Abstract
DNA double-strand breaks (DSBs) threaten genome stability in all kingdoms of life and are linked to cancerogenic chromosome aberrations in humans. The Mre11:Rad50 (MR) complex is an evolutionarily conserved complex of two Rad50 ATPases and a dimer of the Mre11 nuclease that senses and processes DSBs and tethers DNA for repair. ATP binding and hydrolysis by Rad50 is functionally coupled to DNA-binding and tethering, but also regulates Mre11's nuclease in processing DNA ends. To understand how ATP controls the interaction between Mre11 and Rad50, we determined the crystal structure of Thermotoga maritima (Tm) MR trapped in an ATP/ADP state. ATP binding to Rad50 induces a large structural change from an open form with accessible Mre11 nuclease sites into a closed form. Remarkably, the NBD dimer binds in the Mre11 DNA-binding cleft blocking Mre11's dsDNA-binding sites. An accompanying large swivel of the Rad50 coiled coil domains appears to prepare the coiled coils for DNA tethering. DNA-binding studies show that within the complex, Rad50 likely forms a dsDNA-binding site in response to ATP, while the Mre11 nuclease module retains a ssDNA-binding site. Our results suggest a possible mechanism for ATP-dependent DNA tethering and DSB processing by MR.
INTRODUCTION
DNA double-strand breaks (DSBs) are among the most cytotoxic forms of DNA lesions and a threat to genomic integrity in all kingdoms of life. They arise during normal processes of DNA metabolism such as replication but can also be induced by genotoxic chemicals or ionizing radiation (1,2). DSBs are predominantly repaired by two different pathways: homologous recombination (HR) and non-homologous end joining (NHEJ) (3–6). In HR, DNA ends are resected and paired with the sister chromatid to prime repair synthesis, resulting in error-free repair. In contrast, in NHEJ the DNA ends are directly ligated without a template, making NHEJ potentially mutagenic.
The Mre11:Rad50:Nbs1 complex [MRN; Mre11:Rad50:Xrs2 in Saccharomyces cerevisiae; Mre11:Rad50 (MR) in prokaryotes] is an evolutionarily conserved key player in the detection and repair of DSBs (7–9). It functions in both the HR and NHEJ repair pathways and is involved in DSB detection, DNA tethering, damage signaling via ATM, structural organization of repair and checkpoint foci, as well as nucleolytic processing of the DNA ends (8). Mutations in MRN subunits or ATM lead to a defective DSB response and can result in cell death or chromosomal aberrations (10–12). Hypomorphic mutations in the MRN–ATM axis are linked to Ataxia–Telangiectasia (A–T, mutations in ATM), A–T like disorder (A–TLD, mutations in MRE11), Nijmegen breakage syndrome (NBS, mutations in NBS1) and NBS-like disorders (NBSLD, mutations in RAD50 or MRE11) (13–16). These diseases are characterized by neurological disorders and/or cancer predisposition (17). The bacterial homolog of the MR complex is implicated in the processing of DNA ends sealed by hairpin structures, in the repair of DNA interstrand cross-links and also plays a role in removing protein bound to DNA termini (18–20). In polyploid archaea, MR can also time repair by delaying recombination in favor of end-joining (21).
The evolutionarily conserved core MR complex consists of the nuclease Mre11 and the ATP-binding protein Rad50 [SbcCD in bacteria (22)]. In eukaryotes, it also contains Nbs1 or its S. cerevisiae homolog Xrs2 (MRN or MRX complex) which is involved in the recruitment of repair and checkpoint proteins to DSB repair sites (14,16,23,24). MR has a heterotetrameric bipolar structure with M2:R2 stoichiometry (25–27). A head module harbors the Rad50 ATPase domains and a dimer of the Mre11 nuclease and is responsible for the ATP stimulated DNA-binding and DNA processing activities of the complex. Two coiled coil domains of Rad50 of up to 500 Å in length emerge from the head and form long molecular tethers: a Zn-hook motif at the apex of the coiled coil domains mediates the formation of larger MR assemblies upon Zn2+-dependent dimerization (28). The precise molecular function of this Zn-hook-mediated tethering is unknown, but it is critical for the function of the MRX complex in S. cerevisiae (29).
The mechanistic role of ATP binding and hydrolysis by Rad50 in regulating various nuclease activities as well as DSB recognition and DNA-binding of the complex is still poorly understood. Mre11 is a metal-dependent ssDNA endonuclease and 3′–5′ dsDNA exonuclease (30). In addition, MRN/X and MR open hairpins and remove 15- to 25-nt long oligonucleotides from the 5′-end of DSBs, a process that is believed to prime DNA ends for further resection in HR and to remove covalently attached Spo11 at meiotic breaks (31–34). ATP engages the two Rad50 nucleotide binding domains (NBDs) by binding to the Walker A and B motifs from one NBD, and to the signature motif of the opposing NBD, a process that induces a dsDNA-binding conformation in Rad50 and MR (35–37). However, the 3′–5′ dsDNA exonuclease activity of yeast and viral MR complexes was shown to be inhibited in the presence of the slowly hydrolysable ATPγS, whereas the endonucleolytic activity was enhanced or unaffected (38,39). On the other hand, the human complex unwinds or melts duplex DNA ends in a manner that is stimulated by ATP (40,41) and endonucleolytic cleavage of dsDNA to remove short oligonucleotides or bound proteins near the break site requires ATP binding and ATP-dependent engagement of the two Rad50 NBDs (34,40). A coherent mechanistic framework for these activities is lacking at present.
The crystal structure of the bacterial MR complex in nucleotide-free form provided the first insight into the interplay between the nuclease and ATPase subunits of the complex (37). Mre11 forms a nuclease dimer that can bind and bridge dsDNA molecules, and mainly interacts with a C-terminal HLH motif with the coiled coil of Rad50 (37,42). In the absence of ATP, Rad50 NBDs are positioned at the outside of the Mre11 nuclease dimer, allowing access of even obstructed DNA ends to Mre11's active site (37). ATP engages the two NBD domains within the catalytic head leading to a higher affinity DNA-binding conformation. Recently, the structure of archaeal MR bound to ATPγS indicates that in this conformation Rad50 blocks Mre11's dsDNA-binding sites (43).
To better understand the conformational changes associated with ATP binding, we determined the structure of bacterial Thermotoga maritima (Tm)MRNBD trapped in an ATP/ADP bound form. On the basis of our earlier structure of the TmMRNBD complex in the absence of ATP, we can now derive a precise structural framework for ATP-induced conformational changes. We find that ATP binding substantially repositions the Rad50 NBDs and induce a large pivot shift in the coiled coil domains. This raises the possibility that tethering and ATP-dependent DNA-binding are allosterically coupled. Most remarkably, the ATP-induced rearrangement repositions the engaged NBDs into the dsDNA-binding groove of the Mre11 dimer in a similar conformation that was observed in the archaeal system, suggesting that ATP-induced blocking of Mre11's active site is evolutionarily conserved. While this conformation is incompatible with dsDNA binding to Mre11, unwound or single-stranded DNA may still reach Mre11's nuclease sites and could offer a structural explanation how MR targets DNA ends and hairpins.
MATERIALS AND METHODS
Cloning, protein expression and purification
The nuclease deficient Mre11H94Q, F291S:Rad50NBD, D804C, H830C mutant from Thermotoga maritima was engineered by fusing the Rad50 N-terminal (residues 1–190) and C-terminal (residues 686–852) segments with an eight amino acid linker (GGAGGAGG) in a single open reading frame of the modified bicistronic pET28 plasmid (Novagen), and co-expressed with Mre11 (residues L7M-385). Recombinant protein was produced in Escherichia coli Rosetta (DE3) cells (Novagen) grown overnight at 18°C in LB media. Cells were harvested by centrifugation, resuspended in 20 mM Tris pH 7.6, 500 mM NaCl, 5 mM ß-Mercaptoethanol and disrupted by sonication. The cell lysate was cleared by centrifugation and the supernatant was incubated at 60°C for 10 min to remove heat labile E. coli proteins. After Ni–NTA affinity chromatography (Qiagen), the protein was purified by Superdex 200 size-exclusion chromatography (GE Healthcare) in 20 mM Tris pH 7.6, 200 mM NaCl, 5 mM β-Mercaptoethanol. Wild-type MRNBD and Mre11HLH (residues 343–385):Rad50NBD were engineered and purified as described previously (37).
Rad50 disulfide bridging
TmMre11H94Q, F291S:Rad50NBD, D804C, H830C complex was rebuffered with 50 mM Tris pH 7.8, 200 mM NaCl, 1 mM MnCl2, 10 mM MgCl2 using a HiTrap Desalting column (GE Healthcare). The protein was concentrated to 4 mg/ml (21.5 µM), mixed with 5 mM ATP and 50 µM CuSO4 and incubated at 25°C for 2 h. Formation of the disulfide bond (denoted S-S) was analyzed by non-reducing SDS–PAGE.
Crystallization and data collection
Crystals of the nuclease deficient TmMre11H94Q, F291S:Rad50NBD, D804C, H830C (S-S) were grown by the sitting drop vapor diffusion method after mixing 1 µl of protein solution at 4 mg/ml protein concentration with 1 µl of the reservoir solution containing 2.2 M ammonium sulfate and 0.2 M ammonium l-tartrate. Crystals appeared after 9 days at 20°C. Prior to flash freezing in liquid nitrogen, crystals were transferred to reservoir solution supplemented with 10% (v/v) sucrose and 10% (v/v) xylitol. Data from native crystals were collected to a resolution of 2.6 Å at the X06SA beamline (Swiss Light Source). The crystals formed in space group P3221 and contained one molecule of Mre11 and Rad50 in an asymmetric unit (Supplementary Table S1). Data were integrated and scaled with XDS (44).
Structure determination and refinement
The structure of TmMre11H94Q, F291S:Rad50NBD, D804C, H830C (S-S) complex was determined by molecular replacement with Phaser (45) using several fragments of TmMre11HLH:Rad50NBD (PDB entry: 3QF7) and TmMre11core (residues M7-325) (PDB entry: 3THN) as search models. Prior to model building and refinement, we randomly omitted 5% of the reflections for monitoring the free R-value. The model was manually built using COOT (46). After corrections for bulk solvent and overall B-values, the model was refined by iterative cycles of positional and B-factor refinement with PHENIX (47) and manual model building with COOT. Data collection and model statistics are given in Supplementary Table S1. All figures were prepared with PyMOL (DeLano Scientific).
Rad50 dimerization
Dimerization of TmMre11HLH:Rad50NBD was carried out by mixing 54 mg/ml (580 µM) protein with 14 mM of the ATP analog AMPPNP. The dimerization process reached its maximum after 3 days of incubation at 4°C. To quantify the amount of dimerized protein, an aliquot of the mixture was loaded onto a S200 5/150 GL column and analyzed by analytical Gel filtration (GE Healthcare).
Electrophoretic mobility shift Assay
Comparison of the DNA affinity of TmMre11HLH:Rad50NBD (Monomer), TmMre11HLH:Rad50NBD (Dimer) and TmMre11H94Q, F291S:Rad50NBD, D804C, H830C (S-S) was carried out with various concentrations of the protein (0, 0.25, 0.50, 1.0, 2.0 and 3.0 mg/ml, respectively) and 0.025 µg/µl of the ds plasmid DNA ΦX174 RFII (NEB). Comparison of the plasmid ssDNA ΦX174 Virion (NEB) binding activity of TmMre11HLH:Rad50NBD (Dimer) and TmMre11F291S:Rad50NBD, D804C, H830C (S-S) was carried out with 2-fold protein and DNA concentrations. The samples were incubated in 50 mM Tris pH 7.8, 100 mM NaCl, 5 mM MgCl2 at room temperature for 15 min in a total volume of 20 µl. TmMre11H94Q, F291S:Rad50NBD, D804C, H830C (S-S) samples contained additionally 100 µM MnCl2, whereas TmMre11HLH:Rad50NBD (Dimer) contained 1 mM of the ATP analog AMPPNP. The DNA was resolved by electrophoresis on an agarose gel (0.5%) in TAE buffer, stained with ethidium bromide and visualized with a UV-Imaging System (Intas).
DNA-binding assays
Blocking of DNA ends for electrophoretic mobility shift assays (EMSAs) was carried out with 100 nM of 5′ fluorescein labeled ds 40-mer and 14 µM of the single-chain (sc) anti-fluorescein antibody Fv fragment FTIC-E2 (48) in buffer 50 mM Tris pH 7.8, 100 mM NaCl, 5 mM MgCl2, 100 µM MnCl2, 3% glycerol and a total volume of 10 µl. After 10 min incubation various concentrations of the nuclease deficient TmMre11H94Q, F291S:Rad50NBD, D804C, H830C (S-S) protein (0, 1.75, 3.50, 7.0 and 14.0 µM respectively) were added to the samples and incubated for another 10 min. The DNA was resolved by electrophoresis on an agarose gel (0.5%) in TB buffer and visualized with a Typhoon scanner (Amersham Biosciences). To verify complete DNA blocking by the scFv, an aliquot of the DNA-scFv mixture was loaded onto a S200 5/150 GL gel filtration column (GE Healthcare). To verify if TmMre11H94Q, F291S: Rad50NBD, D804C, H830C (S-S) can be trapped onto dsDNA, 50 µM of the protein was incubated for 10 min with 500 nM 5′ fluorescein labeled ds 40-mer in buffer containing 50 mM Tris pH 7.8, 300 mM NaCl, 10 mM MgCl2, 1 mM MnCl2. Subsequently, 5 µM of the fluorescein-binding scFv fragment FITC-E2 was added to the DNA–protein mixture and incubated for another 10 min. The samples were loaded onto a S200 5/150 GL gel filtration column (GE Healthcare) and corresponding elution fractions examined by agarose gel electrophoresis (1%).
RESULTS
Structure of the bacterial MR complex in a trapped ATP/ADP state
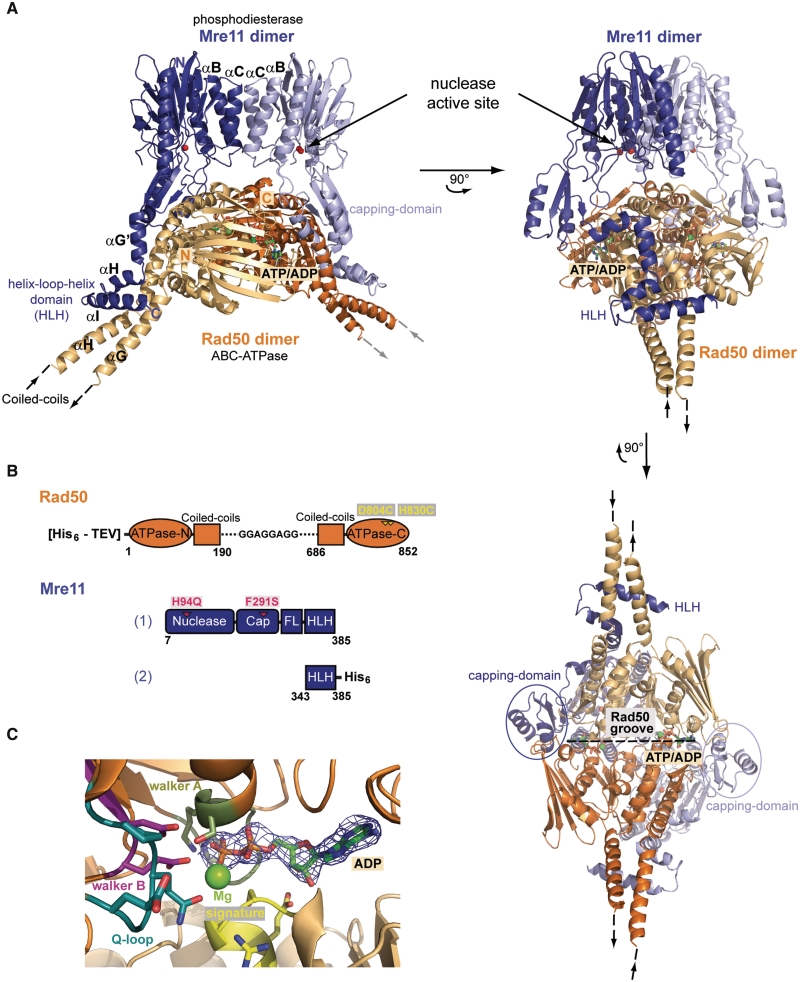
We recently reported the crystal structure of the Thermatoga maritima MR DNA-binding and processing head module [(Tm)MRNBD; Mre11 bound to the Rad50 NBDs] in the absence of ATP (37). Attempts to crystallize this complex in the presence of AMPPNP, ATPγS or other ATP analogs failed, likely because of equilibrium between ATP bound (closed) and free (open) states. To trap TmMRNBD in the ATP bound conformation, we introduced two cysteine residues within the His-switch and D-loop region in the ATPase domain of TmRad50 (D804C and H830C; Supplementary Figure S1). His-switch and D-loops play a role in ATP-dependent dimer formation of NBDs and in positioning of the attacking water molecule (35). Nevertheless, cysteine mutations only slightly decrease ATP hydrolysis activity and the protein remains ∼66% active (Supplementary Figure S1D). While these residues are far apart from each other in the nucleotide free MR complex they have a distance of ∼2.5 Å in the AMPPNP bound TmRad50NBD dimer, perfectly suited for the formation of a disulfide bridge (37). Additionally the mutation F291S, located in an interface between Mre11 and Rad50NBD that stabilizes the open state, was intended to shift the equilibrium from the open to the closed state. Following oxidation, ∼65% of TmMre11H94Q, F291S:Rad50NBD, D804C, H830C (subsequent designated MRNBD (S-S)) became disulfide bridged (Supplementary Figure S1A and S1B). This stabilization enabled us to crystallize and determine the 2.6 Å structure of a trapped ATP/ADP form of TmMR (Figure 1A and B; Supplementary Table S1). Each asymmetric unit contains half a M2RNBD (S-S)2 complex and the complete heterotetrameric complex is generated via a crystallographic 2-fold axis.
Figure 1.
The TmMRNBD (S-S) complex structure in ATP/ADP conformation. (A) Ribbon representation of the TmMRNBD (S-S) heterotetrameric head module complex shown in three different orientations, with Mre11 in blue and Rad50 in orange. Individual domains and important motifs are highlighted and annotated. (B) Constructs of TmMre11 and TmRad50NBD (S-S) used for crystallization. (C) Close up view of the ATP-binding site of TmMRNBD (S-S) with the initially obtained Fo–Fc map (contoured at 2.0σ and colored blue) showing a clearly defined electron density for the ADP ligand.
TmM2RNBD (S-S)2 has a compact ring shape with striking differences to the previously published open ATP free state (Figure 1A). The core of the complex has dimensions of 75 × 80 × 92 Å and consists of the phosphodiesterase and capping domains of the Mre11 dimer as well as the ATP bound, engaged NBDs of Rad50. The NBDs of Rad50 sandwich two nucleotides and Mg2+ in the typical head to tail orientation of lobes I and II (35,37). We noticed that during disulfide bridging or crystallization, the ATP used to engage NBDs for oxidative formation of the disulfide bonds was hydrolyzed to ADP (Figure 1C). The structural similarity of the resulting engaged NBD dimer to the AMPPNP bound dimers of isolated Rad50 NBDs from both T. maritima and P. furiosus conforms that the cross-linking traps an ATP like state (Supplementary Figure S1C).
Surprisingly, the engaged NBD dimer is situated in the groove of the Mre11 dimer that harbors the nuclease active sites (Figure 1A). It contacts the phosphodiesterase domains and is encompassed by the two capping domains. The buried surface between Mre11 and Rad50 comprises a large area of 3052 Å2. However, the new interfaces between capping and phosphodiesterase domains are quite polar and cover a rather small area each. For that reason, they are presumably transient, also because in the presence of ATPγS, a substantial fraction (est. 20–40%) of TmMRNBD is still found in the open state in SAXS experiments (37). The C-terminal HLH domain of Mre11 binds tightly across the base of Rad50NBD coiled coil domains, as seen already in the open form of TmMRNBD. This interface consists mostly of hydrophobic interactions and the preservation of this interaction suggests that this interface is unaltered during the conformational cycle and ‘anchors’ Mre11 and Rad50 in a stable but flexible manner.
In summary, ATP engages both NBDs of Rad50 into a dimer that closely fits into the DNA-binding/active site groove of the Mre11 dimer. This interaction suggests that ATP binding to Rad50 regulates MR by sterically controlling access to Mre11s nuclease and DNA-binding sites.
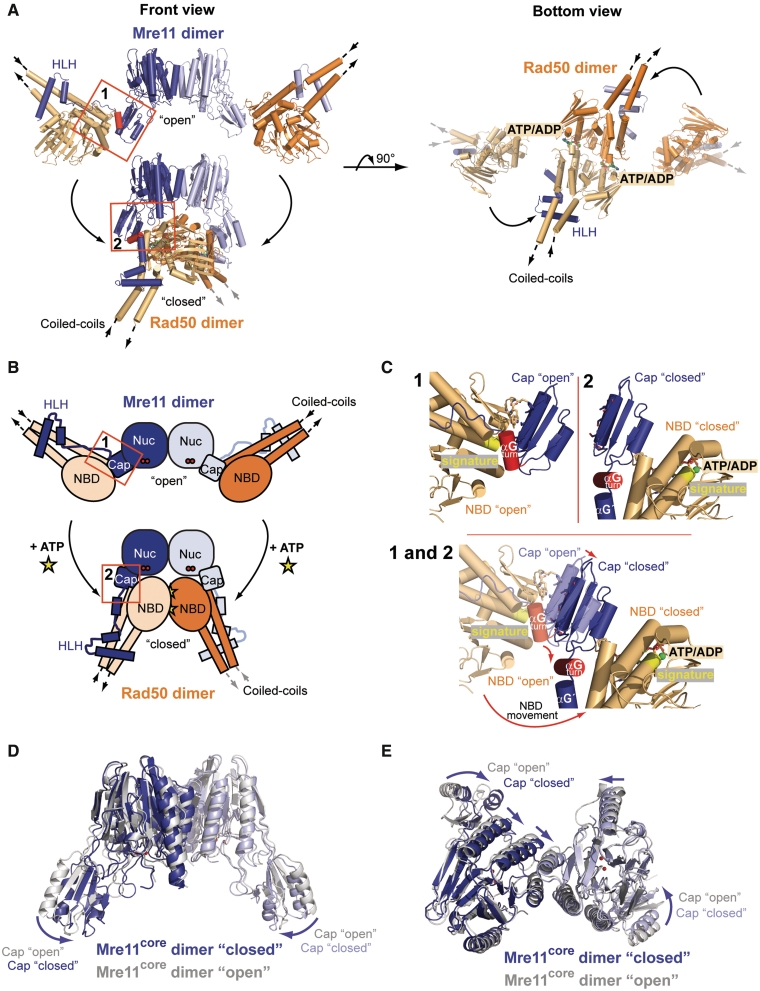
ATP-induced conformational changes
Our results provide a molecular framework for ATP-induced conformational changes by directly comparing the trapped ATP/ADP closed state with the previously open state of TmMRNBD (37) (Figure 2A). The large conformational change within the whole complex induces a globular and compact shape, and we analyzed the structure using previously published small angle X-ray scattering (SAXS) data of TmMRNBD in solution (37) (Supplementary Figure S2). Both scattering intensities and P(r) distribution of TmMRNBD in the presence of ATPγS show characteristic features (e.g. shape of curve and radius of maximum in P(r)) that are qualitatively but not quantitatively preserved in the scattering intensities and P(r) from the crystal structure of ATP/ADP state (Supplementary Figure S2A and S2B). The experimental scattering curves can be interpreted as linear combination of the scattering curves from the two open and closed state crystal structures. Thus, it appears that TmMR exists in equilibrium between two populated states (open and closed), with ATP shifting the equilibrium towards to the closed state.
Figure 2.
Conformational change of the TmMRNBD complex upon nucleotide binding. (A) Comparison of the TmMRNBD complex structures in the open and ATP/ADP-bound conformation shown in two different orientations (colored according to Figure 1A). (B) Schematic representation of the overall domain movement within the complex upon ATP binding. Conformational changes in interface II are highlighted. (C) Close up view of the Mre11 capping domain and interface II in the open (1) and closed (2) TmMRNBD complex. The capping domains move inward by ∼4 Å due to their interaction with the Rad50 NBDs and helix αGturn rotates by roughly 90° from the open to the closed state. (D and E) Superposition of the TmMre11 core domains of the nucleotide bound (blue) and unbound (gray; PDB entry: 3QG5) structures. The arrows indicate the rotation of the capping domain toward the nuclease active site upon Rad50 ATP binding. (E) The bottom view highlights the slight changes in the arrangement of the four-helix bundle between the open and closed conformation (straight arrows).
The open to closed transition is driven by a large swivel movement of the NBDs, which move from the periphery of the MR catalytic head to its center (Figure 2A). Rad50 loses the interaction with the outward facing side of the capping domain but gains a new interaction with the inward facing side. As a result, the angle between the coiled coils is altered from 120° in the open complex to ∼90° in the closed conformation. However, the coiled coils are almost inverted in their orientation with respect to Mre11 and undergo an axial rotation with respect to each other. Such a substantial, large conformational change might be a suitable basis for an ATP-dependent tethering of multiple MR complexes (see ‘Discussion’ section).
We also observe a slight compaction of the Mre11 dimer, driven by the 4 Å inward movement of the capping domains due to their interaction with the Rad50 NBDs and a slight alteration of the Mre11 dimer angle (Figures 2D, E and 3C). Slight to moderate movements of the cap domains in response to DNA have been seen by structural analyses of Mre11 in complex with DNA (49). Cross-linking analysis also indicated that ATPγS binding to Rad50 alters the conformation of Mre11 phosphodiesterases (37). At present, the functional role of these rather moderate conformational changes in Mre11 remains to be investigated. One possibility is that they are involved in dsDNA endonuclease activity, which requires ATP binding to Rad50.
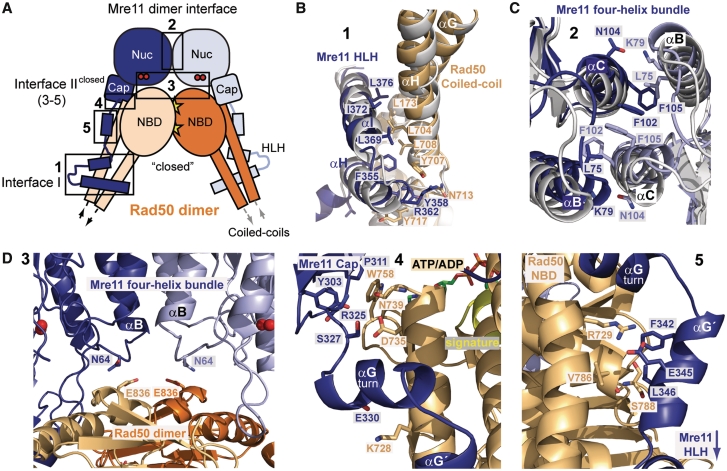
Figure 3.
Details of the TmMRNBD (S-S) interfaces. (A) Denotation of interfaces. (B) Superposition of interface I of the open and closed TmMRNBD in ribbon representation indicate that the interactions remain unchanged (closed complex is colored according to Figure 3A and the open complex is shown in gray). (C) Superposition of the Mre11 homodimer interface (colored according to Figure 3B). (D) Details of the altered and newly formed interface II of the TmMRNBD (S-S) complex in ATP/ADP state (3–5). Selected side chains are shown as color coded sticks and annotated.
Taken together, the conformational changes can be viewed as a rigid body movement of three modules. One module represents the dimer of Mre11's phosphodiesterase and capping domain (Figure 2D and E). The two other modules are both Rad50 polypeptides together with the HLH domains of Mre11. From closer consideration, there are also some notable changes on the secondary structure and subdomain level. The largest conformational rearrangement in this respect is observed in helix αGturn that follows the capping domain in the primary structure of Mre11 (Figure 2B and C). αGturn rotates ∼90° from the open to the closed state. This flexibility helps the relocation of the Rad50 molecules from the outward facing sides of the capping domains to the inward facing sides. In addition, the motif following αG undergoes a substantial structural rearrangement: in the open state, it is a flexible, rather unstructured linker (FL) connecting the HLH and αGturn. Upon relocation of the NBD-HLH module, this motif undergoes a disorder to order transition and forms αG′. As a consequence, a right-angled, loose helix–loop–helix conformation forms at the lateral entry side of Mre11's blocked DNA-binding cleft. It may act as a spring to facilitate and allow conformational changes between the flexible modules, but is also positioned to perhaps function in DNA-binding (see below).
Anchoring and transient interfaces
The two observed conformational states are stabilized by three types of macromolecular interfaces (Figure 3A). The two ‘anchor’ interfaces between the HLH:coiled coil (Interface I) (Figure 3B) and phosphodiesterase:phosphodiesterase (Mre11 dimer interface) (Figure 3C) preserve the Mre112:Rad502 heterotetramer during the conformational cycle and are maintained in the open and closed states. Finally, a third transient interface between the capping domain and NBD and phosphodiesterase and NBD (Interface II) specifically orients Rad50 to Mre11 in open and closed states (Figure 3D).
The Mre11 dimer interface is a four-helix bundle composed of αB and αC of the two Mre11 phosphodiesterase domains as already described for TmMR (37). Comparison of the MRNBD and MRNBD (S-S) dimer interface shows a slight change in the arrangement of the four-helix bundle between the open and closed complex (Figure 3C). The second ‘anchor’ interface between the HLH domain of Mre11 and the coiled coil of Rad50 is largely unaltered between open and closed states (Figure 3B). This interface comprises an area of 1334 Å2 and is composed of mainly aromatic and hydrophobic residues, primarily between αHMre11 and αHRad50 (Mre11:Y351, F352, F355, Y358; Rad50: Y707, F714, Y717, F718). The hydrophobicity in the motifs of interface I is highly conserved in all known homologs. However, it is interesting to note that despite the motif's functional conservation, an additional third Mre11 helix interacts with the Rad50 coiled-coil in archaea (Supplementary Figure S3) (42,43).
Large changes in response to ATP binding are seen in Interface II. The interface between the NBDs and the outward facing side of the capping domain (Interface II open) is disrupted and several new interaction sites are formed (Interface II closed) (Figure 3D). Adjacent to the Mre11 four-helix bundle dimer interface, (αBMre11) residue N64Mre11 contacts E836Rad50, a highly conserved residue in Rad50 (Figure 3D-3). An additional arginine R832Rad50, disordered in our structure, could provide additional interactions with N64Mre11.
The capping domains of Mre11 interact with regions in close proximity to motifs of the ATP-binding site in Rad50 (Figure 3D-4). Residue W758Rad50, situated near the base of a coiled coil helix, forms a small hydrophobic interface with the inward facing side of the capping domain, flanked by several hydrogen bonds. This residue also stabilizes the interaction of the NBDs with the outward facing site of the capping domain β-sheet.
Additionally some interactions are generated, between Rad50 and the newly formed helix αG′ in the course of the disorder to order transition (Figure 3D-5).
DNA affinity of the TmMRNBD complex
A major result of the structural analysis here is that in the ATP bound form Rad50 blocks Mre11's DNA-binding sites (see below). However, previous analysis of the DNA affinity of the disulfide bridged and or AMPPNP bound TmMRNBD compared to TmMRNBD in the absence of ATP revealed increased affinity to single- and double-stranded DNA (37).
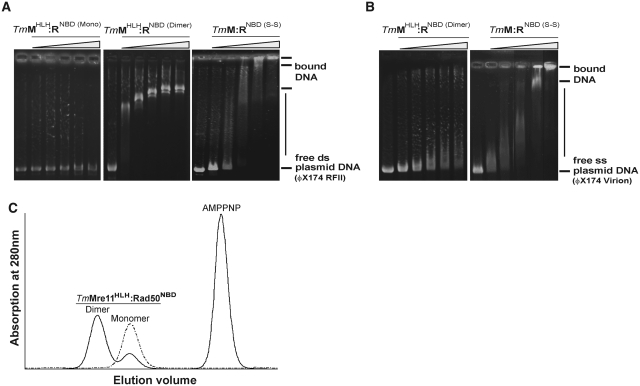
To see if and how the single components TmMre11, TmRad50NBD, or the entire TmMRNBD complex may interact with DNA, we analyzed Mre11HLH:Rad50NBD, lacking the Mre11 ‘core’ domain residues M7-342, in comparison to the whole disulfide bridged MRNBD (S-S) complex (Figure 4A–C). Prior to analysis, Mre11HLH:Rad50NBD was incubated with the non-hydrolysable ATP analog AMPPNP for 3 days at 4°C to allow formation of engaged NBD-NBD dimers (denoted Mre11HLH:Rad50NBD (Dimer)). Subsequent assessment of the dimeric state was achieved by analytical gel filtration. Even though the dimer fraction was varying, a minimum level of 75% of Mre11HLH:Rad50NBD (Dimer) was reached (Figure 4C). Although TmMre11HLH:Rad50NBD (Dimer) lacks the phosphodiesterase as well as the capping domains of Mre11, previously identified as important dsDNA-binding elements (49), it binds plasmid dsDNA (ФX174 RFII) with an affinity that is comparable to that of TmMRNBD (S-S), apart from minor differences in the yield of disulfide bridged protein and dimerization efficiency. We could not detect DNA-binding of the Mre11HLH:Rad50NBD monomer in the absence of ATP as expected (Figure 4A). Thus, the ATP-induced high affinity dsDNA-binding site of TmMR is located in the NBD-HLH module and requires dimerization of this, either by binding of AMPPNP or by disulfide mediated cross-linking of the ATP bound form followed by ATP hydrolysis to ADP.
Figure 4.
Comparison of the DNA affinity of TmMre11HLH:Rad50NBD and TmMRNBD (S-S) using EMSA. (A) While the monomeric TmMre11HLH:Rad50NBD protein shows no binding to ds plasmid DNA (ФX174 RFII), comparable DNA-binding affinities of TmMre11HLH:Rad50NBD (Dimer) (lacks Mre11 catalytic core) and the heterotetrameric TmMRNBD (S-S) complex could be observed. (B) Comparison of the DNA-binding affinity to ss plasmid DNA (ФX174 Virion) revealed a weak interaction between TmMre11HLH:Rad50NBD (Dimer) and DNA whereas TmMRNBD (S-S) showed no difference in binding affinity compared to ds plasmid DNA. Analyzed protein concentrations (0, 0.25, 0.50, 1.0, 2.0 and 3.0 mg/ml respectively). (C) Gel filtration chromatogram of TmMre11HLH:Rad50NBD in the presence (solid line) or absence (dashed line) of AMPPNP. Incubation of monomeric protein with AMPPNP lead to ∼75% dimerized protein.
In contrast to dsDNA-binding, Mre11HLH:Rad50NBD (Dimer) showed almost no binding to single-stranded plasmid DNA (ФX174 Virion). Thus, the binding site on this module is highly specific to dsDNA. However, the disulfide bridged closed complex not only binds dsDNA with high affinity, but also efficiently bound the ssDNA plasmid (Figure 4 B). Thus, the presence of Mre11 nuclease and capping domains created an additional ssDNA-binding site in the complex. Alternatively, short hairpins could form on ФX174 ssDNA which might be recognized by TmMRNBD (S-S) but not by the dsDNA-binding site of TmMre11HLH:Rad50NBD (Dimer). It is therefore conceivable that in the ATP bound closed state the NBD-HLH module dimer is responsible for binding to dsDNA while Mre11 nuclease and capping domains might still bind ssDNA but are blocked from binding dsDNA. These data are consistent with the biochemistry of the bacteriophage MR homolog. Here, the dsDNA exonuclease was inhibited by ATPγS, while ssDNA endonuclease was not (39).
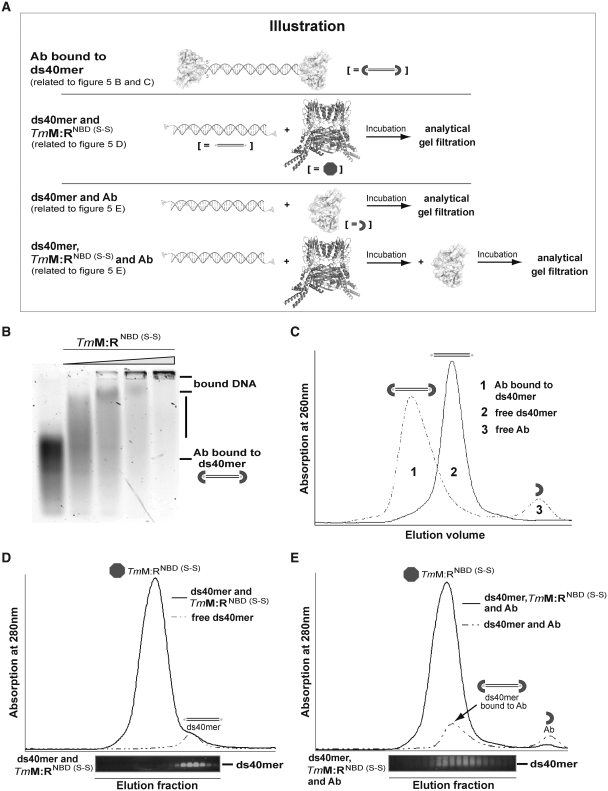
The possibility remains that although the crystal structures reported here and before (37) explains the solution SAXS data, DNA induces an additional conformational change enabling it to bind to both Mre11 nuclease and capping domains and NBD-HLH module. For instance, a relatively moderate structural change could lead to dsDNA being sandwiched between Rad50 NBDs and the phosphodiesterase domains of Mre11. To test this, DNA-binding of MRNBD (S-S) was investigated via EMSA and gel filtration with dsDNA oligos containing a 5′ fluorescein label. A single-chain Fv fragment (scFv) of a fluorescein-binding antibody can be used to block the DNA ends (48) (Figure 5). We first tested whether the cross-linked, trapped ATP bound form can still bind dsDNA with both ends blocked by the scFv fragment FITC-E2 (Figure 5A). DNA affinity of the MRNBD (S-S) complex was tested by adding TmMRNBD (S-S) to the dsDNA with blocked ends and subsequent EMSA (Figure 5B). To exclude incomplete end blocking of the DNA, the interaction between antibody and fluorescein-labeled DNA was validated by analytical gel filtration (Figure 5C). Nevertheless the disulfide bridged, ATP bound complex was still able to bind DNA with blocked ends. Thus, dsDNA is likely not to be encircled by TmMR.
Figure 5.
Details of the DNA-binding mechanism of TmMRNBD (S-S) via the Antibody DNA-binding Assay. (A) Illustration of the Antibody DNA-binding Assay described in Figure 5B–E. (B) EMSA showing that TmMRNBD (S-S) in ATP/ADP state can still bind to antibody blocked dsDNA. Protein concentrations: 0, 1.75, 3.50, 7.0 and 14.0 µM. (C) Gel filtration chromatogram of 5′fluorescein labeled ds40mer in the absence (solid line) and presence (dashed line) of the antibody fragment FITC-E2 verified complete blocking of the respective dsDNA. The presence of antibody scFv shifted the DNA to larger molecular weights (peak 1) compared to DNA alone (peak 2) and free scFv (peak 3). (D) Gel filtration chromatogram of the 5′fluorescein labeled ds40mer in presence (solid line) and absence (dashed line) of TmMRNBD (S-S). Prior to gel filtration protein and DNA were incubated under conditions where in EMSA most of the DNA is shifted by bound protein. The gel filtration retention volumes were subsequently analyzed by agarose gel electrophoresis. The agarose gel lanes are aligned with the respective fractions of the gel filtration elution. (E) Gel filtration chromatograms of the DNA-antibody mixture (dashed line) and the ternary DNA-TmMRNBD (S-S)-antibody complex (solid line). Analysis of the respective elution fractions by agarose gel electrophoresis indicates that the complex could not be trapped on dsDNA.
To further validate this model, fluorescein-labeled dsDNA was first incubated with TmMRNBD (S-S), followed by blocking of the 5′-ends by scFv. Subsequently the DNA–protein complex was analyzed by analytical gel filtration and agarose gel electrophoresis (Figure 5D and E). Since the gel filtration retention volume of DNA bound to scFv and/or MRNBD (S-S) would be shifted in comparison to that of free DNA, it should be possible to detect encircling of MRNBD (S-S) around DNA. However, we did not see any change in retention volume or evidence for a ternary DNA-MRNBD (S-S)-antibody complex. Therefore it is unlikely that TmMRNBD forms a ring around dsDNA (Figure 5B–E). This is also consistent with the previous data that TmMRNBD cannot be cross-linked around plasmid dsDNA (37).
DISCUSSION
We report here the crystal structure of the Thermotoga maritima MRNBD (S-S) (TmMR NBD (S-S)) complex, also known as SbcD:SbcC, trapped in an ATP/ADP state. We have achieved this by introducing two cysteine residues in Rad50NBD, positioned to stabilize the ATP-dependent NBD dimer by forming disulfide bonds across the NBD-NBD interface in the ATP bound, engaged state. Although ATP was hydrolyzed to ADP in the course of disulfide bridging or crystallization, the NBD dimer is structurally similar to the AMPPNP-bound form of the Rad50NBD dimer. This similarity not only emphasizes the relevance of our observations for the ATP-dependent conformational changes of MR, it also suggests that the disulfide bond stabilization could be a general approach to trap ABC enzymes in the ATP bound state for structural studies.
During the preparation of this manuscript, Cho and coworkers (43) reported a related structure of archaeal MRNBD in the ATPγS-bound form. The structure of archaeal MR in the absence of ATP is not known at present. However, the high degree of similarity between the structures of the ATP/ADP state of bacterial MR and the ATPγS-bound state of archaeal MR and our SAXS analysis based on the archaeal Pyrococcus furiosus (Pf)MRNBD complex, suggest that the ATP-induced conformational cycle is an evolutionarily conserved feature of the complex (Supplementary Figure S6A,B).
Together with our previously reported structure of TmMRNBD in the open state, the new structure in the ATP/ADP state allows us to derive the ATP-dependent conformational cycle of the MR complex. ATP induces a remarkably large transition, consistent with the previously proposed ‘clamp’ model of MR (42). In fact, stabilization of MR in open and closed conformations suggests an at least two state switch for the bacterial complex, although we do not want to rule out additional conformations in the presence of DNA. In this model, the capping domains emerge as key site for the transient stabilization of functional states of the MR complex and orients Rad50's NBDs. Although ATP binding can induce a transition to the closed form even in absence of DNA, cross-linking analysis and thermodynamic reasoning argues that formation of the closed state is additionally promoted by dsDNA, because it binds to this state with higher affinity. Thus, we propose a model where dsDNA and ATP cooperatively switch Mre11 from the open to the closed state. Since ATP-dependent engagement of the Rad50 NBDs, as probed by a signature motif mutation, is important to assemble DNA fragments, it is likely that the structure presented here resembles the DNA tethering form of MR (50).
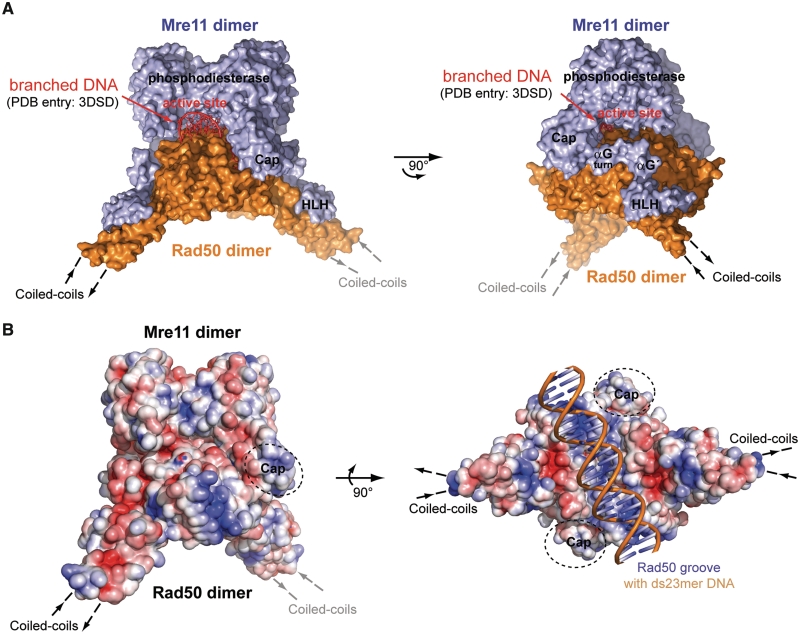
Despite the large structural change in the coiled coils, the most notable result of the closed TmMRNBD (S-S) complex is the blocking of Mre11's dsDNA-binding site by the Rad50 NBD dimer (Figure 6, Supplementary Figure S6C and D). These data are consistent with a variety of biochemical observations. For instance, dsDNA exonuclease activity is inhibited by ATPγS, at least for processive degradation and not single-turnover cleavage of the terminal nucleotide (39). Nevertheless, affinity to dsDNA is increased in TmMRNBD in the presence of AMPPNP and also in the disulfide bridged complex. In summary, it is unlikely that the previously identified dsDNA-binding site of the Mre11 catalytic domain dimer is the dsDNA-binding site of MR in the presence of ATP (49) (Supplementary Figure S6C and D). This is in agreement with the recently reported structure of the archaeal MR complex bound to ATPγS (43). Rather, Mre11's dsDNA-binding site could be important for 3′–5′ exonuclease activity in the open form, for instance to degrade 3′-ends to generate suitable substrates for repair synthesis or overhangs at microhomologies for NHEJ (Figure 7A).
Figure 6.
Surface analysis of TmMRNBD (S-S). (A) Superposition of the molecular surface of TmMRNBD (S-S) and the PfMre11 bound branched DNA (PDB entry: 3DSD) (DNA colored in red; Mre11 and Rad50 are colored in blue and orange respectively; PfMre11 is not shown for clarity). While Rad50 blocks the Mre11 dsDNA-binding site in the ATP-bound state (left hand view), the surface shows holes between Rad50 and Mre11 that are in principle large enough to harbor ssDNA (right hand view). (B) Electrostatic surface potential of TmMRNBD (S-S) in front and bottom view was calculated using APBS tools (55) and indicates positively (blue) charged patches located on the lateral side of Rad50 and along the Rad50 groove. The latter is suitable in size and charge complementary to dsDNA backbones.
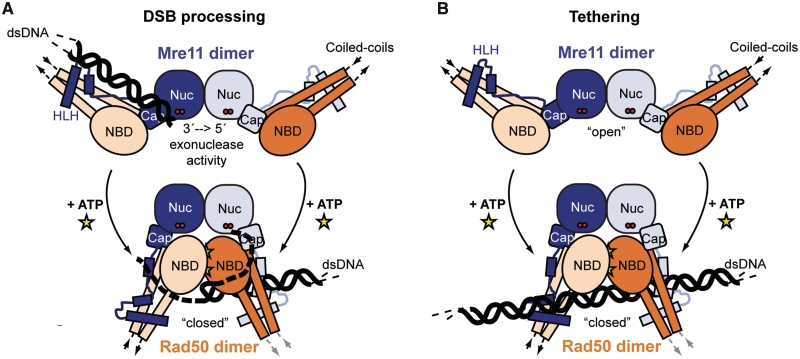
Figure 7.
Proposed clamp Model for DNA Tethering and DSB processing. (A) Proposed model for ATP-controlled DSB processing. ATP driven conformational changes of the MR head module could be responsible for the unwinding of double-stranded DNA and promote endonucleolytic cleavage of single-stranded DNA or hairpins. (B) Hypothetical model for ATP-dependent tethering of dsDNA ends by the MR complex. The extended coiled coil domains of Rad50 undergo an extensive conformational reorientation, which could prevent intramolecular MR interaction after ATP-dependent engagement and allow for intermolecular interaction between different MR complexes via the zinc hook. The proposed DNA-binding groove (Figure 6B) could orient two DNA ends in close proximity to each other.
Alternatively, there may be a further conformation of MR that allows simultaneous dsDNA binding across the Mre11 dimer interface as seen previously and formation of the NBD dimer as seen here (49). In principle, this might be possible, because the interaction of Mre11 with Rad50, apart from the anchoring HLH coiled coil interaction is quite polar and could be modulated or disrupted by DNA. The linker between HLH and capping domain is flexible and can adopt extended (open form) and helical (closed form) conformations, perhaps allowing conformational flexibility between Mre11 and Rad50NBD dimer. However, we failed to crosslink MR around a dsDNA plasmid (37) and do not observe trapping of DNA by blocking the ends with a scFv after binding of the DNA to MR (Figure 5). Since the blocked DNA nevertheless binds to the ATP bound closed MR, it is unlikely that MR encircles dsDNA. This suggests that Rad50 indeed competes with Mre11 in dsDNA-binding in the presence of ATP and at the same time creates an additional dsDNA-binding site.
Although the exact nature of the interaction of TmMR with DNA remains to be determined experimentally, some clues can be observed from the analysis of the molecular surface. A positive patch, complementary in size and charge to dsDNA backbones, is observed at the surface between the protruding coiled coil elements (Figure 6B). Alternatively, positively charged surface patches are located on the lateral side of Rad50. Cho and coworkers identified several positively charged residues in the archaeal MRNBD complex proven to be involved in DNA-binding (41). A superposition of the bacterial and archaeal ATP-bound MRNBD structures identified the according positively charged residues in the MR complex of Thermotoga maritima. Thereby residue R132 and K147 located in the central Rad50 groove correspond to R123 and K144 in M. jannaschii. DsDNA-binding along such an NBD-NBD interface was proposed for the distantly related bacterial recombination enzyme RecF in the RecFOR pathway (51). The residues predicted to connect the DNA-binding sites in Mre11 and in the Rad50 groove namely K897 and R902 in the archaeal complex superpose with K750 and K756 in the bacterial MRNBD complex.
These patches may potentially bind dsDNA but in both cases the DNA is rather far away from the Mre11 nuclease active site, making it at present unclear how DNA-binding and DNA processing are functionally coupled. One explanation is that ATP-dependent engagement of the NBDs could unwind dsDNA ends with one of the ssDNA strands binding between Rad50 and Mre11. While Rad50 blocks the dsDNA-binding site, there are solvent accessible voids between Rad50 and Mre11 that reach the Mre11 active site and are large enough to accommodate ssDNA (Figure 6A). Several loops on Mre11 flanking this hole are unstructured in our crystal structure. Typically, proteins from thermophilic organisms have well folded loops and unstructured loops often indicate unsaturated ligand or DNA-binding sites. A notable structural motif is also seen on the Rad50 side of this cavity. Here, the linker region that connects the capping domain with the HLH domain is folded into a helix. As a result, a right-angled helix–loop–helix structure forms at the lateral entry side to Mre11's nuclease cleft (αGturn -loop- αG′). The equivalent region of S. cerevisiae Mre11 is implicated in DNA-binding (52), thus this transient helix–loop–helix structure might be involved in DNA recognition in the clamp/closed state. A model for the DNA recognition is shown in Figure 7A.
A possible molecular mechanism for ATP-dependent DNA tethering can be seen by comparing the open and the ATP-bound form (Figure 7B). The conformational transition flips and also axially rotates the coiled coil domains of Rad50 with respect to each other. Although the coiled coil domains are flexible (53), the axial rotation of coiled coils and zinc hook domains could prevent intramolecular tethering after ATP-dependent engagement and liberate the zinc-hooks to allow intermolecular tethering of different MR complexes (54). Structural changes between the coiled coils in response to DNA-binding to MRN have been observed by atomic force microscopy (AFM) (54). However, in this study, ATP alone did not result in a conformational change as expected from the structures reported here as well as for the related archaeal system (43). These discrepancies can be explained if both ATP and DNA act cooperatively to induce a structural switch.
While the above mentioned model explains the tethering functions of MR(N) by preventing Mre11-dependent degradation of dsDNA ends, it does not show why 5′–3′ dsDNA nuclease activity by endonucleolytic cleavage at 15–25 nt from the DNA end is promoted by ATP or ATPγS, rather than inhibited (34). In addition, the dsDNA endonuclease activity requires NBD engagement, as shown by a mutation in the signature motif, and requires the Mre11 nuclease active site (50). Furthermore, MRN has been shown to melt/unwind short dsDNA oligonucleotides in the presence of ATP (40). Thus, a possible model is that in the observed ATP-bound state, dsDNA is excluded but ssDNA can still bind to Mre11's nuclease active site (Figure 7A). This would also explain why ssDNA endonuclease activity is not dependent on ATP or inhibited by ATP analogs (39).
Clearly, the interaction of DNA with MR in the light of these new structures needs to be evaluated and it will also be important to clarify how ATP binding and hydrolysis by Rad50 promote endonuclease activity in dsDNA near DNA ends.
ACCESSION NUMBER
PDB ID code 3THO.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Council (SFBs 684 and 646); the German Excellence Initiative (CIPSM), European Commission (IP DNA repair); National Institutes of Health (NIH) (U19AI83025). Funding for open access charge: NIH (U19AI83025).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Hopfner lab, especially Christian Schiller for kindly providing us with unpublished data, Matthew Bennett and Gregor Witte for helpful discussion, and the staff of the Swiss Light Source (Villingen), German Electron Synchrotron (Hamburg), European Synchrotron Radiation Facility (Grenoble) and Advanced Light Source (Berkeley) for support with data collection and analysis. We thank Prof. Andreas Plückthun for providing the fluorescein scFv and the Max-Planck-Crystallization Facility (Martinsried) for crystal screening setups.
REFERENCES
- 1.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Progr. Nucleic Acid Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 2.Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 3.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Ann. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 6.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Ann. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 7.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol.=Biochimie et Biologie Cell. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 8.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat. Rev. Mo., Cell. Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair. 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 11.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am. J. Hum. Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 15.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 16.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 17.Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 18.Zahradka K, Buljubasic M, Petranovic M, Zahradka D. Roles of ExoI and SbcCD nucleases in ‘reckless’ DNA degradation in recA mutants of Escherichia coli. J. Bacteriol. 2009;191:1677–1687. doi: 10.1128/JB.01877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 20.Darmon E, Eykelenboom JK, Lincker F, Jones LH, White M, Okely E, Blackwood JK, Leach DRE. coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol. Cell. 2010;39:59–70. doi: 10.1016/j.molcel.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmas S, Shunburne L, Ngo HP, Allers T. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 2009;5:e1000552. doi: 10.1371/journal.pgen.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 23.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 24.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 26.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DE, Trujillo KM, Sung P, Erickson HP. Structure of the Rad50 x Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 2001;276:37027–37033. doi: 10.1074/jbc.M106179200. [DOI] [PubMed] [Google Scholar]
- 28.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 29.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 30.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 31.Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PloS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 36.Raymond WE, Kleckner N. RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA-binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 39.Herdendorf TJ, Albrecht DW, Benkovic SJ, Nelson SW. Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J. Biol. Chem. 2011;286:2382–2392. doi: 10.1074/jbc.M110.178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4:737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- 42.Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATP{gamma}S complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 45.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 48.Honegger A, Spinelli S, Cambillau C, Pluckthun A. A mutation designed to alter crystal packing permits structural analysis of a tight-binding fluorescein-scFv complex. Protein Sci. 2005;14:2537–2549. doi: 10.1110/ps.051520605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol. Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007;26:867–877. doi: 10.1038/sj.emboj.7601537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 53.de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DR, Kanaar R, Wyman C. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J. Mol. Biol. 2004;339:937–949. doi: 10.1016/j.jmb.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 55.Baker NA. Poisson-Boltzmann methods for biomolecular electrostatics. Methods Enzymol. 2004;383:94–118. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.