Abstract
Homologous recombination (HR) is a major mechanism utilized to repair blockage of DNA replication forks. Here, we report that a sister chromatid exchange (SCE) generated by crossover-associated HR efficiently occurs in response to replication fork stalling before any measurable DNA double-strand breaks (DSBs). Interestingly, SCE produced by replication fork collapse following DNA DSBs creation is specifically suppressed by ATR, a central regulator of the replication checkpoint. BRCA1 depletion leads to decreased RPA2 phosphorylation (RPA2-P) following replication fork stalling but has no obvious effect on RPA2-P following replication fork collapse. Importantly, we found that BRCA1 promotes RAD51 recruitment and SCE induced by replication fork stalling independent of ATR. In contrast, BRCA1 depletion leads to a more profound defect in RAD51 recruitment and SCE induced by replication fork collapse when ATR is depleted. We concluded that BRCA1 plays a dual role in two distinct HR-mediated repair upon replication fork stalling and collapse. Our data established a molecular basis for the observation that defective BRCA1 leads to a high sensitivity to agents that cause replication blocks without being associated with DSBs, and also implicate a novel mechanism by which loss of cell cycle checkpoints promotes BRCA1-associated tumorigenesis via enhancing HR defect resulting from BRCA1 deficiency.
INTRODUCTION
Homologous recombination (HR) promotes genome stability through the precise repair of DNA double-strand breaks (DSBs) and other lesions that are encountered during normal cellular replication (1). Although DNA structures caused by replication arrest are the primary substrate for HR in mitotic mammalian cells (2), the precise functions of breast cancer suppression gene BRCA1 in HR, particularly under replication stress, remain largely unknown.
The repair mechanisms utilized by HR are different, depending on the nature of the DNA structure (3–5). Two ended DNA DSBs can be caused directly by ionizing radiation (IR) or by restriction enzymes. One end DNA breaks can occur indirectly as a result of discontinuities of replication or when stalled replication forks are resolved by endonucleases such as Mus81 (6,7). However, one ended DNA DSBs could subsequently progress to two ended DSBs due to a new origin fire under conditions of replication stress in mammalian cells (8). Alternatively, ssDNA gaps are created without DSBs generation during replication arrest (9). Two ended DNA DSBs in mammalian cells trigger HR repair by short gene conversion (10), whereas spontaneous HR or HR induced by replication inhibition triggers a crossover event (2,11). In addition, our recent publication demonstrated that phosphorylation of RPA2, one subunit of ssDNA binding protein replication protein A (RPA), is specifically required for HR in response to replication arrest but is not essential for the HR induced by DSBs from I-Sce-I overexpression, further supporting the notion that HR mechanisms triggered by replication arrest differ from those involved in repairing classical two ended DSBs (12).
The HR induced by two ended DSBs is initiated by a generation of 3′-ended single-strand DNA (ssDNA). CTIP plays a critical regulatory role in ssDNA resection, along with the Mre11 complex (13). Through the action of recombination mediator/comediator proteins, the RAD51 proteins displace RPA from ssDNA and form a RAD51 nucleoprotein filament (14). Holliday Junction (HJ) intermediates resulting from RAD51 filament-dependent DNA strand invasion and exchanges can be subsequently resolved by gene conversion (non-crossover) or crossover. However, non-crossover products is generated if invaded ssDNA undergoes synthesis-dependent strand annealing (SDSA) (15). The role of BRCA1 in HR induced by DNA DSBs has been demonstrated previously (16–18). Athough the precise molecular mechanisms by which BRCA1 promotes HR are not clear it has been suggested that BRCA1 might act as a mediator/comediator, which facilitates displacement of RPA from ssDNA (19). In addition, a recent study revealed that BRCA1 functions in HR by promoting ssDNA resection via association with CTIP (20). Particularly, several groups suggested a crosstalk between 53BP1 and BRCA1 in ssDNA resection by demonstrating that 53BP1 inhibits HR in BRCA1-deficient cells via a blocking resection of DNA breaks (21–23).
The HR mechanisms required for repairing the lesions caused by replication blockage remain poorly understood in mammalian cells. It appears that similar to the RuvABC complex in Escherichia coli (9), the endonuclease Mus81 in mammalian cells contributes to replication restart by promoting HR via facilitation of one-ended DSB generation (7,9). Interestingly, the one ended DSBs are converted to two-ended DSBs due to new origins firing following replication blockage, which are repaired by RAD51-mediated HR (8). However, HR-mediated repair of DNA DSBs following replication collapse do not contribute to restart of stalled replication forks (8). Although the mechanisms causing this difference have not been identified, it has been well established that HR repair following replication arrest is stimulated by collapsed DNA replication forks when DSBs are generated (8,24).
The observation that HR defective cells are highly sensitive to agents that cause replication blocks without being associated with DSBs suggested that HR is also important for the repair of lesions caused by stalled replication forks (25,26). ssDNA is produced when replication forks are stalled. In yeast, ssDNA-mediated HR is a mechanism to repair stalled DNA replication forks (27). In addition, the substrate for spontaneous sister chromatid recombination is more likely to be an ssDNA gap formed at a stalled replication fork than a DSB (28). ssDNA gap repair in E. coli requires RecA, a human equivalent of RAD51, dependent HR in which gene conversion or crossover products are produced (9,29). In E. coli, ssDNA resection is required to enlarge the ssDNA gap for RAD51-dependent HR (30). Studies in mammalian cells have shown the existence of ssDNA gaps during stalled DNA replication (31,32) but it is not clear how these lesions are repaired in mammalian cells. The observation that BRCA1 is required for subnuclear assembly of RAD51 and survival following treatment with a DNA damaging agent that does not cause obvious DNA DSBs (25,33) suggests that BRCA1 is involved in HR upon replication fork stalling. However, this idea has not been tested.
An emerging concept in the field is the continuous crosstalk between HR and DNA damage checkpoint pathways. DNA replication checkpoint suppresses HR by preventing the formation of recombination foci in yeast (34–36). In mammalian cells, the ATR- and Chk1-dependent checkpoints prevent excessive formation of DNA DSBs, an important substrate for HR, during replication arrest (37,38). It seems that similar control mechanisms exist in both prokaryotic and eukaryotic cells. However, the observation that defects in ATR or Chk1 led to a deficient HR, using HR reporter, suggested that ATR/Chk1 promotes HR (39,40). Therefore, it has not been clear how ATR regulates HR upon replication arrest in mammalian cells
Hydroxyurea (HU) has been used to cause either replication fork stalling or collapse by altering periods of HU treatment (8). We determined the roles of BRCA1 in HR-mediated repair in response to stalled or collapsed DNA replication forks via different periods of HU treatment by measuring sister chromatid exchange (SCE) levels. Here, we report that SCE occurs before and after the presence of any measurable DNA DSBs. BRCA1 plays a dual role in both types of SCE formation via regulation of RAD51 recruitment. However, the role of BRCA1 in SCE-associated HR in response to replication fork collapse is suppressed by ATR. In addition, we report that BRCA1 depletion leads to decreased RPA2-P upon fork stalling but has no effect on RPA2-P following fork collapse. We conclude that BRCA1 plays a dual role in HR in two distinct HR repair upon replication fork stalling and collapse.
MATERIALS AND METHODS
Plasmids, cell lines, infections and transfections
MCF-7, H1299 and MDA-MB-231 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were grown in DMEM supplemented with 10% FBS in a humidified atmosphere of 5% CO2 and 95% air at 37°C. BRCA1 short hairpin RNAs (shRNA) were generated based on lentiviral pLKO.1-puro vector (Sigma, St Louis, MO, USA). The oligonucleotides BRCA1 is forward 5′ CCGGCCCTAAGTTTACTTCTCTAAAC TCGAGTTTAGAGAAGTAAACTTAGGGTTTTTG 3′ and reverse 5′ AATTCAAAAACCCTAAGTTTACTTCTCTAAACTCGAGTTTAGAGAAGTAAACTTAGGG—3′. The oligonucleotides of a second shRNA BRCA1 is forward 5′ CCGGGCCCACCTAATTGTACTGAATCTCGAGATTCAGTACAATTAGGTGGGCTTTTTG and reverse AATTCAAAAAGCCCACCTAATTGTACTGAATCTCGAGATTCAGTACAATTAGGTGGGC. The shRNA sequence targeting non-coding regions of BRCA1 were annealed and cloned into the AgeI and EcoRI sites of the Lentivirus transfer pLKO.1-puro vector. The ATRsh and CTIPsh were purchased from Sigma. All the DNA plasmid transfections were performed using Lipofectamine2000 according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA, USA).
Comet assay
The Neutral Comet Assay was performed using the Comet Assay kit from Trevigen (Gaithersburg, MD, USA) following manufacturer's instructions. The lyses occurred at 4°C for 30 min. Comets were analyzed using CometScore software (TriTek, Sumerduck, VA, USA) (41).
SCE assay
The exponential cells were treated by HU (Hydroxyurea, 2 mM, Sigma) for 6-h or 18-h, and further incubated with BrdU (5 μg/ml, Sigma) for another 50–60 h. The colcemid (0.1 μg/ml, Gibco) was added into the cells for 4-h incubation before fixation onto the slides. Hoechst dye (Invitrogen) and Giemsa (Gibco) were used for SCE staining according to a standard procedure (42).
Immunoblotting and Immunofluorescence analysis
The experiments were performed according to our previous publications (12,41,43).
Real time PCR
CTIP forward primer: ACAGCTGAGGGAACAGCAGAAA. CTIP reverse primer: TCTG CTGGAGTTGTT CAGAAAGC. The second primer of CTIP is forward CTACGTCCACGTGAAAGTTTGG and reverse CCGGATCTATACTCCACTGGAT. Quantitative real time PCR was performed according to the standard procedure. Experiments were carried out in triplicate for each data point.
HR assay
HR was measured in MCF7-pDR-GFP cells according to previous publications (12,24,44). The cells with or without ATR depletion were transfected with I-Sce-I expression vector pCMV3xnls-1-SceI or a control vector or construct pGFP, containing the full-length GFP cDNA, as a calibration control for GFP-positive cells (45). For each analysis, 20 000 cells were processed and each experiment was repeated three times.
Fluorescence in situ hybridization for chromosome aberration analysis
Fluorescence in situ hybridization (FISH) was performed using pan-telomeric peptide nucleic acid (PNA) probes. Telomere (C3TA2)3-specific probe directly labeled with Cy3 fluorescent dyes were obtained from Applied Biosystems (Foster City, CA, USA). The cells with or without HU treatment were processed with FISH analysis 24-h later according to previous publication (12,41).
Antibodies
Anti-BRCA1 (Santa Cruz, D-9), 1:200 dilutions for immunofluorescence staining or western blotting; RAD51 (H92, Santa Cruz Technology), 1:300 dilutions for immunofluorescence; RPA2-P (Bethyl Laboratories),1:200 for immunofluorescence and at 1:1000 for immunoblotting; Anti-phospho-H2AX (Upstate, 05-636),1:500 dilutions for immunostaining; Anti-ATR (Santa Cruz, G4),1:1000 for immunoblotting; Monoclonal antibody RPA2 (EMD Bioscience Inc.), 1:200 for immunofluroscence staining or western Blot; Rabbit-anti-human Phospho-Chk1 (Ser317, Cell Signaling), 1:1000 for immunoblotting; Actin (Sigma), 1:5000 dilutions for immunoblotting; BrdU (Sigma), 1:100 dilution for the immunostaining; anti-HA (covance) was used for immunoblotting at 1:1000. Secondary antibodies were goat-anti-mouse IgG–HRP conjugated and goat-anti-rabbit IgG–HRP conjugated both at 1:5 000 dilutions for immunobloting. Other secondary antibodies for immunofluorescence, goat–anti-mouse IgG Alexa Fluor 594 and chicken-anti-rabbit IgG Alexa Fluor 488 (Molecular Probe), were used at a concentration of 1:300 and 1:500, respectively.
RESULTS
Frequencies of SCE increased following DNA replication fork stalling or collapse
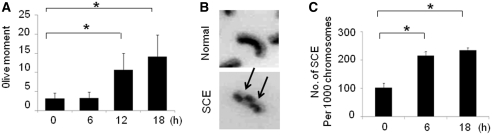
HU depletes the cells of dNTPs which initially results in stalled replication forks that after prolonged treatment collapse into DNA DSBs (8). We found no evidence of DNA DSBs following 6-h HU treatment whereas DNA breaks were obvious in MCF7 cells under 18-h of continuous HU treatment by COMET assays (Figure 1A), consisting with previous reports that prolonged HU treatment could lead to collapse of stalled replication forks and formation of DNA DSBs (8,24). Next, we determined if stalled or collapsed DNA replication forks stimulate HR by cytogenetic SCE assay. SCE results from DNA cross over between two essentially identical DNA duplexes that are generated when a chromosome undergoes DNA replication (Figure 1B). We found that the rate of SCE increased in MCF7 cells treated with HU for 6 h compared to control cells without HU treatment (Figure 1C), indicating that SCE-associated HR occurs following DNA replication fork stalling before DNA DSBs creation. We also found an increased frequency of SCE following 18-h HU treatment when detectable DSBs are created compared to untreated cells. Strikingly, the SCE rate is not obviously elevated following 18-h HU treatment in comparison to that associated with cells treated with HU for 6-h (Figure 1C). To confirm our result, we determined the frequencies of SCE before and after DNA DSBs generation following HU treatment in a second cell line MDA-MB-231. No obvious DNA DSBs are observed following 2- to 6-h HU treatment by Comet assay whereas significant DNA DSBs are detected following 18-h HU treatment in MDA-MB-231 cells (Supplementary Figure S1A). We found that the frequency of SCE induced by 4-h HU treatment in MDA-MB-231 is elevated compared to untreated cells. Similarly, the rate of SCE produced by DSBs following 18-h HU treatment did not produce a dramatic increase compared to that associated with 4-h HU treatment (Supplementary Figure S1B). In summary, the data illustrated in Figure 1 and Supplementary Figure S1 suggest that stalled replication forks can efficiently activate SCE before any detectable DSBs during replication arrest.
Figure 1.
SCE occurs following stalled or collapsed DNA replication forks. (A) The Comet assay was conducted in MCF7 cells treated with 2 mM HU. At least 150 cells were analyzed for each treatment. Results were expressed by Olive moment. P-values were calculated by Student's t-test (*P < 0.01). (B) Representative SCEs. Arrows indicate the discontinuity in the staining pattern of the metaphase chromosomes due to SCEs. (C) The levels of SCE in MCF7 cells with or without HU treatment. In brief, cells treated with 2 mM HU for indicated periods of time were grown in the presence of bromodeoxyuridine (BrdU) for 50–60 h and mitotic cells were prepared according to a standard procedure (see ‘Materials and Methods’ section). Histograms show the frequency of SCE per 1000 chromosomes with at least 40–50 metaphase cells being counted. The data shown is the result from three independent experiments. P-values were calculated by Student's t-test (*P < 0.01).
It should be noted that the term of stalled replication forks here represents a broader implication. A transient phase when the structure and integrity of a replication fork has been disrupted but a DNA DSB has not been created is also included.
The SCE induced by replication fork collapse is specifically inhibited by ATR
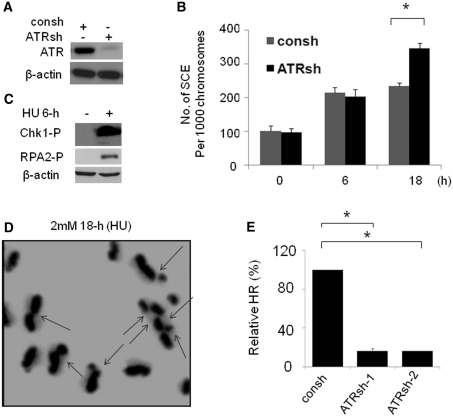
ATR-induced S phase cell cycle checkpoint has been suggested to suppress HR by stabilizing replication forks and preventing the generation of DNA DSBs (35). To test our hypothesis that SCE induced by collapsed replication forks would be blocked by an ATR-dependent cell cycle checkpoint, we assessed SCE in cells depleted of ATR. ATR was efficiently downregulated by lentiviral transduction with a shRNA (ATRsh) (Figure 2A). We found that ATR depletion has no effect on the SCE induced by stalled replication forks since a similar frequency of SCE following 6-h HU treatment was observed in cells with a proficient or deficient ATR (Figure 2B). The lack of effect of ATR depletion on SCE induced by replication fork stalling is not due to a lack of activation of cell cycle checkpoint, since phosphorylation of Chk1 and RPA2, two important downstream factors of ATR, are induced in the cells treated with 6-h HU (Figure 2C). Conversely, we found that SCE frequency is elevated in cells depleted of ATR compared to those observed in control cells following 18-h HU treatment (Figure 2B). This observation is in agreement with previous reports showing that ATR suppresses HR protein foci (35,46). A representative metaphase following 18-h HU treatment in cells depleted of ATR is shown in Figure 2D. Although loss of ATR/Chk1 resulted in increased DNA DSBs following replication arrest (8,35), the increased frequency of SCE in cells depleted of ATR is unlikely to be a consequence of increased amount of DSBs, due to the following observation. We found that ATR knockdown by two independent shRNAs leads to a significant reduction in HR induced by DNA DSBs resulting from I-Sce-I endonuclease overexpression using a DR-GFP HR reporter (12,45) (Figure 2E), consistant with a previous report that ATR kinase dead expression leads to inefficient HR using the same HR report (40). Our result was further confirmed by a second cells line, H1299 cells. With chromosomal integration of DR-GFP reporter (Supplementary Figure S2A and B) (12), we found that depletion of ATR impaired HR induced by I-Sce-I overexpression compared to control cells. We do not think that the differences in frequencies of HR in cells with or without ATR depletion are caused by a variation of I-Sce-I expression because similar levels of HA-I-Sce-I expression were observed under experimental conditions (Supplementary Figure S2C). Given the fact that HR detected by DR-GFP HR reporter is the event mediated by short tract gene conversion (47), our result suggests that ATR promotes HR involved in short tract gene conversion caused by DNA DSBs. We could not exclude the possibility that gene conversion mediated by long track is also regulated by ATR. In summary, we conclude that SCE-associated HR following replication fork collapse is specifically suppressed by ATR whereas SCE-associated HR induced by replication fork stalling is independent of ATR. In addition, HR involved in short gene conversion is promoted by ATR.
Figure 2.
ATR suppresses the SCE induced by replication fork collapse. (A) ATR knockdown by shRNA targeting ATR (ATRsh). Lysates were prepared from MCF7 cells after 72-h infection with ATRsh or control shRNA (consh). (B) The SCEs induced by collapsed DNA replication forks are suppressed by ATR. At 48-h after infection with ATRsh, MCF7 cells were used for SCE assay, which is performed as described in Figure 1C. The data shown is the result from three independent experiments. P-values were calculated by Student's t-test (*P < 0.01). (C) Chk1 phosphorylation (Chk1-P) and RPA2-P (anti-RPA2-p4/8 antibody) are increased in cells treated with 2 mM HU for 6-h. Actin was detected as a load control. (D) Representative metaphase prepared in MCF7 cells treated with 18-h HU following ATR depletion by ATRsh. The chromosomes with SCE(s) were indicated by arrows. (E) ATR-deficient cells show significant reductions in HR mediated by short tract gene conversion. HR induced by I-Sce-I was measured by dual-color flow cytometric detection of GFP-positive cells. The relative HR frequencies in cells depleted of ATR are shown in comparison to cells with intact ATR expression. Results are means from three independent experiments, with standard errors shown. P-values were calculated by Student's t-test (*P < 0.01).
The role of BRCA1 in facilitating SCE induced by replication fork collapse is enhanced by ATR deficiency
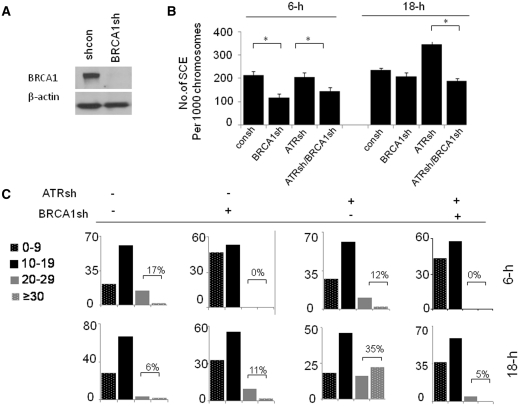
We next determined how BRCA1 regulates SCE following replication arrest. We first established lentiviral (pLKO.1–puro vector)-based constructs which target BRCA1 (BRCA1sh) (Figure 3A). We found that BRCA1 depletion leads to a similar decrease in the frequency of SCE in cells with proficient or deficient ATR following 6-h HU treatment (Figure 3B). These results indicate that BRCA1 promotes HR following replication fork stalling independent of ATR. In contrast, BRCA1 differentially regulates SCE produced by replication fork collapse in cells proficient or deficient in ATR. We found that, in cells with a proficient ATR, BRCA1 knockdown has no obvious effect on SCE following 18-h HU treatment compared to control cells. However, in cells depleted of ATR, BRCA1 depletion results in a decrease in the SCE rate compared to control cells (Figure 3B). Similar results are obtained by further categorizing the frequency of SCE into four classes according to the number of SCE per metaphase (i.e. ≤9 versus 10–19 versus 20–29 versus ≥30 count per metaphase) (Figure 3C). In addition, our observation is further confirmed in MDA-MB-231 cells. We found that BRCA1 knockdown causes a dramatic decrease in the rate of SCE induced by 18-h HU treatment when ATR is depleted. In contrast, BRCA1 knockdown leads to a decrease in SCE formation following 4-h HU treatment independent of ATR depletion (Supplementary Figure S3). Therefore, the data presented in Figure 3 and Supplementary Figure S3 suggests that BRCA1 promotes SCE-associated HR upon replication fork stalling, which is independent of ATR. In contrast, BRCA1 plays a minor role in SCE-associated HR upon replication fork collapse unless ATR is depleted. The differential regulatory roles of ATR in BRCA1 dependent SCE following replication fork stalling or collapse support the notion that the molecular mechanism by which BRCA1 promotes SCE associated HR following replication fork stalling or collapse are distinct.
Figure 3.
The role of BRCA1 in promotion of SCE induced by replication fork collapse is enhanced by ATR depletion. (A) BRCA1 knockdown via shRNA targeting BRCA1 (BRCA1sh) in MCF7 cells. (B) The roles of BRCA1 in SCE upon replication fork stalling and collapse are differentially regulated by ATR depletion. The MCF7 cells were infected by consh, BRCA1sh, ATRsh or both of BRCA1sh and ATRsh. Forty-eight hours after infection, the cells were treated with 2 mM HU for indicated periods of time. The frequencies of SCE per 1000 chromosomes in cells treated with HU are demonstrated. At least 40–50 metaphase cells are counted per group. Results are means from three independent experiments, with standard errors shown. P-values were calculated by Student's t-test (*P < 0.01). (C) The histogram shows the fraction of metaphases with 1–9 versus 10–19 versus 20–29 versus ≥30 SCEs count.
BRCA1 and RAD51 proteins are localized in ssDNA and DNA DSB regions following DNA replication fork stalling or collapse
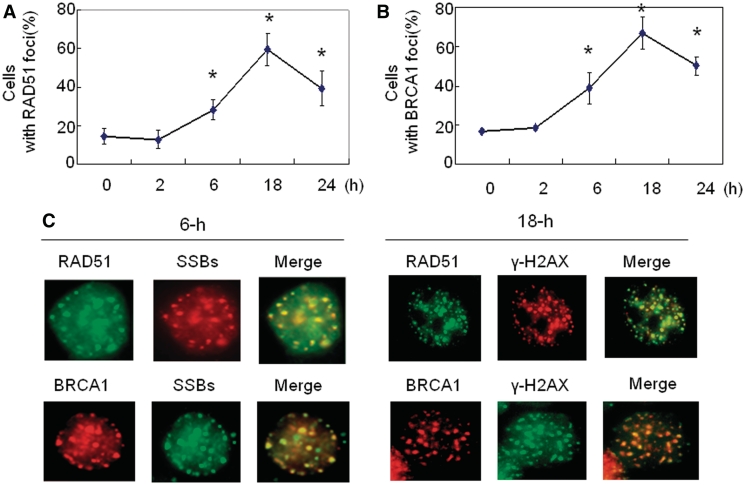
RAD51 plays a key role in controlling SCE in DT40 cells. In addition, overexpression of RAD51 leads to an increased SCE event in Chinese hamster V79 cells (48,49). The RAD51 form foci in response to DNA damage and proteins accumulate at sites of ssDNA, suggesting that RAD51 foci are sites of recombinational DNA repair (50,51). Therefore, we next analyzed the Rad51 foci upon replication arrest. We found that the percentage of cells with RAD51 foci did not show an obvious increase following 1- or 2-h HU treatment in MCF7 cells (Figure 4A), which is similar to that observed in U2OS cells (8). Interestingly, RAD51 form foci following 6-h HU treatment although no obvious DSBs are present at this time point (Figure 1A), which is consistent with a previous report indicating that RAD51 foci is independent of DSBs (7). The formation of Rad51 foci following 6-h HU treatment is concurrent with ssDNA generation since we have reported ssDNA regions are generated at the same condition (12). RAD51 foci became more evident following 18-h HU treatment compared to that observed in cells treated with HU for 6 h (Figure 4A) although the frequency of SCE in cells treated with 18-h HU did not display an obvious increase in comparison to that associated with the cells treated with HU for 6-h (Figure 1C). The main reason for this difference is due to the fact that RAD51 foci were determined right after HU treatment. However the SCE assay was conducted following 50–60 h BrdU labeling and the cells have to go through two cycles before the SCE assay. Therefore, only the cells that went through two cell cycles can be counted for SCE. A fraction of cells containing DSBs-induced RAD51 foci following 18-h HU treatment might die before SCE assay due to an intact ATR signaling. Alternatively, multiple subtypes of HR exist following replication collapse, all of which may involve strand invasion and exchange steps in which RAD51 protein form foci. However, not all of these pathways necessarily result in SCE. For example, SDSA involves strand exchange and invasion but only non-crossover (gene conversion) products are formed (15).
Figure 4.
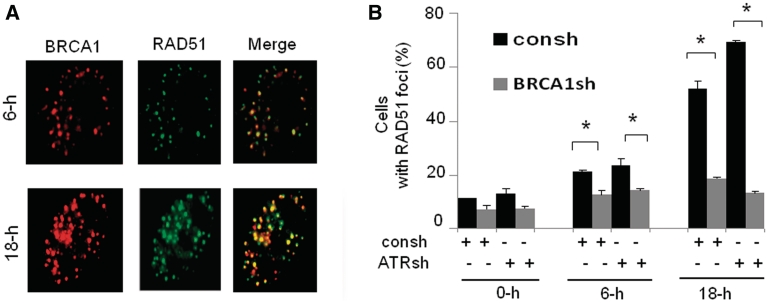
BRCA1 and RAD51 proteins are localized at DNA ssDNA or DSBs region in response to stalled or collapsed DNA replication forks. (A and B). Kinetics of RAD51 (A) or BRCA1 (B) focus formation. The percentage of cells with more than 10 nuclear foci was calculated. In each experiment, 200 nuclei were counted per data point. Error bars indicate standard errors from three independent experiments (*P < 0.01). (C) BRCA1 and RAD51 colocalizes at the ssDNA region in response to replication fork stalling (left panel). The protocol for ssDNA detection has been described in a previous publication (12,51). Cells treated with 6-h HU were co-stained by anti-BrdU antibody and antibody against RAD51 or BRCA1. Representative foci images in response to 6-h HU treatment were shown. BRCA1 and RAD51 protein colocalizes at DSBs region in response to replication fork collapse (right panel). The cells co-stained with anti-γ-H2AX antibodies and antibodies against RAD51 or BRCA1. Representative foci images in response to 18-h HU treatment were shown.
In addition, the dynamic curve of BRCA1 foci in response to HU is similar to that of RAD51 (Figure 4B). We found that both BRCA1 and RAD51 proteins are co-localized with ssDNA regions following 6-h HU treatment (Figure 4C, left panel), and both proteins are co-localized with γ-H2AX foci following replication fork collapse (Figure 4C, right panel). Although γ-H2AX also form foci in the absence of DNA DSBs, it is most likely that H2AX foci following 18-h HU treatment represent DNA DSBs because they are detected by a comet assay at the same condition (Figure 1A). In summary, we conclude that BRCA1 and RAD51 proteins are recruited to ssDNA and DSBs regions following replication fork stalling and collapse, which supports the notion that these two proteins function in HR-mediated repairs upon replication fork stalling or collapse.
The function of BRCA1 in promotion of RAD51 recruitment upon replication fork collapse is enhanced by ATR depletion
The role of BRCA1 in HR repair was initially proposed based on its association with RAD51 (17). To clarify the relationship between BRCA1 and RAD51 following replication fork stalling and collapse, we first determined the co-localization of these two proteins in response to stalled and collapsed DNA replication forks. BRCA1 is co-localized with RAD51 in the cells treated with 6- or 18-h HU, respectively (Figure 5A). To elucidate the molecular mechanisms behind SCE defects in cells with BRCA1 knockdown, we determined how BRCA1 regulates RAD51 foci in cells with or without ATR depletion. We found that ATR depletion has no obvious effect on RAD51 foci formation following 6-h HU treatment. In contrast, ATR depletion leads to a significant increase in RAD51 foci formation following 18-h HU treatment (Figure 5B), indicating that ATR inhibits RAD51 foci induced by replication fork collapse. This result is consistent with a previous report that ATR-deficient cells have increased RAD51 foci in response to replication stress in MEF cells (46). We found that in cells with or without depletion of ATR, the fraction of cells containing RAD51 foci induced by 6-h HU treatment markedly decreased in cells with BRCA1 knockdown. However, we found that BRCA1 knockdown leads to a greater decrease in RAD51 foci formation induced by 18-h HU treatment in cells depleted of ATR compared to the control cells expressing intact ATR (Figure 5B). Altogether, the results presented in Figure 5 suggests that the role of BRCA1 in facilitating RAD51 foci upon replication fork stalling is independent of ATR but the role of BRCA1 in promoting RAD51 recruitment upon replication fork collapse is enhanced by ATR depletion, in line with the observation illustrated in Figure 3B that the role of BRCA1 in SCE are differentially regulated by ATR depletion. We conclude that BRCA1 plays a role in efficient RAD51 recruitment in response to replication fork stalling and collapse. The deficiency in RAD51 recruitment due to BRCA1 depletion upon replication fork collapse was amplified by ATR depletion. This data further support the idea that molecular mechanisms by which BRCA1 promotes HR following replication fork stalling and collapse are different.
Figure 5.
The roles of BRCA1 in promoting RAD51 recruitment induced by replication fork collapse are enhanced by ATR depletion. (A) BRCA1 and Rad51 colocalizes in response to replication fork stalling and collapsed. (B) The roles of BRCA1 in Rad51 recruitment upon replication fork stalling and collapse are differentially regulated by ATR depletion. Exponentially growing MCF7 cells were first infected with both of BRCA1sh and ATRsh. Forty-eight hours after infection, the cells treated with HU were fixed at indicated time points. Then the fixed cells were detected for RAD51 by immunostaining. The data are from three independent experiments. Y-axis represents the percentage cell with positive foci. Cells were scored positive when 10 nuclear foci were visible. Results are means from three independent experiments, with standard errors shown. P-values were calculated by Student's t-test (*P < 0.01).
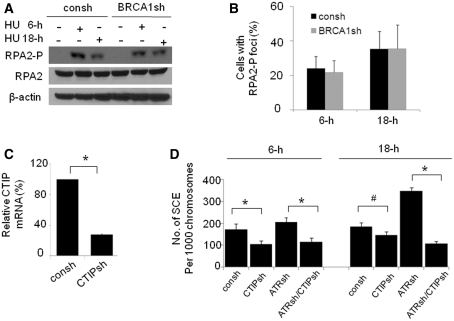
BRCA1 depletion leads to decreased RPA2-P in response to replication fork stalling but has no effect on RPA2-P upon replication fork collapse
We have reported that RPA2-P is required for HR in response to replication arrest (12). BRCA1 is important for ssDNA resection (20). Thus, we next determined RPA2-P levels in cells with or without BRCA1 knockdown. We found that BRCA1 depletion leads to decreased RPA2-P levels by immunoblotting in cells following 6-h HU treatment, suggesting that BRCA1 promotes ssDNA resection following replication fork stalling (Figure 6A). However, BRCA1 depletion appears to have no obvious effect on RPA2-P foci formation because a similar fraction of cells containing RPA2-P foci was observed in cells proficient or deficient in BRCA1 in response to 6-h HU treatment (Figure 6B). The discrepancy between RPA2-P protein levels and RPA2-P foci is probably caused by the fact that RPA foci formation requires several kilobases of ssDNA, and thus the alteration in RPA2-P coated ssDNA may not be detected by immunofluorescence staining (8). Interestingly, we found that BRCA1 depletion has no obvious effect on both RPA2-P protein levels and foci formation following 18-h HU treatment (Figure 6A and B), indicating that the molecular mechanism by which BRCA1 promotes HR upon replication fork stalling and collapse are distinct. To further test our hypothesis that the ssDNA resection is required for BRCA1-dependent HR following replication fork stalling and collapse, we determined how CTIP regulates SCE. CTIP was efficiently downregulated by shRNA targeting CTIP (Figure 6C). We found that CTIP knockdown leads to decreased frequency of SCE following replication fork stalling, which is independent of ATR. In contrast, the CTIP knockdown leads to an obviously decreased SCE frequency in cells depleted of ATR after 18-h HU treatment although it has a minor effect on SCE formation in cells with intact ATR expression (Figure 6D). This result is similar to that observed in cells with BRCA1 knockdown (Figure 3B), indicating that CTIP functions in the same pathway as BRCA1. In summary, the data illustrated in Figure 6 suggests that BRCA1 depletion leads to a decreased RPA2-P following replication fork stalling but has no obvious effect on RPA2-P following replication fork collapse. In addition, BRCA1 promotes ssDNA resection along with CTIP in response to replication fork stalling and collapse.
Figure 6.
BRCA1 depletion leads to decreased RPA2-P protein levels following replication fork stalling but has no effect on RPA2 protein levels following replication fork collapse. (A) Immunoblot analysis of RPA2-P expression in MCF7 cells with or without BRCA1 depletion before and after 2 mM HU treatment for 6-h or 18-h. Proteins were visualized by immunoblot with anti-RPA2-p4/8, or anti-RPA2 antibody. (B) RPA2-P foci in cells with or without BRCA1 depletion. Fixed cells were stained by anti-RPA2-p4/8 antibody. Cells were scored positive when 10 nuclear foci were visible. Data were collected from three independent experiments, and the mean with standard errors was calculated. P-value was calculated by Student's t-test (P > 0.05). (C) CTIP was downregulated by shRNA targeting CTIP (CTIPsh). The CTIP mRNA was amplified by real-time quantitative PCR. mRNA CTIP in cells with or without CTIPsh infection were presented as a relative value compared to control cells infected with consh. Data were collected from three independent experiments. P-values were calculated by Student's t-test (*P < 0.01). (D) CTIP knockdown results in a more profound defect in SCE induced by 18-h HU treatment when ATR is depleted compared to cells with intact ATR expression. The SCE assays were performed as described in Figure 1C. Results are means from three independent experiments, with standard errors shown (*P < 0.01, #P < 0.05). P-values were calculated by Student's t-test.
BRCA1 is critical for maintaining genomic integrity following replication fork stalling
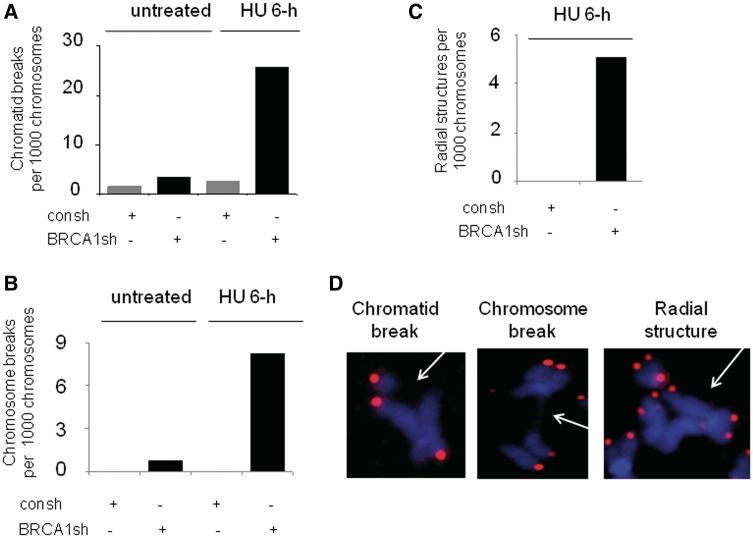
We have found that BRCA1 is required for SCE following replication fork stalling before DSBs are produced (Figure 3B). We anticipate that BRCA1 knockdown leads to accumulated ssDNA following replication fork stalling. However, BRCA1 depletion appears to have no obvious effect on RPA2 foci formation induced by 6-h HU treatment (Supplementary Figure S4). This is most likely caused by the following two reasons. First, as we discussed previously, the immunofluorescence may not be able to detect the accumulated ssDNA due to the limitation of this assay. Second, unrepaired ssDNA upon replication fork stalling is not stable and will be targeted by nucleases, which subsequently leads to DNA DSBs (9). Therefore, the unrepaired ssDNA due to a BRCA1 defect are most likely converted into DSBs. To test this hypothesis, we determined the effect of BRCA1 knockdown on chromatid breaks and chromosome breaks following replication fork stalling, using FISH assay. As expected, cells depleted of BRCA1 by shRNA displayed higher frequencies of chromatid and chromosome breaks after 6-h HU treatment although no detectable breaks are found in control cells (Figure 7A and B). Most importantly, in BRCA1 deficient cells, 6-h HU treatment leads to increased radial structures (Figure 7C) which are generated by the increased frequency of chromatid breaks and subsequent inter-chromatid fusions. The representative chromosome abnormalities induced by 6-h HU treatment in cells with BRCA1 knockdown are demonstrated in Figure 7D. Moreover, both the frequencies of chromatid breaks (Figure 7A) and chromosome breaks (Figure 7B) are not obviously increased in control cells treated with 6-h HU treatment compared to the untreated cells, further confirming the results obtained using a comet assay that no detectable DSBs are present in cells with 6-h HU treatment (Figure 1A). The result from a second repeat was also presented in Supplementary Figure S5. In summary, we concluded that function of BRCA1 in HR induced by replication fork stalling is critical for maintaining genomic stability following replication fork stalling.
Figure 7.
BRCA1 knockdown leads to increased frequencies of chromosome aberrations following replication fork stalling. (A–C) Frequencies of chromatid breaks (A) and chromosome breaks (B) and radial structure (C) in MCF7 cells. For each sample 40–50 metaphases were analyzed. The data shown are from one of two independent experiments. (D)The representative metaphase spreads following 6-h HU treatment in MCF7 cells depleted of BRCA1 are shown. FISH using telomeric probe reveals the pink color. Chromosomes were stained with DAPI (blue).
DISCUSSION
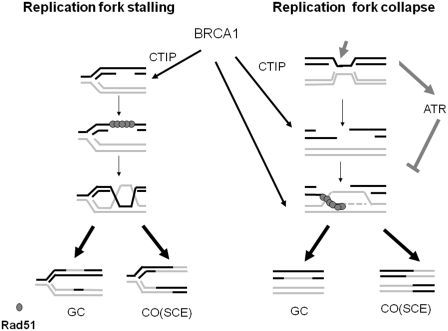
Previous studies have revealed that BRCA1 functions in HR induced by DNA DSBs. However, how BRCA1 functions in HR in response to replication blockages remains poorly understood. Here, we report the existence of two distinct SCE-associated HR repair pathways. One occurs when replication forks stall before DSB generation and the other take place after DSB creation when replication fork collapse. ATR specifically suppresses SCE-associated HR induced by replication fork collapse but has no role on the SCE-associated HR following replication fork stalling. BRCA1 promotes these two distinct HR pathways but the role of BRCA1 in promotion of SCE-associated HR upon replication fork collapse is suppressed by ATR (Figure 8).
Figure 8.
A dual role of BRCA1 in two distinct HR mediated repairs in response to replication arrest. BRCA1 promotes ssDNA gap repair by HR independent of ATR. In addition, BRCA1 facilitates the repair of DNA DSBs following replication fork collapse, which is suppressed by ATR. BRCA1 perhaps functions with CTIP in promoting ssDNA resection at stalling replication forks. However, BRCA1 promotes HR following replication fork collapse via multiple mechanisms, in which BRCA1 acts as a HR mediator/comediator and also facilitates ssDNA resection along with CTIP.
Both stalled and collapsed DNA replication forks stimulate SCE-associated HR in mammalian cells
Consistent with earlier reports (8,24) we found that HR-mediated repair takes place when replication fork progress collapses. However, unprecedentedly, our present study shows that SCE-associated HR efficiently occurs before DSBs are detectable (Figure 1), which is different to the previous reports that replication arrest induced HR is triggered when DNA DSBs are generated using HR reporter assays (8,24). Our results provide molecular basis for the observation that BRCA1 deficiency leads to an enhanced sensitivity to chemotherapeutic drugs that cause replication blockage without being associated with DSBs (25,33). Recombination-related processes have a central function in the recovery of stalled or collapsed replication forks in bacteria (9). The importance of recombination-mediated fork restart mechanisms remains to be established in eukaryotic cells. Thus, it would be interesting to know whether the role of BRCA1 in HR upon replication fork stalling or collapse in mammalian cells contributes to the replication fork recovery.
ATR differentially controls SCE upon replication fork stalling or collapse
We found that SCE-associated HR upon replication fork collapse is blocked by ATR, an essential regulator for cell cycle checkpoints during replication arrest. However, SCE-associated HR occurring following replication fork stalling is independent of ATR (Figure 2). The question raised by our study is why ATR differentially controls SCE-associated HR during replication fork stalling and collapse. The primary means of repairing ssDNA gaps following replication fork stalling is by HR, which uses complementary DNA from the sister chromatid (52,53). This process is inherently error-free compared to other mechanisms, such as error-prone translesion polymerases (53). In contrast, DSB creations are involved during repair of collapsed DNA replication forks by HR, which may cause a consequence of genomic instability due to illegitimate recombination, such as non-homologous end joining. Therefore, HR repair in response to replication fork collapse is suppressed by ATR. Alternatively, a lack of substrates for ATR during replication fork stalling may be one of the reasons. HR repression by ATR operates at least in part, by impeding the resection of cutting free DNA ends (35). There are not obvious free DNA DSB ends present at the site of replication fork stalling. Hence, it is not surprising that SCE-associated HR following forks collapse before DSBs are generated is not affected by the ATR status.
Our data revealed that ATR promotes DSBs induced HR involving short track gene conversion (Figure 2E, Supplementary Figure S2) although ATR suppresses SCE-associated HR following replication fork collapse. These results indicate that subtypes of HR are differently regulated by ATR. HR is a typical error-free repair mechanism. However, inappropriate HR events can have deleterious consequences by causing loss of heterozygosity (LOH) and chromosome translocations. Therefore, HR must be tightly regulated (15). Although error free HR involving gene conversion is favorably operated in unchallenged cells (10), it may not be the case when the cells lose ATR signaling. SCE-associated HR is perhaps a backup mechanism to repair DNA DSBs when the cells with a defective checkpoint signaling are challenged by replication stress. To repair overwhelmingly increased DNA DSBs due to loss of checkpoint signaling, all potential HR mechanisms, including SCE-associated HR, would be activated, particularly when gene conversion-mediated HR is compromised. Hence, SCE-associated HR is more evident when ATR signaling is defective. Alternatively, loss of checkpoint signaling by disruption of ATR leads to a defect in phosphorylation of downstream factor(s) controlling the balance between gene conversion- and crossover-associated HR. Future studies would be required to address the molecular mechanisms associated with differential regulation by ATR in subtypes of HR following replication fork collapse.
BRCA1 deficiency leads to an enhanced HR defect when ATR is hindered
Our study demonstrates that BRCA1 plays a role in SCE-associated HR following replication fork stalling (Figure 3), which contributes to maintenance of genomic stability (Figure 7). Our work provides the evidence supporting the observation that defective BRCA1 leads to be highly sensitive to agents that cause replication arrest without being associated with DSBs. In addition, BRCA1 also promotes SCE upon replication fork collapse when DSBs are generated. However, this mechanism is suppressed by ATR (Figure 3). The function of BRCA1 in SCE-mediated HR following replication fork collapse is amplified when upstream DNA checkpoint signaling is hindered. This finding is of utmost importance because it provides a novel mechanism by which loss of cell cycle checkpoints promotes BRCA1-associated tumorigenesis. Mutations in checkpoint pathways can enhance the likelihood of malignant transformation by permitting the continued growth of cells with genomic abnormalities (54). There is no exception for BRCA1-associated tumorigenesis (55–57). ATR was found downregulated in BRCA1 mutation carriers (58). Thus, it is possible that loss of ATR or other cell cycle checkpoint proteins amplify the phenotypes due to BRCA1 mutation in addition to permitting survival of cells with a BRCA1 mutant. This could be an alternate molecular mechanism explanation of why mutations in a second gene required for cell cycle checkpoints is required for BRCA1-associated cancer development.
BRCA1 promotes SCE-associated HR upon replication fork stalling and collapse via distinct mechanisms
Of note, our results point to distinct molecular mechanisms by which BRCA1 facilitates SCE- associated HR in response to replication fork stalling or collapse based on the following three reasons (Figures 4–6). First, the DNA substrates for SCE-associated HR are different, which are associated with or without DSBs, respectively. Second, the functions of BRCA1 in SCE-associated HR following replication fork stalling and collapses are differentially regulated by ATR deficiency. Third, BRCA1 differentially regulates RPA2-P before or after the presence of DSBs in response to replication arrest, in which BRCA1 depletion leads to an impaired RPA2-P in response to replication fork stalling but has no obvious impact on RPA2-P induced by fork collapse (Figure 6A). The defect of RPA2-P induced by fork stalling in cells with a defective BRCA1 could be caused by an impaired ssDNA resection. RPA2-P occurs when RPA binds to elongated ssDNA and is dependent on the length of ssDNA (59). BRCA1 perhaps functions in ssDNA resection at the stalled replication forks along with CTIP. Although direct evidence indicating that BRCA1/CTIP promotes ssDNA resection at the site of replication fork stalling without being associated with free DNA end has not been reported, it has been demonstrated that the ssDNA gap is required for enlargement by the 5′–3′ exonuclease before HR in bacteria (30). In contrast to a sole role in DSBs end resection by BRCA1 upon replication fork stalling, BRCA1 perhaps functions in HR when replication fork collapses via multiple mechanisms. BRCA1 acts as a mediator/comediator of HR to promote displacement of RPA from ssDNA in addition to enhancing ssDNA resection (Figures 7 and 8). The likely scenario following replication fork collapse is that BRCA1 deficiency is supposed to incur a reduced level of RPA2-P due to impairment in ssDNA resection. However, on the other hand, BRCA1 also functions as a mediator/comediator of RAD51. The defect in BRCA1 could also cause an increase in RPA2-P levels due to a failure of displacement of RPA from ssDNA. This bidirectional regulation by BRCA1 depletion may not necessarily cause an obvious alteration in RPA2-P levels upon replication fork collapse (Figures 6 and 8).
The question as to why BRCA1 plays a profound role in SCE-associated HR in cells with ATR deficiency following replication fork collapse has not been made clear. One possibility is that, as we discussed above, the SCE-mediated repair pathway may became more important when the cells lacking checkpoints are challenged by replication arrest. Therefore, the role of BRCA1 in SCE-associated HR is not evident unless ATR is depleted. Alternatively, loss of ATR may provide substrates for efficient function of BRCA1 in HR. BRCA1 promotes repair of DSBs following replication forks collapse via multiple mechanisms whereas BRCA1 promotes HR following replication fork stalling solely via the facilitation of ssDNA resection (Figure 8). Last, ATR plays a direct role in BRCA1-dependent SCE following replication fork collapse via phosphorylation of BRCA1. The ATR gene has been linked with BRCA1 by the evidence that ATR phosphorylates BRCA1 (60). However, it is unclear how ATR-dependent phosphorylation of BRCA1 alters BRCA1 activity in HR under replication stresses, which would be an interesting question to be addressed in the future.
In conclusion, two distinct SCE-associated HR occur following replication fork stalling and collapse. SCE-associated HR upon replication fork collapse when DSB is created is suppressed by ATR. BRCA1 promotes SCE-associated HR upon replication fork stalling regardless of ATR deficiency but the role of BRCA1 in SCE-associated HR upon replication fork collapse is enhanced by ATR defect. Our study provides a novel mechanism by which loss of cell cycle checkpoints promotes BRCA1-associated tumorigenesis via amplification of HR defect.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Cancer Institute (grant R01CA154625); a seed grant and startup fund from the Department of Radiation Oncology, Washington University School of Medicine, the Dr Joseph Roti Roti and Stephanie Pagano Fund for Mesothelioma Research; American Cancer Society (IRG-58-010-51); Wendy Will Case Cancer Fund (to J.Z.); Independent Innovation Foundation of Shandong University IIFSDU (to Z.F.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We apologize to those whose work was not cited due to the limited space. We thank Dr Wei Shi for the technique assistance for the BRCA1 shRNA preparation.
REFERENCES
- 1.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- 3.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA repair. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Ann. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 5.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 8.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nature Rev. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi W, Feng Z, Zhang J, Gonzalez-Suarez I, Vanderwaal RP, Wu X, Powell SN, Roti Roti JL, Gonzalo S, Zhang J. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis. 2010;31:994–1002. doi: 10.1093/carcin/bgq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 15.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell. Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 16.Moynahan ME, Cui TY, Jasin M. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 17.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schild D, Wiese C. Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res. 2010;38:1061–1070. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30:3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadyk LC, Hartwell LH. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics. 1993;133:469–487. doi: 10.1093/genetics/133.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mozlin AM, Fung CW, Symington LS. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics. 2008;178:113–126. doi: 10.1534/genetics.107.082677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grompone G, Sanchez N, Dusko Ehrlich S, Michel B. Requirement for RecFOR-mediated recombination in priA mutant. Mol. Microbiol. 2004;52:551–562. doi: 10.1111/j.1365-2958.2004.03997.x. [DOI] [PubMed] [Google Scholar]
- 30.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Svoboda DL, Vos JM. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc. Natl Acad. Sci. USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordeiro-Stone M, Makhov AM, Zaritskaya LS, Griffith JD. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 1999;289:1207–1218. doi: 10.1006/jmbi.1999.2847. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 34.Barlow JH, Rothstein R. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J. 2009;28:1121–1130. doi: 10.1038/emboj.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alabert C, Bianco JN, Pasero P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009;28:1131–1141. doi: 10.1038/emboj.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meister P, Taddei A, Vernis L, Poidevin M, Gasser SM, Baldacci G. Temporal separation of replication and recombination requires the intra-S checkpoint. J. Cell Biol. 2005;168:537–544. doi: 10.1083/jcb.200410006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zachos G, Rainey MD, Gillespie DA. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22:713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Ma Z, Vanderwaal RP, Feng Z, Gonzalez-Suarez I, Wang S, Zhang J, Roti Roti JL, Gonzalo S, Zhang J. The mTOR inhibitor Rapamycin Suppresses DNA double-strand break repair. Radiat Res. 2011;175:214–224. doi: 10.1667/rr2323.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayani J, Squire J. Current Protocals in Cell Biology. Hoboken, New Jersey: John Wiley& Sons, Inc; 2004. [Google Scholar]
- 43.Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, Powell S, Zhang J. Disassembly of MDC1 foci is controlled by ubiquitin-proteasome-dependent degradation. J. Biol. Chem. 2008;283:31608–31616. doi: 10.1074/jbc.M801082200. [DOI] [PubMed] [Google Scholar]
- 44.Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, Powell S. Disassembly of MDC1 foci is controlled by ubiquitin-proteasome-dependent degradation. J. Biol. Chem. 2008;283:31608–31616. doi: 10.1074/jbc.M801082200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nature Struct. Mol. Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 46.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnaudeau C, Helleday T, Jenssen D. The RAD51 protein supports homologous recombination by an exchange mechanism in mammalian cells. J. Mol. Biol. 1999;289:1231–1238. doi: 10.1006/jmbi.1999.2856. [DOI] [PubMed] [Google Scholar]
- 50.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith KC, Wang TV, Sharma RC. recA-dependent DNA repair in UV-irradiated Escherichia coli. Journal Photochem. Photobiol. 1987;1:1–11. doi: 10.1016/1011-1344(87)80002-7. [DOI] [PubMed] [Google Scholar]
- 53.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Molecular Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 54.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 55.Cao L, Kim S, Xiao C, Wang RH, Coumoul X, Wang X, Li WM, Xu XL, De Soto JA, Takai H, et al. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J. 2006;25:2167–2177. doi: 10.1038/sj.emboj.7601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holstege H, Joosse SA, van Oostrom CT, Nederlof PM, de Vries A, Jonkers J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009;69:3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nature Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 58.Kote-Jarai Z, Williams RD, Cattini N, Copeland M, Giddings I, Wooster R, tePoele RH, Workman P, Gusterson B, Peacock J, et al. Gene expression profiling after radiation-induced DNA damage is strongly predictive of BRCA1 mutation carrier status. Clin. Cancer Res. 2004;10:958–963. doi: 10.1158/1078-0432.ccr-1067-3. [DOI] [PubMed] [Google Scholar]
- 59.Liu JS, Kuo SR, Melendy T. Phosphorylation of replication protein A by S-phase checkpoint kinases. DNA Repair. 2006;5:369–380. doi: 10.1016/j.dnarep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 60.Chen J. Ataxia telangiectasia-related protein is involved in the phosphorylation of BRCA1 following deoxyribonucleic acid damage. Cancer Res. 2000;60:5037–5039. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.