. For time-based measurements, a stable baseline was obtained, the cuvette was removed from its holder to add ParA, ParB, DNA and/or nucleotide and the sample contents were mixed rapidly before repositioning the cuvette (dead time ≈5 s). For competition assays, the scan was paused, competitor was added, the cuvette was repositioned, and measurement was resumed. Unless indicated, all intensity measurements are given in arbitrary units (AU); the light scattered prior to component addition was subtracted from the data presented. Light scattering data from the competition assays were fit to a multi-parameter exponential decay function using the software package, SigmaPlot V10.0 (Systat Software Inc.).
RESULTS
ParA and ParB interact with DNA to form a large complex
DNA binding by ParA and ParB play important roles in P1 plasmid partition. Both site-specific and non-specific binding of ParB onto and around parS are required for the formation of the partition complex on the plasmid while nsDNA binding by ParA results in its nucleoid co-localization (7,33,34). These different DNA-binding activities led us to investigate the influence of ParA and ParB interactions with DNA, and we first used sucrose gradient sedimentation to measure changes in DNA migration caused by protein binding.
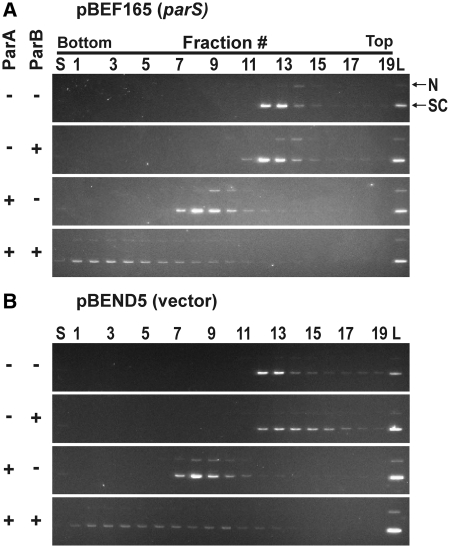
We used a plasmid containing parS, pBEF165 (∼3 kb) and chose sedimentation conditions in which free DNA migrated only a few fractions into the sucrose gradient (Figure 1A) while free protein remained at the top fraction (see below). We observed that ParA, ParB and ATP had a dramatic effect on the migration of DNA in the sucrose gradient (Figure 1A). Under these conditions, plasmid DNA migrated to the bottom of the gradient and was spread over several fractions. The broad distribution indicates that the complex is heterogeneous compared to free DNA. This large complex required both ParA and ParB. ParB alone had no effect on DNA migration. At these concentrations, ParB binds both specifically and non-specifically to DNA (32), so we infer that ParB–DNA complexes are forming but are not large enough to have a noticeable effect on DNA migration in a sucrose gradient. Consistent with this interpretation, the presence of integration host factor (IHF), an E. coli protein that increases the affinity of ParB's site-specific binding activity to parS (32), did not affect the ability of ParB (or ParA and ParB) to alter DNA migration (data not shown). ParA alone (with ATP) formed a complex with DNA that sedimented in between free DNA and the ParA–ParB–DNA complex (Figure 1A). DNA topology did not affect protein binding in this assay as the results were identical on linearized plasmid DNA (data not shown).
Figure 1.
ParA and ParB form large complexes with plasmid DNA. Complexes formed on (A) pBEF165 (parS) and (B) pBend5 (vector) plasmid DNA. ParA and/or ParB (as indicated on the left of each panel) were incubated with DNA and ATP, and then the reaction mixture was separated on a 5–20% sucrose gradient as described in ‘Materials and methods’ section. Fractions were collected from the bottom of the tube, along with the sucrose at the very bottom of the tube (S). A portion of the reaction mixture load (L) and the gradient fractions were examined by electrophoresis in a 0.8% agarose gel stained with ethidium bromide. Only the region of each gel containing DNA is shown in each panel. SC, supercoiled plasmid; N, nicked plasmid.
We next asked if the parS site was essential for protein–DNA complex formation. We repeated the above experiment with pBEND5, the plasmid vector for pBEF165 (Figure 1B). The ParA–ParB–DNA complex and the ParA–DNA complex both formed on pBEND5 and showed similar migration in sucrose gradients as those formed on pBEF165. We conclude that the parS-specific DNA-binding activity of ParB is not necessary to form the ParA–ParB–DNA complex observed by sedimentation. However for reasons presented later, we think that both non-specific and specific-DNA-binding activities of ParB are playing roles in activity of the complex.
Complex formation is regulated by the nucleotide bound state of ParA
We asked how the nucleotide-bound state of ParA affected the reaction. ParA has a nsDNA-binding activity that requires ATP (7), and a site-specific DNA-binding activity to the par operator that prefers ADP over ATP (31,35). The latter is necessary for ParA's transcriptional role as a repressor of the par operon. The ATP form of ParA is necessary for its partition activities (13,35). We repeated the sedimentation experiment with ADP. In contrast to ATP, ADP was unable to support ParA–ParB–DNA complex or ParA–DNA complex formation (Figure 2A). The requirement for ATP indicates that these complexes play a role in partition and not in the repressor function of ParA.
Figure 2.
Adenine nucleotide regulation of protein–DNA interactions. ParA and ParB at the indicated concentrations (in µM) were incubated with pBEF165 DNA and the indicated nucleotide (at 0.5 mM), and the products were examined by density gradient sedimentation in (A) and (C) 5–20% sucrose, and (B) in 10–40% sucrose.
We examined the ability of ATPγS, a non-hydrolyzable analog of ATP, to support complex formation. ATPγS promoted the formation of ParA–ParB–DNA complexes that were larger than those formed with ATP; these complexes sedimented to the bottom of the tube, even at higher sucrose concentrations (Figure 2B). We were able to resolve the complexes formed with ATPγS by reducing the concentrations of ParA and ParB below those necessary to obtain similar complexes formed with ATP (Figure 2C, and see below). The simplest interpretation of these observations is that ATP binding is necessary to form the protein–DNA complex, which is then disassembled by ATP hydrolysis. We further examine this interpretation below.
In contrast to ParA–ParB–DNA complex formation, ATPγS was unable to support ParA binding to DNA in the absence of ParB (Figure 2C), which is consistent with previous studies showing that the ability of ParA on its own to interact with nsDNA requires its ATP-bound conformation (7). The nucleotide specificity for complex formation was identical on pBEND5; that is, in the absence of parS (data not shown).
ParA, ParB and DNA form a large nucleoprotein complex detected by light scattering
We complemented the sucrose-gradient sedimentation assays with a solution-based method called 90o offline light scattering, which allowed us to examine the reaction in real time and without the use of separative steps. The method is a measure of turbidity that tests whether components in solution can assemble into large structures.
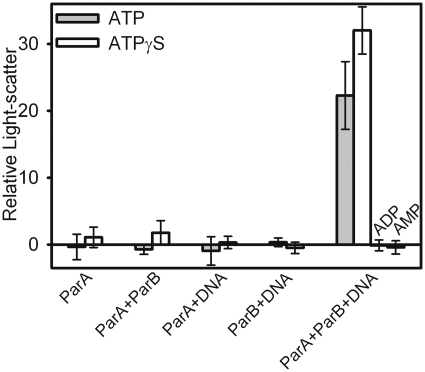
As in the sucrose gradient assay, we first identified the partition components that were necessary to support an increase in light scattering intensity. ParA, ParB and nsDNA were incubated in different combinations, and then changes in light scattering were measured after 30 min (Figure 3). Sonicated salmon sperm DNA was first used as a nsDNA substrate to determine if assembly could occur in the absence of a parS site as shown in the sedimentation assays. Here, a significant increase in light scattering was observed only when ATP was added to ParA, ParB and DNA together (Figure 3). No changes in light scattering were observed in the absence of any one component. ParA–ATP and ParB have nsDNA-binding activities alone; however these nucleoprotein complexes were not detectable under the conditions tested here, and we conclude that these complexes are too small to elicit a significant change in light scattering. Together, the results show ParA–ATP interacts with ParB and DNA to form a large nucleoprotein complex in solution.
Figure 3.
ParA–ParB–DNA complex assembly measured by steady-state light scattering. Mixtures containing 1 µM ParA, 2 µM ParB and 0.1 mg of sonicated salmon sperm DNA/ml were pre-incubated with 1 mM adenine nucleotide as indicated for 30 min at 23°C, and changes in light scattering were measured at 467 nm. The light scattering prior to nucleotide addition (20 ± 2 AU for all samples) was subtracted from the raw data to obtain ‘Relative Light scatter’.
The nucleotide requirements for the large complex detected by light scattering were identical to those defined by sedimentation experiments. An increase in light scattering was observed only in the presence of ATP or ATPγS (Figure 3). ADP and AMP did not elicit an increase in light scatter. The maximum change in light scattering was greater with ATPγS than with ATP. Therefore, ATP-binding is required for ParA to interact with ParB and DNA to produce a significant increase in light scattering in solution. We conclude that sedimentation and light scattering are measuring assembly of the same complex.
A conformational change in ParA limits the rate of ParA–ParB–DNA complex formation
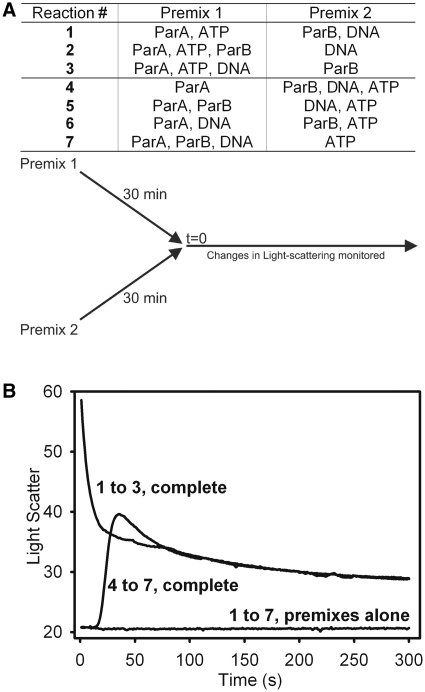
We next examined the kinetics of and order of assembly of the components in the ParA–ParB–DNA complex, using order-of-addition experiments and the light scattering assay. In the time-based assays, we were able to start, pause and stop the scan to add components or competitors. The ‘dead-time’ required for mixing the sample, inserting the cuvette and starting or resuming a scan was 5 ± 2 s. Components were assembled into two ‘premixes’ which were incubated for 30 min and then combined at ‘time zero’ (Figure 4A). ParA was always in premix 1, and all possible combinations of the other components yielded seven conditions. As shown later, complex assembly was influenced by DNA length, therefore for these experiments a nsDNA substrate of uniform length (pBEND5) was used. Only two patterns emerged, either a hyperbolic or a sigmoidal curve, which are summarized in Figure 4B (all curves are shown in Supplementary Figure S1). The difference depended only on the presence of ATP in premix 1; that is, whether ParA was pre-incubated with or without ATP. A hyperbolic curve resulted when ParA was pre-incubated with ATP; the initial time point showed a light scattering signal ∼3-fold greater than the background signal (produced by the premixes) (Figure 4B, reactions 1–3). This pattern indicated that complex assembly initiated during the ∼5 s dead-time (prior to starting the scan), and therefore, the hyperbolic decrease in light scattering is considered as a rapid disassembly of the pre-steady state over accumulation of the complex to steady state.
Figure 4.
Order-of-addition effects on the kinetics of complex assembly. (A) The components, 1 µM ParA, 2 µM ParB, 20 µg/ml pBend5 DNA and 1 mM ATP, were combined in all combinations of two ‘premixes’ such that premix 1 always contained ParA, which yielded seven reaction conditions. For each reaction, each pair of premixes was incubated (separately) for 30 min at 23°C, and then combined, and light scattering was measured after ∼5 s and then followed in real-time. (B) The curves summarize the changes in light scattering for reactions 1–3, 4–7, or each premix alone (the individual curves are provided in Supplementary Figure S1). ‘Light Scatter’ represents raw light scattering intensities.
When ATP was in premix 2 (Figure 4B, reactions 4–7), a sigmoidal increase in light scattering resulted, showing that this order of complex assembly required a long lag phase, which lasted ∼15 s. The lag phase was followed by an exponential increase in light scattering that peaked at a 2-fold greater intensity than the light scattering produced prior to mixing all components (Figure 4B). This maximal change in light scattering was transient and dropped to a more stable state. The hyperbolic and sigmoidal curves aligned after ∼75 s, and we conclude that both methods of complex assembly eventually reached the same steady state complex-size distribution (Figure 4B). All premixes alone showed no significant change in light scattering over a 5 min time course (Figure 4B and Supplementary Figure S1).
Together, the results show that ATP binding by ParA is a prerequisite event in forming the ParA–ParB–DNA complex. We have shown previously that ATP binding by ParA can be detected in two steps; a quick initial but less stable binding followed by a slower more stable nucleotide association (7). The initial ATP-binding step is too fast to be responsible for the slow sigmoidal kinetics of complex assembly (7). Therefore, we think that the slow rate-limiting step is a result of a slow conformational change in ParA that follows stable ATP-binding but precedes hydrolysis.
The parS site stabilizes the ParA–ParB–DNA complex
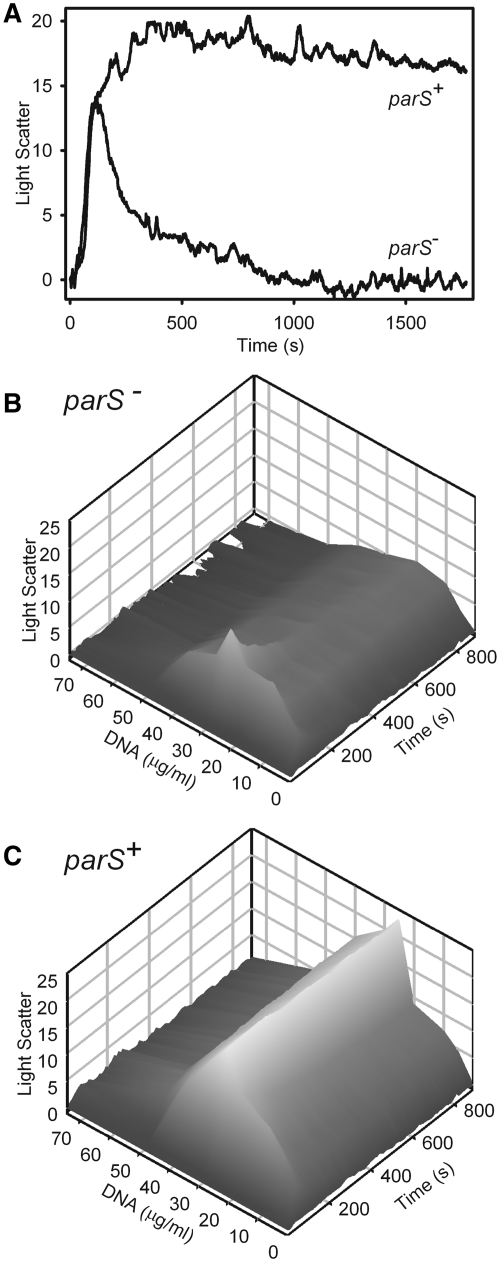
We re-examined the influence of parS using the light scattering assay, comparing pBEF165 (parS) with pBEND5 (vector). The rapid rise in both curves representing assembly kinetics was identical for both plasmids (Figure 5A). However, the plasmid with parS displayed a greater extent of light scattering that was relatively stable 30 min after addition of ATP. Without parS, disassembly occurred more rapidly. When this comparison was performed over a DNA concentration range between 10 and 75 µg/ml, parS supported a light scattering signal that was significantly more stable than the signal supported by nsDNA (Figure 5B and C). Together, the data suggest that while nsDNA can support the ParA–ParB–DNA complex, site-specific interactions between ParB and parS add significantly to complex stability.
Figure 5.
ParA–ParB–DNA complexes are stabilized by parS. (A) Light scattering intensities of ParA–ParB–plasmid DNA complexes were measured as a function of parS (pBEF165 versus pBend5) and DNA concentration. Samples containing 1 µM ParA, 2 µM ParB and 50 µg/ml plasmid DNA were pre-incubated for 30 min, ATP was added to 1 mM, and changes in light scattering were then monitored over time. (B) Samples were assembled with pBend5 DNA (parS−) as in (A) except over a range of DNA concentrations. (C) As in (B) except the plasmid DNA was pBEF165 (parS+).
We also tested the effect of DNA length on complex size using linear DNA fragments of varying size while keeping the mass of DNA constant. The size of the complexes correlated with the length of the DNA substrate used in both gradient sedimentation and light scattering assays (Supplemental Figure S2). Therefore, the DNA is a key component whose length determines the size of the complexes formed.
ParA, ParB and DNA levels influence complex assembly rate and size
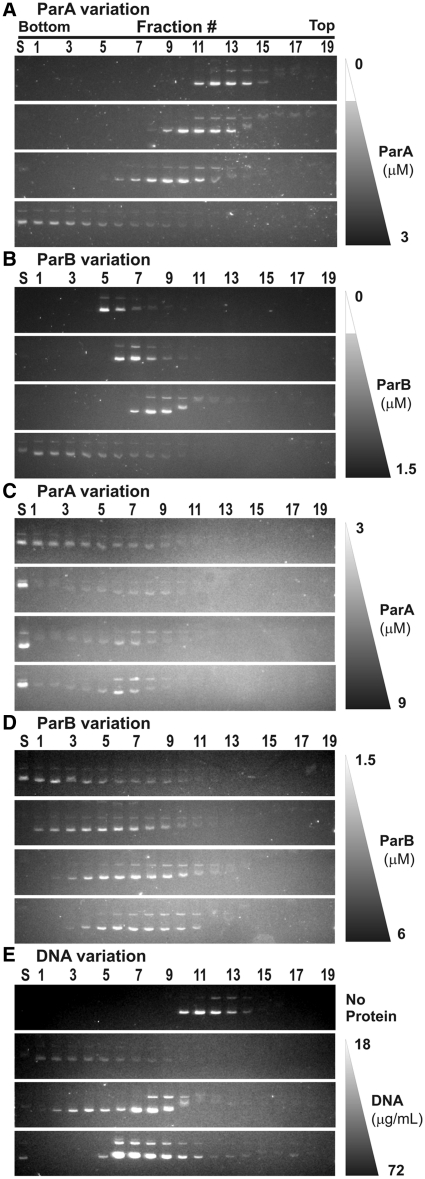
We next measured the effect of the concentration of each component on complex size and kinetics using both sedimentation and light scattering assays. First, gradient sedimentation showed that the absolute and relative concentrations of ParA and ParB affected the size and distribution of the complexes. We varied one component (ParA, ParB, or pBEF165 DNA) while keeping the others constant (Figure 6). Decreasing ParA concentration produced smaller ParA–ParB–DNA complexes (Figure 6A). At low concentrations, ParB actually decreased the size/mobility with respect to ParA–DNA complexes, and a critical concentration of ParB was necessary to form the large ParA–ParB–DNA complexes that sedimented towards the bottom of the gradient (Figure 6B). We believe the simplest explanations for this behavior are that a minimal cooperativity of ParB–DNA binding is necessary to form the large complex, and that below this concentration ParB inhibits ParA–DNA interactions by stimulating its ATPase activity. When ParA concentration alone was increased >3 µM, most of the DNA still pelleted to the bottom of the tube, although some DNA migrated as ParA–DNA complexes (Figure 6C). Increasing ParB concentration caused the ParA–ParB–DNA complex size to decrease (Figure 6D), suggesting that high levels of ParB with respect to ParA disrupt or prevent the formation of the ParA–ParB–DNA complex. Finally, we observed that increasing the DNA concentration decreased the average size of the complexes, from which we conclude that a higher stoichiometry of proteins to DNA produces larger complexes. These results indicate that both the absolute concentration of ParA and its concentration relative to ParB have the greatest effect on complex formation in this assay. In vivo, the concentrations of ParA and ParB are also in the low (∼1–3) µM range, and perturbations in the ratio of ParA to ParB do disturb partition (2,4).
Figure 6.
Protein concentration dependence of complex assembly. In each set of experiments (A–E), one component (ParA, ParB, or pBEF165 DNA) was varied as indicated, and the other two components were present at fixed concentrations. When fixed, ParA was at 3 µM, ParB at 1.5 µM and pBEF165 DNA at 18 µg/ml. ATP was included in all experiments at 0.5 mM. (A) ParA was varied from top to bottom as: 0, 0.75, 1.5 and 3 µM; (B) ParB: 0, 0.375, 0.75 and 1.5 µM; (C) ParA: 3, 4.5, 6 and 9 µM; (D) ParB: 1.5, 3, 4.5 and 6 µM. In (E) DNA and protein were (top to bottom): 18 µg/ml DNA with no protein, and then 18, 36 and 72 µg/ml DNA with standard concentrations of ParA and ParB.
Using the light scattering approach we investigated the protein and DNA requirements in real-time (Figure 7 and Supplementary Figure S3). ATP was added last, which allowed us to monitor two parameters of complex assembly: the start, measured as the ‘lag time’ before light scattering intensity increased, and the ‘extent’, measured as the light scattering signal 30 min after the addition of ATP (Figure 7A). First, increasing ParA concentration decreased the lag time, showing that assembly initiates more rapidly at higher ParA concentrations (Figure 7B). At 1 µM ParA (1:2 ratio of ParA:ParB), the lag-phase lasted ∼ 35 s. Above 4 µM ParA (2:1 ratio of ParA:ParB), the light scattering increased almost immediately, with little or no lag-phase. Like ParA, increasing the ParB concentration decreased the lag-phase, but it did not fall <40 s even at 8 µM ParB (1:8 ratio of ParA:ParB) (Figure 7C). Notably, 40 s is the approximate lag-time when using 1 µM ParA (Figure 7B). The simplest explanation is that the ability of ParB to stimulate ParA–ParB–DNA complex assembly is limited by the amount of ParA present, supporting the idea that ParA is the major protein determinant of the complex assembly rate. Unlike ParA and ParB, increasing the DNA concentration increased the lag-time almost linearly (Figure 7D). Therefore, a high protein:DNA ratio initiates assembly more rapidly, suggesting that complex assembly is cooperative, which is further supported by the sigmoidal kinetics of the reaction.
Figure 7.
ParA, ParB and DNA concentration effects on the kinetics of complex assembly. (A) Changes in light scattering were monitored immediately after the addition of ATP. Lag time was obtained by extrapolating the slope of the exponential phase of the curve to the x-intercept. ‘Extent’ of light scattering was obtained 30 min after ATP-addition (steady state). (B) ParA (C) ParB and (D) DNA concentration effects on the lag time of assembly. (E) ParA, (F) ParB and (G) DNA concentration effects on the extent of light scattering at steady-state. The curves for all titrations are provided in Supplementary Figure S3.
We also compared the extent of light scattering. At least 0.5 µM ParA (1:4 ratio of ParA: ParB) was necessary to detect a significant increase in light scattering (Figure 7E). With increasing ParA concentrations, the extent of complex assembly continued to increase almost linearly and did not level off, consistent with the idea that ParA is the limiting component in this complex. Varying ParB concentration showed that the extent of light scattering was greatest with 1 µM ParB (1:1 ratio of ParA:ParB) (Figure 7F). Unlike ParA, ParB concentrations >2 µM decreased light scattering, and at 10 µM ParB (1:10 ratio of ParA:ParB) no change in light scattering was observed. This trend was also found when varying the DNA concentration (Figure 7G). Overall, the concentration ratio that provided the greatest increase in the extent of light scattering was ≥1 ParA dimer(s): 2 ParB dimers: 120 bp of DNA. Higher ParA continued to increase light scattering whereas higher ParB or DNA decreased it. The finding that ParA concentration is directly correlated to complex size, even at sub-stoichiometric concentrations of ParB, indicates that ParA is the major player in complex assembly.
This conclusion was further supported by the observation that ParA, and ParA with ParB, were able to significantly protect the DNA from limited DNase I digestion (Supplementary Figure S4A). Protection required ATP, and implied that ParA was spreading along the DNA. However, protection was not complete. At higher DNase I levels, degradation of the DNA prevented (if added before ATP) or dismantled (if added after ATP) the complex as measured by light scattering (Supplementary Figure S4B).
We then used the sedimentation approach to measure the amount of protein that co-migrated with DNA (Supplementary Figure S5). From immunoblots of pooled fractions, ∼50 dimers of each ParA and ParB per DNA molecule were observed in the ParA–ParB–DNA complex fractions. Free protein remained at the top of the gradient. To a first approximation, 100 protein dimers do not have sufficient mass to increase the mobility of an individual DNA molecule to the extent observed here. We compared the migration of ParA–ParB–DNA complexes with that of very large DNA molecules, and found that the protein–DNA complexes migrated further in the gradients than (free) DNA molecules over 10 times the size of pBEF165 (Supplementary Figure S6). We suspected that the migration of ParA–ParB–DNA in sucrose gradients with ATP was reflecting both assembly and disassembly of a very large complex consisting of multiple DNA molecules bridged together, resulting in the heterogeneity of complex size observed here.
The observation that these complexes were large enough to scatter light suggested we might be able to visualize them by light microscopy. ParA–ParB–DNA complexes were assembled as above, cross-linked with glutaraldehyde, isolated by centrifugation onto a glass coverslip through a 5% sucrose cushion, and stained for DNA (Supplementary Figure S7). Under these conditions, complexes formed with ATPγS yielded large interconnected mats of DNA (Supplementary Figure S7A and B). An equivalent amount of DNA applied directly to the slide was not directly visible (Supplementary Figure S7F), implying that the increase in fluorescence was due to many DNA molecules held together by protein. ATP supported smaller complexes, while ADP supported none (Supplementary Figure S7C–E). Although the pictures provide a low resolution image of the protein–DNA complexes, they are consistent with ParA and ParB bridging multiple molecules of DNA. Furthermore, the difference between images with ATP, ATPγS and ADP is consistent with the role of ATP binding and hydrolysis in assembly and disassembly measured biochemically.
The ParA–ParB–DNA complex is dynamic
The observation that ATPγS promoted formation of larger complexes than those with ATP suggested ATP-hydrolysis is coupled to complex disassembly. A dynamic complex that assembles and disassembles with ATP binding and hydrolysis predicts that the complex should be reversible and recyclable. We examined this prediction in several ways. First, we monitored complex assembly using limiting nucleotide concentrations (100 µM) in real time by light scattering. Following addition of ATP, the light scattering signal rose to a transient maximum and then dropped to initial values within 1 h (Figure 8A). ATPγS however, produced a higher peak of light scattering and a slow rate of decay. These results mirror those from the sucrose gradient experiments, and we conclude that ATP hydrolysis is necessary for complex disassembly. The rate of light scattering decay with ATP is similar, although not identical, to the rate of ATP hydrolysis, so other processes may also contribute to disassembly (Supplementary Figure S8). ADP, as expected, was unable to support complex formation.
Figure 8.
Complex assembly is reversible and recyclable. (A) Samples containing 1 µM ParA, 2 µM ParB and 0.1 mg/ml sonicated salmon sperm DNA were pre-incubated for 30 min at 23°C. Adenine nucleotide was added to 100 µM and changes in light scattering were monitored over time. (B) As in (A) except at 1 h post-ATP addition, the scan was paused, an additional 100 µM ATP was added, and monitoring was resumed. (C) ADP competition on assembled complexes. Complexes were assembled with ParA–ATP or ParA–ATPγS as in (A) and at t = 0, 2 mM ADP was added where indicated, and monitoring was resumed. The curves were normalized to the light scattering of the sample just prior to competition.
Next, we asked whether the decay in light scattering in the ATP reaction represented disassembled complexes that can be regenerated by a second round of ATP binding by ParA. The ParA–ParB–DNA complex was assembled with ATP and allowed to decay, and then the ATP supply was replenished (Figure 8B). Following the second ATP addition, the signal immediately increased to the previous maxima of light scattering and then dropped back to initial values with kinetics that were similar to the first round of disassembly. Therefore, disassembled ParA subunits in solution can rebind ATP and can be recycled in complex assembly. This result also rules out the possibility that the decrease in light scattering in the presence of ATP represents protein aggregation rather than disassembly of the complex due to ATP depletion.
Finally, we examined the stability of ParA–ParB–DNA complexes by challenging them with ADP, which cannot support complex formation and would thus render disassembly irreversible and unidirectional. ParA–ParB–DNA complexes were first formed with limiting ATP or ATPγS and then challenged with a saturating concentration of ADP (2 mM) (Figure 8C). When complexes were assembled with ATP, addition of ADP resulted in rapid disassembly with a rate of 2.6/min. Complexes formed with ATPγS were more stable but still sensitive to ADP; they decayed at a rate of 0.54/min, ∼5-fold slower than the decay of ATP complexes. We conclude that ParA–ATP turnover within the complex is rapid compared to ATP hydrolysis-mediated disassembly. This observation implies that ParA–ATP subunits associated with the ParA–ParB–DNA complex can exchange with ParA subunits in solution.
DISCUSSION
The ParA–ParB–DNA complex represents dynamic interactions between plasmid and nucleoid
Using a combination of sedimentation, light scattering and fluorescence microscopy, we have identified a large, heterogeneous protein–DNA complex that requires the presence of both ParA and ParB, and is supported by ATP or ATPγS, but not by ADP. The nucleotide requirements are identical to those we have previously shown to support an interaction of ParA with ParB bound to parS (13). We found that complex assembly and disassembly are modulated by ATP binding and hydrolysis. The complex formed on nsDNA, but was more stable in the presence of parS. Fluorescence microscopy revealed large networks of DNA in the complex, and we conclude that bridging of DNA molecules contributes to the large size and high sedimentation mobility of the complex. As we discuss below, we believe that this complex represents a dynamic interaction between the plasmid and the bacterial nucleoid, and refer to it as the nucleoid–adaptor complex or ‘NAC’.
P1 ParA as well as other Type I partition ATPases possess an ATP-dependent nsDNA-binding activity that is essential for partition in vivo (7,14,15,26). Cell biology studies show that ParA co-localizes with the bacterial nucleoid, and that plasmid positioning coincides with the bacterial nucleoid in a ParA-dependent fashion (5,6,10–12). Based on biochemical and conformational studies of P1 ParA, as well as its structural and sequence similarities with bacterial MinD protein, we have presented a model in which P1 plasmids dynamically associate with the bacterial nucleoid via ParA (7). The interactions of ParB with ParA, including ParB stimulation of ParA ATPase activity, set-up dynamic patterns of ParA that in turn position plasmids. In this way, the bacterial nucleoid serves as a matrix support for plasmid partition (as the bacterial membrane serves for MinD localization). The model predicts that ParB/parS partition complexes should dynamically associate with nsDNA in a ParA-dependent fashion, and we believe that the ParA–ParB–DNA complexes, or NAC, identified in this study represent these interactions in vitro.
Composition of NAC
The formation of NAC is absolutely dependent on ParA, ParB, DNA and ATP. Both ParA and ParB are DNA-binding proteins, so we cannot yet dissect the exact nature of the protein–DNA and protein–protein interactions present in NAC. However the properties of the complex and its requirements here provide several clues as to the composition. The sigmoidal shape of the light scattering signal and the longer lag time for complex assembly at lower concentrations of ParB (Figure 7C and F) imply that NAC assembly is favored when protein binding is continuous and cooperative, and that a minimal patch of ParB is necessary to nucleate assembly. This conclusion is further supported by the increase in lag time and decrease in complex size with increasing DNA concentration (Figures 6F, 7D and G). The simplest interpretation of these observations is that the nucleation of bridged complexes involves a highly cooperative process and that at high DNA concentration, protein molecules would be more spaced out and less amenable to bridging.
ParA and the length of the DNA were the main limiting components for the size of the complex. When DNA mass was held constant but length was varied, the extent of light scattering was directly related to DNA size (Supplementary Figure S2), consistent with a bridged network being limited by the size of a DNA scaffold. At optimal ParB levels, ParA is limiting, from which we conclude that ParA is intimately involved in the bridging process; i.e. more ParA, more bridging. This may be direct, via a ParA–DNA interaction (see below), or indirect, via remodeling ParB to promote its bridging activity, or both. As ParB concentration is raised above an approximately equimolar ratio of ParB:ParA, the size of NACs decreases (Figures 6E and 7F), which suggests that ParB is inhibiting ParA action above this ratio. One simple explanation may be the increased stimulation of ParA's ATPase activity by more ParB; however, the answer may turn out to be more complicated as the conformations of these proteins are further examined under these conditions.
The observation that both ATP and ATPγS support NAC assembly raises the question whether ParA is binding directly to DNA. In the absence of ParB, ParA's nsDNA-binding conformation, called ParA–ATP*, is dependent on ATP and no other adenine nucleotide, including ATPγS, will support it (Figure 2) (7). Two explanations are possible (and are not mutually exclusive): first, ParB may allow ATPγS to support ParA's nsDNA-binding activity, or second, ParA with either ATP or ATPγS promotes bridging by ParB bound to DNA. In either case, a ParB–ParA interaction is necessary to form NAC.
These findings raise several questions concerning the number of ParA forms that function in partition as well as how the conformational transitions of ParA are linked. By measuring changes in ParA tryptophan fluorescence under pre-steady state conditions, we have previously shown that ParA undergoes a series of conformational changes upon binding adenine nucleotide, which occur at different time scales: nucleotide binding by a ParA monomer (milliseconds), ParA dimerization (∼20 s) and an ATP-specific conformational change necessary for ParA to bind DNA (ParA–ATP*; ∼1 min with DNA present) (7). Our current data are most consistent with the conclusion that the rate limiting step in NAC assembly is ParA dimerization for several reasons. The lag time for NAC assembly was ∼ 20 s. ParA dimerization and NAC assembly are both strongly ParA concentration dependent. Finally, ATPγS supported NAC and not the ParA–DNA complex, which argues against the idea that the ATP-specific conformational change is a prerequisite to NAC assembly. However, dimerization is not sufficient for NAC assembly as ADP supports dimerization (31) but not NAC (Figures 2 and 3). Therefore, ParB must influence ParA conformation in ways that enable ATP- or ATPγS-bound dimers but not ADP-bound dimers to join NAC. We refer to this conformation of ParA as ParA–ATPC, the ParA component of the complex.
The role of polymerization
Other models for ParA action have proposed that ParA filaments (which we call ‘self-supporting’ filaments) are nucleated by partition complexes and that ParA filament growth away from these complexes pushes or pulls plasmids apart (10,11,21,22). The observation that the length of the DNA limits NAC size however suggests that any polymerization here is limited by the length of the DNA, which is most consistent with polymerization along DNA rather than away from a DNA site. The partial protection of DNA from DNase I (Supplementary Figure S5A) and sensitivity of the light scattering signal to DNase I (Supplementary Figure S5B) are consistent with this idea. Indeed, these possibilities are not mutually exclusive as some amount of spreading and polymerization by ParA and/or ParB across DNA may be necessary for bridging to occur. In addition, because large complexes are necessary to scatter 467 nm light, small polymers of ParA may not be detectable (with or without the other components) with this approach. For example, one possible explanation for the sigmoid nature of the light scattering signal (Figure 4) is that small polymers of ParA must form first to seed the assembly of large visible complexes.
Several Type I partition ATPases have been observed to form filament bundles in vitro by EM and polymerize by light scattering assays (9,21,22,24). However, the experimental conditions and requirements for polymerization activity vary, and a common set of properties has yet to emerge. For example, F plasmid SopA forms small filaments with ATP but not ATPγS, and polymerization is stimulated by SopB but inhibited by nsDNA (22). Polymerization of the ParAs from plasmids TP228, pTAR, pVT745 and pB171 is stimulated by cognate ParBs and partition site DNA, but the effect of nsDNA and DNaseI sensitivity, for example, have not been reported (36). ParAs from T. thermophilus (Soj), pSM19035 (δ) and Vibrio cholerae (ParA2) polymerize across a DNA matrix in an ATP-dependent fashion (24,26,37).
Like Type I ParAs, MinD forms dynamic patterns on an intracellular surface. ParAs pattern the nucleoid, whereas MinD patterns the membrane. In vitro, ATP-bound MinD forms ParA-like filaments that are stimulated by phospholipid vesicles and regulated by MinE (38,39). ParA and MinD show weak ATPase activities that are cooperatively stimulated by accessory proteins (ParB and MinE, respectively) and by the biological surfaces to which they pattern (DNA and phospholipid, respectively). When fluorescently labeled MinD and MinE were visualized on a membrane coated flow cell, the continual chasing of MinD by MinE produced a variety of dynamic patterns (40,41). Consistent in all patterns were regions of MinD–MinE co-localization, which were interpreted as membrane-bound copolymers critical for the system dynamics. We find it an attractive possibility that nucleoid-associated ParA and NAC work together in ways that are similar to how membrane-associated MinD interacts with MinD–MinE copolymers in their respective transport mechanisms. We predict that polymerization on DNA, rather than as self-supporting filaments, will emerge as the common theme among Walker-type partition ATPases; however, the differences in experimental systems and details need to be resolved before a common mechanism can be established.
A model for the adaptor complex
We think that the kinetics and protein requirements measured here are most consistent with the following model of complex composition. First, ParB binds to DNA and results in spreading from the initial binding site. If parS is present, this spreading will occur primarily around parS, but since ParB binds DNA non-specifically, it will bind elsewhere. These cores of ParB interact with ParA–ATP, generating ParA–ATPc, which in turn promotes extensive bridging that is manifested in vitro by a significant increase in sedimentation and light scattering. ATP hydrolysis by ParA results in release of ParA–ADP and the DNA. The complexes are more stable in the presence of parS because ParB affinity is higher for parS than for nonspecific DNA. The exact nature of the complex in vivo is likely not the extensive network of DNA as seen here because of the presence of the bacterial nucleoid. Instead, we envision that a ParB–parS partition complex (i.e. the plasmid) will be attached to the nucleoid via the ATP-dependent bridging activities measured here, and that this attachment is dynamic as ParB–ParA–DNA interactions are both assembled and disassembled with ATP binding and hydrolysis. First, ParA–ATP* is bound to the bacterial nucleoid. ParB and the partition complex interact with the nucleoid via ParA, and then this interaction creates ParA–ATPC/ParB/parS, or NAC, which is the complex that maintains the association between the plasmid and the nucleoid track. ATP hydrolysis by ParA–ATPC is then stimulated by ParB causing ParA–ADP subunits to dissociate and diffuse away from the nucleoid. However, because the partition complex contains many molecules of ParB, an association between the nucleoid and the partition complex is maintained but shifts towards the ParB–ParA–ATPc contacts that have not yet been disrupted by ATP hydrolysis. New ParA–ATPc contacts are then made with ParB where the nucleoid-bound ParA–ATP* concentration is higher, which initiates movement in that direction. As a result of the lower concentration of nucleoid-bound ParA in the wake of NAC movement, the initial drift toward one stochastically chosen direction enforces the continued movement towards the same direction.
Several recent biochemical and cell biology observations in P1 and other plasmid systems support the models for NAC assembly and its interactions with ParA bound to the bacterial nucleoid. The ParA- and ParB-like proteins of the plasmid pSM19035 have been observed in DNA-binding assays to form dynamic complexes that are involved in the bridging of both plasmid and nsDNA (42). The dynamics of P1 ParA and plasmid DNA were simultaneously visualized in living cells (6). Two populations of ParA-associated nucleoprotein complexes were observed; one that diffusely co-localized with the nucleoid and another that densely co-localized with plasmids of limited mobility. We believe the former represents the ParA–DNA complex and the latter represents NAC. Fitting with our model, plasmid movement was only observed when the dense ParA foci disappeared from the plasmids. When the plasmids were finished migrating to opposite cell halves, their fixation were coupled to the reappearance of the co-localized ParA foci. In vivo, F, pB171 as well as P1 plasmids have been shown to oscillate over the nucleoid, apparently ‘chasing’ their cognate partition ATPase (6,10,11). The DNA binding and nucleotide hydrolysis characteristics of the NAC complex provide a framework for the interactions necessary for a partition complex to chase its ATPase across the cell.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
FUNDING
Canadian Institutes of Health Research (MOP-37997 to B.E.F.); University of Toronto Fellowships (to J.C.H., A.G.V.). Funding for open access charge: Canadian Institutes of Health Research (MOP-37997).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Andrew Wilde for his help and advice with the fluorescence microscopy assays, and for the use of his microscope. The authors also thank Kiyoshi Mizuuchi and Ling-Chin Hwang for comments and suggestions concerning this study.
REFERENCES
- 1.Hayes F, Barilla D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 2.Surtees JA, Funnell BE. Plasmid and chromosome traffic control: how ParA and ParB drive partition. Curr. Topics Dev. Biol. 2003;56:145–180. doi: 10.1016/s0070-2153(03)01010-x. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher MA, Funnell BE. ParB-DNA structures reveal DNA-binding mechanism of partition complex formation. Nature. 2005;438:516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 4.Dunham TD, Xu W, Funnell BE, Schumacher MA. Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 2009;28:1792–1802. doi: 10.1038/emboj.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sengupta M, Nielsen HJ, Youngren B, Austin S. P1 plasmid segregation: accurate re-distribution by dynamic plasmid pairing and separation. J. Bacteriol. 2010;192:1175–1183. doi: 10.1128/JB.01245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatano T, Niki H. Partitioning of P1 plasmids by gradual distribution of the ATPase ParA. Mol. Microbiol. 2010;78:1182–1198. doi: 10.1111/j.1365-2958.2010.07398.x. [DOI] [PubMed] [Google Scholar]
- 7.Vecchiarelli AG, Han YW, Tan X, Mizuuchi M, Ghirlando R, Biertümpfel C, Funnell BE, Mizuuchi K. ATP control of dynamic P1 ParA–DNA interactions: a key role for the nucleoid in plasmid partition. Mol. Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes K, Moller-Jensen J, Jensen RB. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 9.Ebersbach G, Ringgaard S, Moller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol. Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- 10.Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 2007;64:1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc. Natl Acad. Sci. USA. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdmann N, Petroff T, Funnell BE. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc. Natl Acad. Sci. USA. 1999;96:14905–14910. doi: 10.1073/pnas.96.26.14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouet J-Y, Funnell BE. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 1999;18:1415–1424. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hester CM, Lutkenhaus J. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc. Natl Acad. Sci. USA. 2007;104:20326–20331. doi: 10.1073/pnas.0705196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castaing J-P, Bouet J-Y, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 2008;70:1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- 16.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu. Rev. Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 17.van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 20.Orlova A, Garner EC, Galkin VE, Heuser J, Mullins RD, Egelman EH. The structure of bacterial ParM filaments. Nat. Struc. Mol. Biol. 2007;14:921–926. doi: 10.1038/nsmb1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barilla D, Rosenberg MF, Nobbmann U, Hayes F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouet J-Y, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 23.Lim GE, Derman AI, Pogliano J. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl Acad. Sci. USA. 2005;102:17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratto F, Cicek A, Weihofen WA, Lurz R, Saenger W, Alonso JC. Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 2008;36:3676–3689. doi: 10.1093/nar/gkn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. (corrigendum: J. Mol. Biol. 1232:1013, 1993) [DOI] [PubMed] [Google Scholar]
- 26.Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer – a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonard TA, Moller-Jensen J, Lowe J. Towards understanding the molecular basis of bacterial DNA segregation. Phil. Trans. R. Soc. B. 2005;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raskin DM, de Boer PAJ. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z Ring. Annu. Rev. Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 30.Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 2006;356:850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- 31.Davey MJ, Funnell BE. The P1 plasmid partition protein ParA. A role for ATP in site-specific DNA binding. J. Biol. Chem. 1994;269:29908–29913. [PubMed] [Google Scholar]
- 32.Funnell BE. The P1 partition complex at parS: the influence of Escherichia coli integration host factor and of substrate topology. J. Biol. Chem. 1991;266:14328–14337. [PubMed] [Google Scholar]
- 33.Davis MA, Austin SJ. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 1988;7:1881–1888. doi: 10.1002/j.1460-2075.1988.tb03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surtees JA, Funnell BE. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 2001;276:12385–12394. doi: 10.1074/jbc.M009370200. [DOI] [PubMed] [Google Scholar]
- 35.Fung E, Bouet J-Y, Funnell BE. Probing the ATP-binding site of P1 ParA: partition and repression have different requirements for ATP binding and hydrolysis. EMBO J. 2001;20:4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machon C, Fothergill TJG, Barilla D, Hayes F. Promiscuous stimulation of ParF protein polymerization by heterogeneous centromere binding factors. J. Mol. Biol. 2007;374:1–8. doi: 10.1016/j.jmb.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 37.Hui MP, Galkin VE, Yu X, Stasiak AZ, Stasiak A, Waldor MK, Egelman EH. ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA. Proc. Natl Acad. Sci. USA. 2010;107:4590–4595. doi: 10.1073/pnas.0913060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suefuji K, Valluzzi R, RayChaudhuri D. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl Acad. Sci. USA. 2002;99:16776–16781. doi: 10.1073/pnas.262671699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu ZL, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl Acad. Sci. USA. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc. Natl Acad. Sci. USA. 2010;107:8071–8078. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soberón NE, Lioy VS, Pratto F, Volante A, Alonso JC. Molecular anatomy of the Streptococcus pyogenes pSM19035 partition and segrosome complexes. Nucleic Acids Res. 2011;39:2624–2637. doi: 10.1093/nar/gkq1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.