Abstract
Eukaryotic chromosome ends have a DNA–protein complex structure termed telomere. Integrity of telomeres is essential for cell proliferation. Genome-wide screenings for telomere length maintenance genes identified several components of the transcriptional regulator, the Mediator complex. Our work provides evidence that Mediator is involved in telomere length regulation and telomere heterochromatin maintenance. Tail module of Mediator is required for telomere silencing by promoting or stabilizing Sir protein binding and spreading on telomeres. Mediator binds on telomere and may be a component of telomeric chromatin. Our study reveals a specific role of Mediator complex at the heterochromatic telomere and this function is specific to telomeres as it has no effect on the HMR locus.
INTRODUCTION
Telomeres are the DNA–protein complex structure at the eukaryotic linear chromosomal ends, which are essential for chromosomal complete replication and genome stability (1). Telomeres are featured by tandem repeats of G-rich sequence running from 5′ to 3′ toward the chromosomal ends. In budding yeast Saccharomyces cerevisiae, two kinds of repetitive telomere-associated sequences, X and Y′ elements are present at subtelomeric region. X element is a heterogeneous sequence found at virtually all telomeres (2). Y′ element is found in one to four tandem copies, immediately internal to the TG1–3 repeats, on about two-thirds of yeast telomeres (3,4). Telomeres are elongated by a specialized reverse transcriptase, telomerase (5). Yeast telomerase consists of the template-containing RNA subunit Tlc1, protein catalytic subunit Est2 and two regulatory subunits Est1 and Est3 (6,7). In vivo, telomeres are subjected to replication-coupled shortening and telomerase-mediated elongation, and thus maintained within a particular length range of 350 ± 75 bp. Telomere integrity is essential for cell proliferation and survival. Telomere shortening to a critical point may trigger checkpoint activation and cell-cycle arrest, and result in cellular senescence.
Previous genome-wide screenings for telomere length maintenance genes (TLM genes) have revealed ∼270 candidate genes including the known telomerase components (8–10). These genes are associated with several different cellular processes, including DNA and RNA metabolism, chromatin modification and transcription. Notably, the identified TLM genes include 10 genes that encode components of the Mediator complex.
Mediator complex is a multi-subunit transcriptional regulator (11–14). Yeast Mediator complex consists of 25 subunits which form four sub-complexes, named head, middle, tail and CDK modules (15–17). Mediator mainly functions as a bridge and conveys the regulatory information from promoter-bound regulatory proteins (such as activators or repressors) to the RNA polymerase II-containing transcription machinery (18). The head and middle modules are in contact with RNA polymerase II (19–21), and the tail module is thought to be the recruitment module because it recognizes and binds to multiple activators (22–25), while the CDK module is associated with negative regulation of a subset of genes (26). The different subunits of Mediator seem to play either negative or positive roles in telomere length regulation, while interestingly, mutation of subunits within the same modules shows similar trend of telomere length change (8,9), suggesting that the molecular mechanism by which Mediator affects telomere replication is complicated.
Eukaryotic genome is organized into domains of euchromatin and heterochromatin that are transcriptionally active or repressed, respectively. In S. cerevisiae, heterochromatin is restricted to three domains, telomeres, HM mating loci (HML and HMR) and the rRNA-encoding DNA (rDNA) region (27). The heterochromatic telomeres can repress the expression of adjacent genes, a phenomenon called telomere position effect (TPE) (28). Heterochromatin formation at yeast telomeres is initiated by the TG1–3 sequence-specific binding protein Rap1 (Repressor/activator protein 1) (29). Rap1 can recruit one of the silent information regulators (Sir proteins), Sir4, independent of the other Sir proteins, Sir2 and Sir3 (29–31). Sir4 then recruits Sir3 and Sir2 through protein–protein interactions (29,32–36). Sir2 is a NAD-dependent histone deacetylase that can deacetylate H4K16 and to a less extent H3K9 and K14 (37,38). Deacetylation by Sir2 may cause compaction of chromatin and initiates a cascade in which more Sir3 and Sir4 are recruited by interacting with hypoacetylated N-terminus of H4 and then more Sir2 binding and histone deacetylation occur (39–41).
Before identified as components of Mediator complex, transcription factors Gal11 (Med15) and Hrs1 (Med3) have been reported to affect TPE, and deletion of either MED15 or MED3 caused reduced telomere silencing (42,43). Both Med15 and Med3 were later proved to be subunits of Mediator complex and components of the tail module (11,44). Given that lack of certain Mediator subunit causes telomere length change (8,9), we hypothesize that the effect of Mediator on telomere maintenance could be direct. In this work, we report that Mediator tail module appears to be required for maintaining telomeric heterochromatin and telomere silencing. Deletion of tail subunits can cause reduced Sir protein binding which results in elevated acetylation of H4K16 and compromised silencing of telomeres. Our results reveal a specific role for Mediator complex other than its canonical function in transcriptional regulation.
MATERIALS AND METHODS
Yeast strains and plasmids
The Mediator mutants are from a collection of S. cerevisiae systematic deletion strains from EUROSCARF (Frankfurt, Germany) in which each open reading frame has been replaced with the KanMX4 module that confers resistance to G418. These strains are in the BY4741 background (MATa his3Δ leu2Δ met15Δ ura3Δ). The isogenic strain BY4742 (MATα his3Δ leu2Δ lys2Δ ura3Δ) was used for genetic analysis. All the yeast strains used in this work are listed in Supplementary Table S1. Yeast strains were constructed either by transformation with a lithium acetate procedure or by genetic cross (mating and tetrad dissection). The plasmids for gene deletion were constructed based on the pRS series (45), and polymerase chain reaction (PCR)-based 13Myc-tagging of proteins was based on the plasmid pFA6a-13Myc-HIS3MX6 (46).
Telomere Southern blot
Genomic DNA was extracted and digested with Pst I and then subjected to telomere Southern blot as described previously using the TG1–3 probe (47). Telomere length was quantified with Multi Gauge V3.0 software (Fujifilm).
TPE assay
Yeast strains were cultured in YPD medium at 30°C overnight and suspended in sterile water at a concentration of OD600 ∼0.3, and serial dilutions were plated onto Yeast Complete (YC) medium with or without 0.15% 5-FOA. Plates were incubated at 30°C for 2 days and photographed.
Quantitative real-time-PCR
Cells from 4-ml fresh cultures (OD600 ∼0.7) were harvested and digested with Zymolyase 20 T (MP Biomedicals, LLC) to obtain spheroblasts. RNA was extracted with RNeasy mini kit (Qiagen) and DNA was digested on-column with RNase-free DNase (Qiagen). One microgram RNA was used as template for the reverse transcription of 20 µl cDNA. Gene expression level was analyzed by real-time PCR and normalized to that of ACT1. The wild-type value is arbitrarily set to 1. At least three biological replicas were done for all the mRNA analysis and statistical analyses were calculated using Student's t-test (*P < 0.05 and **P < 0.01).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed as described previously except some small modifications (48). Fifty-milliliter fresh culture in YPD media (OD600 ∼ 0.7) was crosslinked with formaldehyde for 20 min and quenched with glycine for 5 min. Cells were harvested and washed with cold TBS and lysis buffer. Cells were suspended in 500 µl lysis buffer [50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1 mM phenylmethylsulfonyl fluoride (PMSF) and with Roche complete] and beaded to remove cell walls. Sonication was then performed six times for 15 s at high power (320 W) with 15-s intervals on Bioruptor (Diagenode). After sonication, samples were centrifuged for 20 min at 13 200 rpm at 4°C, and the supernatant was recovered as cell extracts (CE). Ten microliters of bovine serum albumin (BSA; 25 µg/µl) and 1–2-µg antibodies were added to 400 µl CE. After incubation on a rotator at 4°C for 4 h, 20 µl protein G Sepharose beads (GE healthcare) was added and IPs were carried out overnight at 4°C on a rotator. Beads were washed twice with 0.5 ml lysis buffer, twice with 0.5 ml wash buffer 1 (lysis buffer with 0.5 M NaCl), twice with 0.5 ml wash buffer 2 [10 mM Tris (pH 8), 0.25 M LiCl, 0.025% SDS, 0.5% dodium deoxycholate and 1 mM EDTA) for 4 min on a rotator at room temperature and finally rinsed once in 0.5 ml TE buffer (10 mM Tris pH 8, 1 mM EDTA), removing all supernatant. To reverse the bead–antibody interaction, beads were eluted in 150 µl TE-SDS 1% for 30 min at 65°C. Part of the eluate (10 µl) was analyzed by western blot to determine IP efficiencies. Reversal of the crosslinks was performed in a final volume of 120 µl of TE-SDS 1%. In parallel, 5 µl of CE was processed similarly as the input DNA. Samples were incubated for 8 h at 65°C and protein was digested by adding 10 µl proteinase K (20 µg/µl) and 110 µl TE buffer for 2 h at 37°C. DNA was purified with Qiaquick PCR purification kit (Qiagen), and eluted in 120 µl EB buffer. ChIP results were analyzed by Real-time PCR (Tiangen Realmaster kit, Eppendorf realplex). Each pair of primers was tested for linear range and amplification efficiency. For ChIP of Myc-tagged proteins (Mediator subunits or Sir proteins), binding ratio is normalized to an isogenic no-tag control strain without Myc-tagging, and for that of H4K16 acetylation, normalization is against the H4K16R mutant strain which has no acetylation at this site (Figure 3A). In ChIP assay of H4, an isogenic wild-type strain treated with anti-Myc antibody is used as the ‘no-tag’ control for normalization. For binding of Mediator, an internal control was included which is located in the POL1 gene which has little Mediator binding (48), while for that of Sir proteins, H4K16 acetylation and H4, ACT1 was used where Sir binds little and H4K16 acetylation or H4 level is relatively stable. The ChIP of Myc-tagged Mediator in temperature-sensitive rap1-2 mutants were similar except several differences at the beginning: after growing at 25°C to mid-log phase in fresh YPD medium, the cells were shift to 37°C and cultured for 1.5 h or stayed at 25°C as control. The crosslinking and quenching steps were kept at 37°C or 25°C, respectively, before harvesting cells for the following processes. All the ChIP assays were performed for at least three independent repeats and the statistical significances were indicated. Sequences of primers used in this work are available upon request.
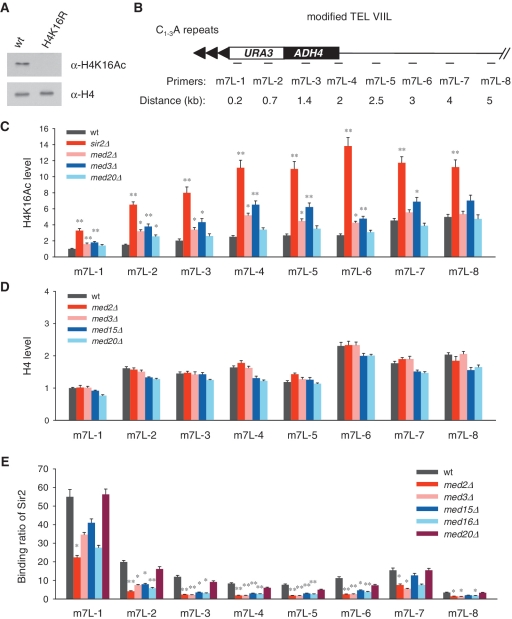
Figure 3.
H4K16 acetylation increases and Sir2 binding is reduced at modified telomere VIIL in Mediator tail mutants. (A) Specificity of anti-H4K16Ac antibody was verified. Chromatin of wild-type and H4K16R mutant cells was extracted and subjected to western blot. (B) Schematic diagram of the modified telomere VIIL (m7L) and the locations of primers used in ChIP assay. (C) ChIP results of H4K16 acetylation level on telomere m7L. The wild-type value at the most distal locus (m7L-1) is arbitrarily set to 1 where the acetylation level is supposed to be the lowest. The statistical significances of difference in H4K16 acetylation between mutants and wild-type cells are provided (*P < 0.05 and **P < 0.01). (D) H4 level is detected by ChIP assay. The wild-type value at m7L-1 is set to 1. (E) Binding of 13Myc-tagged Sir2 on m7L was detected by ChIP assay using anti-Myc antibody.
Chromatin extraction
Cells in 50 ml YPD cultures (OD600 ∼0.5) were harvested and washed successively with 20 ml phosphate-buffered saline (PBS) and 5 ml Zymolyase buffer (50 mM Tris pH 7.5, 10 mM MgCl2, 1 M Sorbitol), and suspended in 300 µl Zymolyase buffer containing Zymolyase at a final concentration of 10 µg/µl. Cells were incubated at 30°C with rotation for 90 min to obtain spheroblasts. Spheroblasts were collected by centrifuged for 5 min at 2000g at 4°C and washed twice with 1 ml Zymolyase buffer. Spheroblasts were then suspended in 500 µl EBX buffer (20 mM Tris pH 7.4, 100 mM NaCl, 0.25% Triton X-100, 5 mM DTT, 50 mM Na-butyrate and protease inhibitors) with 0.5% Triton X-100 to lyse the outer cell membrane. Samples were kept on ice for 10 min with gentle mixing and then layered on top of 1 ml NIB buffer (20 mM Tris pH 7.4, 100 mM NaCl, 1.2 M sucrose, 5 mM DTT, 50 mM Na-butyrate and protease inhibitors) and centrifuged for 15 min at 13 000g at 4°C. The glassy white pellets were suspended in 500-µl EBX buffer with 1% Triton X-100 to lyse the nuclear membrane. Samples were kept on ice for 10 min with gentle mixing. Chromatin was obtained by centrifuging for 10 min at 16 000 g at 4°C. Chromatin was then washed for three times with 1 ml EBX buffer and finally suspended in appropriate volume of PBS. Samples were incubated at 100°C for 5 min and centrifuged at top speed for 5 min. The supernatant was used for SDS-PAGE and western blot analysis.
Western blot
Chromatin extracted from different strains were separated on 18% SDS–polylyacrylamide gel electrophoresis (PAGE) gels and transferred to nitrocellulose membrane (Whatman). Amounts of chromatin were normalized with antibody against histone H4 (ab10158, Abcam), and acetylation of H4K16 was detected with antibody specifically against this modification (39929, Active motif).
RESULTS
Yeast Mediator complex is involved in telomere length regulation
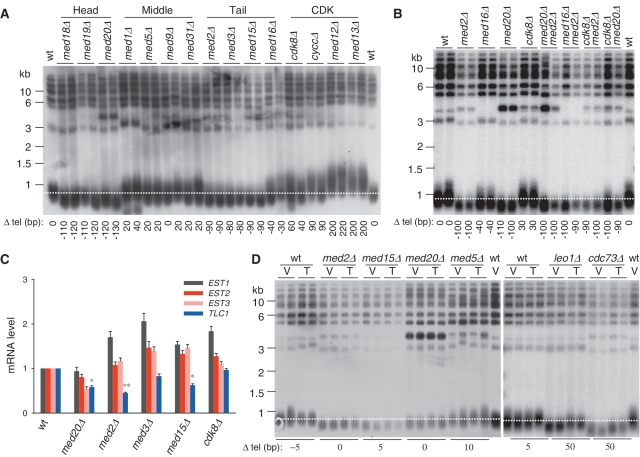
Med18 and Med20 of the head module, Med1 and Med5 of the middle module, Med3 and Med15 of the tail module and all four subunits (Cdk8, CycC, Med12 and Med13) of the CDK module were previously identified as telomere length regulator in genome-wide screenings for telomere length maintenance genes (8,9). To further characterize the role of Mediator in telomere length regulation, we examined telomere length in all the 15 viable Mediator mutants using telomere-specific Southern blot assay. Deletion of subunits of the head or tail module caused modest telomere shortening, and deletion of the CDK-module subunits resulted in telomere lengthening, while mutation of the middle-module subunits had only marginal effect on telomere length (Figure 1A). Overall, these results are consistent with those reported in the large-scale screenings (8,9). Since disruption of different modules causes distinct telomere length phenotype, we then characterized the epistasis among head, tail and CDK modules by analyzing telomere length in double mutants of med20Δmed2Δ (head/tail), med16Δmed2Δ (tail/tail, as a control), cdk8Δmed2Δ (CDK/tail) and cdk8Δmed20Δ (CDK/head). Interestingly, the CDK/head or CDK/tail double mutants exhibited shorter telomeres as the head or tail single mutants (Figure 1B), suggesting that the CDK module functions upstream of the head or tail module. The head/tail double mutants displayed shorter telomeres as the head or tail single mutants (Figure 1B), suggesting that the head and tail modules regulate telomere length in one genetic pathway.
Figure 1.
Mediator is involved in telomere length regulation. (A) The telomere length of all 15 viable Mediator mutants was examined by telomere Southern blot. The telomere lengths of the tested strains were quantified and the changes of telomere length compared with wild-type strain were indicated at the bottom of the corresponding lanes. (B) Telomere length of double mutants of different modules was monitored as in (A). (C) Expression of genes encoding telomerase components in Mediator mutants was detected by qRT-PCR. Statistical analyses in this study were calculated using Student's t-test (*P < 0.05 and **P < 0.01). (D) Telomere length analysis of the Mediator mutants overexpressing TLC1 with the CEN plasmid pRS316-TLC1. V: vector (pRS316), T: Tlc1 (pRS316-TLC1). The telomere length changes indicated at the bottom denote the differences between the mean values of two clones with or without TLC1 overexpression respectively.
Because Mediator is a transcription regulator, it is possible that Mediator affects transcription of telomerase components or other telomere length maintenance genes. However, genome-wide expression profile previously shown by Holstege group revealed that deletion of Mediator subunits does not affect the expression of the well-known telomere-related genes (49). To verify the results of the genomics study (49), we re-examined the expression levels of telomerase components (Est1, Est2, Est3 and Tlc1) and some other telomere binding factors (Cdc13, Rap1, Ku70, Ku80, Rif1 and Rif2) by real-time fluorescent quantitative PCR (qRT-PCR) in Mediator mutants (50–53). Only Tlc1 expression was modestly downregulated in the head and tail mutants with short telomeres, including med20Δ, med2Δ and med15Δ (Figure 1C, Supplementary Figure S1A and S1B; and data not shown). It was reported that the abundance of Tlc1 is essential for maintaining telomere length (54), and deficiency in Tlc1 level in some mutants, such as Paf1C (polymerase II-associated factor complex) mutants, can cause shortened telomere which can be rescued by overexpression of Tlc1 (55). To determine whether the lower expression of Tlc1 in Mediator mutants accounts for the shorter telomeres, we overexpressed Tlc1 with the CEN plasmid pRS316-TLC1 and analyzed the telomere length in the mutants. Unexpectedly, overexpression of TLC1 could not rescue the shorter telomeres in either tail- or head-module mutants, although overexpression of TLC1 partially restored the telomere length in Paf1C mutants, the leo1Δ and cdc73Δ mutants (55) (Figure 1D). Therefore, less Tlc1 expression is unlikely a major cause of shorter telomeres seen in Mediator head and tail mutants. Telomere length is the combinational outcome of regulations of telomerase activity, telomere protection and/or telomere structure. There should be some other factors or pathways functioning in conjunction with Mediator in the telomere length regulation (see ‘Discussion’ section).
Mediator tail subunits are required for telomeric silencing and functions through Sir pathway
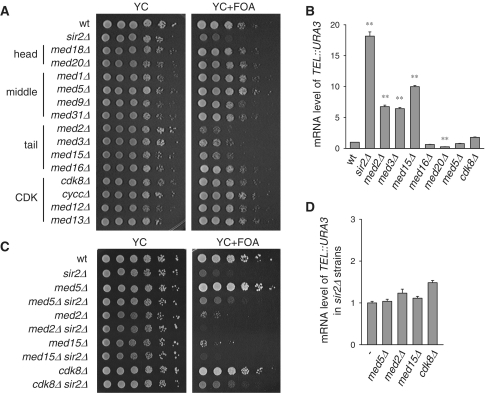
Med3 and Med15 have been reported to affect telomere structure (42,43). To investigate whether the other subunits of Mediator complex are also involved, we examined telomere position effect by dotting assay in the 14 viable Mediator mutants (Figure 2A). The non-essential MED19 gene of head module was not included because of severely slow growth of the med19Δ mutant. All the Mediator mutants were constructed in the UTΔU background as previously described by Gottschling et al. (28): a URA3 reporter gene was inserted at chromosome VII left end, adjacent to the telomere repeat sequence. The modified telomere bears a URA3 gene inserted between telomeric TG1-3 repeats and ADH4 gene, and ∼15 kb sequence that contains an X element on the chromosome from the telomere end to part of the open reading frame of ADH4 gene is deleted. TPE was assessed by detecting expression of telomeric URA3 gene, whose product can convert 5-FOA into a toxic compound and cause reduced cell viability. In wild-type cells, URA3 gene is efficiently silenced which confers resistance to 5-FOA. Deletion of Sir2 disrupts telomeric heterochromatin and thus the sir2Δ mutant is sensitive to 5-FOA and serves as a positive control. For all the 14 mutants, med2Δ, med3Δ and med15Δ mutants of tail module showed great reduction of telomere silencing, while the other mutants exhibited a comparable TPE level to wild-type cells (Figure 2A), suggesting that these Mediator tail subunits are important for telomere silencing.
Figure 2.
Mediator tail module is required for telomere position effect and acts through Sir2. (A) Telomere position effect (TPE) of 14 Mediator mutants. Cells were grown in YPD and diluted to the same concentration. Five-fold serial dilutions of each strain were spotted to YC plates with or without 0.15% 5-FOA as indicated. (B) The expression of telomeric URA3 (TEL::URA3) reporter gene detected by qRT-PCR, and normalized to that of ACT1 gene. (C) TPE of Mediator single mutants and double mutants of Mediator and SIR2. (D) qRT-PCR results of expression of double mutants of Mediator and SIR2.
Recent study argues that 5-FOA sensitivity dose not fully reflect the true silencing state in some mutants (56). The metabolic changes caused by the drug may partially account for the phenotype. We then validated the TPE phenotype by analyzing the expression of telomeric URA3 gene with qRT-PCR assay. Expression of URA3 gene is significantly elevated in the Mediator tail mutants (med2Δ, med3Δ and med15Δ), but not in other module mutants (med20Δ, med5Δ and cdk8Δ) (Figure 2B). This result consistently indicates that these Mediator tail subunits are required for telomere heterochromatin structure maintenance. To exclude the possibility that Mediator affects the basal transcription of URA3 gene, we integrated the URA3 reporter gene at a euchromatic TRK2 gene locus, and examined its expression in both wild-type and the Mediator mutants. The mRNA level of URA3 was not changed significantly upon disruption of Mediator subunits (Supplementary Figure S1C).
At telomeres, heterochromatin forms in a Sir-dependent manner. Double deletion of Mediator and SIR2 phenocopied sir2Δ single mutation (Figure 2C and D), indicating that Mediator affects TPE through SIR pathway. qRT-PCR assay revealed that the expression of the major players in heterochromatin formation, like Rap1 and Sir2-4, was not affected in Mediator mutants (Supplementary Figure S1A). The expression of other telomere factors, including Ku70/80 and Rif1/2, changed little, either (Supplementary Figure S1B).
H4K16 acetylation is increased in Mediator tail mutants
Hypoacetylation is a key feature of heterochromatin (57–59). We detected the level of acetylated H4K16 in Mediator mutants using antibodies against acetylated H4K16. The specificity of the antibody is verified by western blot (Figure 3A). Mutation of H4K16 to arginine (R) abolished acetylation at this site and the antibody recognized no acetylation signal in this mutant. Telomeric H4K16 acetylation is monitored by ChIP assay. The primers were designed to detect H4K16 acetylation at different distances from the telomere end, from 0.2 to 5 kb on the modified telomere VIIL (m7L) in UTΔU strain (Figure 3B). In wild-type cells, acetylation of H4K16 is absent at the telomere end, and increases at the regions toward the centromere. In sir2Δ mutant, the acetylation level of H4K16 at telomere VIIL is significantly elevated. Accordingly, in Mediator tail mutants, the telomeric H4K16 acetylation is upregulated (Figure 3C). As a control, the abundance of histone H4 on this telomere is also examined by ChIP and no significant change of H4 level is observed in the tested Mediator mutants (Figure 3D).
Mediator tail subunits affect Sir2 binding at telomeres
At telomeres, histone deacetylase Sir2 and acetyltransferase Sas2 have opposing roles in regulating heterochromatin spreading (60). Sir2 deacetylates H4K16 and the hypoacetylation promotes spreading of heterochromatin. Sas2 is the catalytic subunit of SAS-I histone acetyltransferase complex which comprises two additional subunits Sas4 and Sas5. Sas2 acetylates H4K16 and facilitates the formation of a barrier against Sir binding and heterochromatin spreading (60,61). The counteracting of these two proteins forms a gradient of H4K16 at telomeres and establishes equilibrium between spreading of heterochromatin and euchromatin. The elevated acetylation of H4K16 on telomere may be caused by lost equilibrium between Sir2 and Sas2 at telomere region. To test this, we detected Sas2's telomeric binding in Mediator mutants by ChIP assay, and surprisingly no significant binding of Sas2 was detected in either wild-type or Mediator mutants (data not shown) (62), suggesting that the elevated H4K16 acetylation may not be a result of increased Sas2 activity at telomeres. qRT-PCR assay showed that in Mediator tail mutants, expression of SAS2, SAS4 or SAS5 was comparable to that in wild-type cells (Supplementary Figure S2A). This result argued against the possibility that deletion of Mediator tail subunits upregulates transcription of SAS-I components to increase H4K16 acetylation. In addition, deletion of Mediator subunits had no effect on the global acetylation of H4K16 revealed by analyzing the total chromatin by western blot (Supplementary Figure S2B).
To examine whether Mediator affects Sir proteins’ binding on telomeres, we performed ChIP assay with anti-Myc monoclonal antibody in wild-type strain and the Mediator mutants that express 13Myc-tagged Sir2. In wild-type cells, Sir2 bound abundantly at the position of 0.2 kb on modified telomere VIIL, and tended to bind less with increasing distance from telomere end, and its binding extended to about 5 kb (Figure 3E). Deletion of the tail subunits, Med2, Med3 and Med15, but not Med20 of the head module, caused reduced Sir2 binding at the telomere end, and Sir2 protein could hardly spread to over 1 kb (Figure 3E). This result was consistent with the reduced TPE in these mutants (Figure 2A and B). Interestingly, the med16Δ mutant, which exhibited no telomere silencing defect in 5-FOA assay (Figure 2A), also showed a significant reduction of Sir2 binding at the modified telomere VIIL, similar to the other three tail-module mutants (Figure 3E), suggesting that every subunit in the tail module is important for the maintenance of telomere heterochromatin structure. Consistently, telomere binding of Sir3 and Sir4 showed the same pattern as that of Sir2 in Mediator tail mutants (Supplementary Figure S3A and S3B). Thus, the tail module of Mediator is required for telomere heterochromatin maintenance.
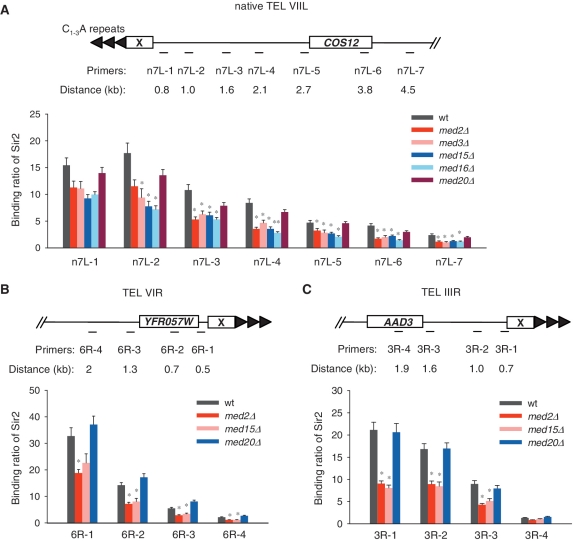
The recent study by Takahashi et al. (63) pointed out that a modified telomere could differ from a natural one. We then examined the association of 13Myc-tagged Sir2 with natural telomeres VIIL, VIR and IIIR in wild-type and Mediator mutant strains (64,65) (Figure 4A–C). The natural telomere VIIL (n7L) contains a ∼0.75 kb X element adjacent to the TG1-3 repeats at the chromosome end (Figure 4A). The designed primers map to a region from ∼0.8 to ∼4.5 kb from the end of n7L. Telomere VIR is one of the most strongly silenced telomeres bearing a X element but not Y’ elements (64) (Figure 4B). Telomere IIIR is another silenced telomere (65) (Figure 4C). The ChIP results showed that Sir2 binding was reduced in Mediator tail mutants at these natural telomeres (Figure 4A–C). These results supported a role of Mediator in promoting or stabilizing Sir protein binding and spreading on telomeres.
Figure 4.
Sir2 binding on native telomeres is decreased. (A–C) ChIP assays detecting Sir2 binding on native telomeres VIIL (n7L) (A), VIR (6R) (B) and IIIR (3R) (C). Primers mapped to each telomere are indicated on the top schematic diagrams.
Mediator complex localizes at telomeres independently of Sir2 or Rap1
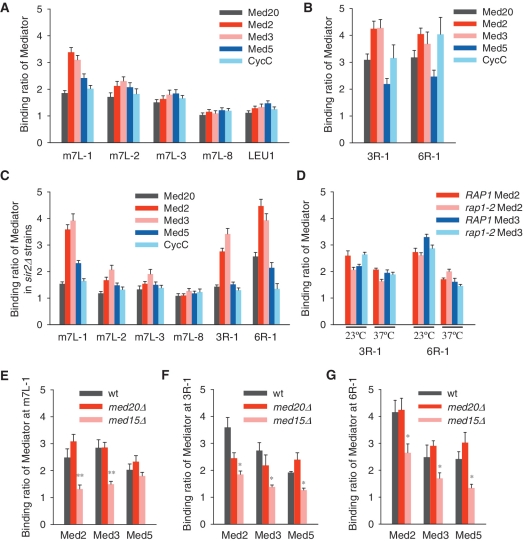
Mediator appears to play roles in both telomere length and telomere structure regulation. To investigate whether Mediator complex affects telomeres directly, we tagged five subunits of Mediator, including Med20 of the head module, Med5 of the middle module, Med2 and Med3 of the tail module and CycC of the CDK module, with 13Myc epitope respectively, and performed ChIP assay. The ChIP results revealed that all the tested Mediator subunits interacted with the modified telomere VIIL directly (Figure 5A). The binding ratio of Mediator decreased with the increase of the distance from the telomere end, and reached to background-level at 5-kb region (Figure 5A). We also observed a direct association of Mediator with two natural telomeres VIR and IIIR (Figure 5B). Because the modified telomere VIIL and native telomeres VIR and IIIR are all silenced, we wondered whether the Sir proteins on these telomeres are required for the binding of Mediator. Deletion of SIR2 did not cause dissociation of Mediator with telomeres (Figure 5C), indicating that Mediator association with telomeres is independent of Sir2 or the silencing state of telomeres.
Figure 5.
Mediator is associated with both modified and natural telomeres independently of Sir2 or Rap1. (A) Binding of 13Myc-tagged Mediator subunits on telomere m7L. The LEU1 locus serves as a negative control where little Mediator binds (48). (B) Binding of Mediator on native telomeres 3R and 6R. (C) Binding of Mediator on telomeres m7L, 3R and 6R in sir2Δ strains. (D) Telomeric binding of Mediator subunits Med2 and Med3 in RAP1 or rap1-2 strains at 23°C or 37°C were detected by ChIP assay. (E–G) ChIP results of Mediator binding in wild-type, med20Δ or med15Δ cells on telomeres m7L (E), 3R (F) and 6R (G).
Both Sir proteins and Rap1 are the most abundant components of telomere heterochromatin. Besides a specific role on telomere, Rap1 binds to the upstream-activating-sequence (UAS) of many genes, including ribosomal protein genes, the MATα genes and several glycolytic enzyme genes, and functions as a transcription activator or repressor which plays either positive or negative roles in transcriptional regulation in different context respectively (66,67). Thus, it is tempting to speculate that Rap1 might be responsible for recruiting Mediator to telomeres. Since Rap1 is encoded by an essential gene, we used a temperature-sensitive allele rap1-2 (A458T), which harbors a point mutation in the DNA-binding domain, to assess whether Rap1 is essential for Mediator's telomeric binding. The DNA-binding activity of Rap1-2 protein is severely compromised at a restrictive temperature of 37°C (68). At the permissive temperature of 25°C, Mediator subunits Med2 and Med3 bind robustly at telomeres IIIR and VIR in both RAP1 and rap1-2 cells (Figure 5D). At the nonpermissive temperature of 37°C, the binding ratio of Mediator subunits at these telomeres in rap1-2 mutant is comparable to that in RAP1 strain (Figure 5D). These results suggest that Rap1 is not responsible for telomere recruitment of Mediator.
The association of Mediator complex to telomeres is bridged by the tail module
To address which module in Mediator complex is responsible for its interaction with telomeres, we performed ChIP assay in med20Δ (head subunit) or med15Δ (tail subunit) strains. Deletion of MED20 has little effect on chromosomal VIIL, VIR and IIIL telomere binding of either Med2 and Med3 of tail module, or Med5 of middle module. In striking contrast, deletion of MED15 significantly reduced binding of the tested Mediator subunits at telomeres (Figure 5E–G). Notably, the binding ratio of Med5 on modified telomere VIIL remains unchanged in med15Δ cells, and the reason for that is unclear (Figure 5E). These results were highly consistent with telomere silencing status observed in med20Δ and med15Δ mutants, respectively (Figure 2A and B), and led us to propose that the Mediator tail module mediates Mediator–telomere interaction.
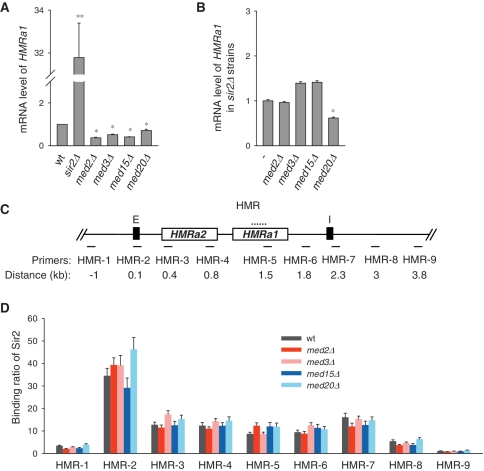
Mediator does not affect heterochromatin structure at the HMR locus
In S. cerevisiae, the HM mating type loci, HMR and HML locating near the opposite ends of chromosome III, are two well-characterized silent domains (69). A pair of silencers that flanks the HMR and HML loci respectively serves as a cis-element in promoting the heterochromatin structure formation at HM loci (70,71). For HMR, HMR-E silencer resides on the left side, and is important for the initiation of Sir proteins binding and silencing at HMR, while the other silencer on the right side, the HMR-I silencer, is thought to help strengthening silencing (72). We wondered whether Mediator also played a role in the heterochromatin maintenance of HM mating type loci. Expression of the endogenous HMRa1 gene was not increased, but rather slightly reduced in the Mediator mutants (Figure 6A). It could be a result of increased silencing or compromised transcription. We examined the expression of HMRa1 in sir2Δ medΔ double mutants, and found out that deletion of Mediator tail-module subunit hardly affects the expression of HMRa1 when the silencing state is disrupted by sir2 deletion, and the med20 deletion slightly reduces the mRNA level (Figure 6B). These results suggest that Mediator does not affect the transcription of HMRa1 when it is not silenced.
Figure 6.
Mediator does not affect heterochromatin at the HMR locus. (A) Expression of endogenous HMRa1 gene was examined. (B) Mediator does not affect expression of HMRa1 in sir2Δ strains. (C) Schematic diagram of HMR region. Primers for ChIP assay were indicated. (D) Sir2 binding at HMR region in Mediator mutants.
To validate that Mediator may not contribute to the heterochromatin structure at the HMR locus, we examined the binding of Sir2 at HMR locus in wild-type cells and Mediator mutants. Primers were designed to cover a region from 1 kb upstream to 4 kb downstream of HMR-E silencer (Figure 6C). In wild-type cells, Sir2 binds abundantly at the HMR regions including both the HMR-E and HMR-I silencers (Figure 6D), and this result is consistent with what was previously reported (72). In the Mediator mutants, Sir2 binding at HMR regions has no significant change (Figure 6D). Thus, Mediator is not involved in heterochromatin maintenance at the HMR locus.
DISCUSSION
Mediator has been well studied in transcriptional regulation (18,73). In this work, we reported that yeast Mediator is involved in telomere length regulation, and the Mediator tail module is required for the maintenance of the heterochromatin structure of telomeres. Deletion of individual tail subunits causes reduced silencing, likely by disrupting the hypoacetylation state of histone H4K16 established by Sir2. Our work revealed a specific role of Mediator in telomere heterochromatin maintenance, which appears to be different from its canonical function in transcriptional regulation. Recently, a novel role of Mediator was discovered in mammalian cells that links Mediator to epigenetic regulation. In nonneuronal cells, Mediator establishes a repressed state of neuronal genes by facilitating recruitment of the methyl-transferase G9a, which methylates H3K9, in conjunction with the RE1 silencing transcription factor (REST) (74). Therefore, it remains possible that Mediator functions in other cellular process than those that have been disclosed.
Because Mediator is responsible for both basal transcription and regulated transcription by activators or repressors, one may argue that Mediator binds to telomeres to exert its transcriptional function. However, our data do not support this idea. Mediator binds to the regions proximal to telomere sequence (Figure 5A and B), where no open reading frames or transcriptional initiation elements exist. Consistent with our results, previous genome-wide ChIP-on-chip analyses show that Mediator binds at telomere region where RNA polymerase II is absent (48). In addition, in the double deletion mutant of SIR2 and Mediator subunit, the mRNA expression of telomeric URA3 marker gene was comparable to that of sir2Δ cells (Figure 2C and D). Thus, Mediator is unlikely to function on telomeres as a traditional transcription regulator.
Mediator participates in both telomere length and structure regulation. How Mediator affects telomere length remains elusive. A simple model would be that Mediator affects telomerase activity to regulate telomere length. However, this model cannot fully explain the shorter telomeres in head- and tail-module mutants and longer telomeres in CDK-module mutants (Figure 1A). An alternative model is that the telomere length change is attributed to abnormal telomere structure. Several lines of evidence seem to support this model. Deletion of Sir3 or Sir4 causes telomere heterochromatin collapse and telomere length shrinking (40,75). In the histone H4K12R mutant, the over-condensation of telomere heterochromatin affects telomere replication, and results in shorter telomeres, and this defect is rescued by SIR2 deletion (76). In the mutants of the Mediator tail module, telomere-associated Sir proteins are dramatically reduced, and accordingly telomeres are shorter (Figure 1A, 3E, 4 and Supplementary Figure S3). Therefore, it is tempting to speculate that deletion of Mediator tail subunits reduces Sir proteins’ binding at telomeres and the disrupted heterochromatin structure causes telomere shortening, although this notion calls for more and further evidence to support.
The tail module of Mediator is the only module that appears to be required for telomere silencing. Paradoxically, the subunits of the other modules also show telomeric binding as the tail module. We could not tell whether Mediator functions on telomeres as a whole complex or the tail module acts as an independent sub-complex. ChIP assay shows that deletion of the head subunit Med20 does not affect telomeric binding of the middle or tail module, while deletion of tail subunit Med15 dissociates the tested Mediator subunits from telomeres (Figure 5E–G). Consistently, the med20Δ mutant has no silencing defect while the med15Δ mutant displays significantly reduced silencing (Figure 2A and B). Therefore, we favor the argument that the tail-module functions as a ‘sub-complex’ on telomeres. In other words, the tail module per se directly contributes to the maintenance of telomere structure, although it recruits the other modules to telomeres through module–module interactions.
Our results show that Mediator is associated with telomeres in the absence of Sir or Rap1. Very recently, it was reported by Zhu et al. (77) that Mediator could bind to histone tail peptides and the Mediator–H4 histone peptide interaction is specifically inhibited by acetylation of H4K16. These results suggest that histone may be the key factor responsible for the recruitment of Mediator to telomeres. Besides, when our work is undergoing, another work by Zhu et al. (78) coincidently pointed out that purified Mediator complex could compete with Sir3 for binding to nucleosomes in vitro, suggesting that Mediator interacts with chromatin directly. Consistent with our results, Zhu et al. (78) showed that Mediator affects telomere silencing and cellular life span. They found that deletion of Med5, a subunit residing at the junction between the middle and tail module, could change the balance between Sir2 and Sas2 proteins at telomere X element, and then cause increased H4K16 acetylation near telomeres and derepression of subtelomeric genes. In our work, we focused on the tail module and observed similar phenotypes in the tail mutants to that of med5Δ mutant reported in their work. However, we did not observe a significant reduction of telomere silencing in med5Δ mutant by 5-FOA assay or qRT-PCR (Figure 2A and B). We examined TPE in YPH499 strain background, and deletion of MED5 has little effect either (data not shown). We could not fully explain this discrepancy. Perhaps it was due to the different locus of URA3 marker gene inserted in telomere VIIL. The modified telomere VIIL utilized in our work lacks the subtelomeric X and Y′ elements. Deletion of Mediator tail module significantly reduced Sir2 binding at this telomere (Figure 3E). At the natural telomeres, including the unmodified VIIL, VIR and IIIR that contain the subtelomeric X elements, Mediator tail module appears to function in the same way (Figure 4A–C). Thus, we propose that Mediator tail module functions on telomeres independently of either the X or the Y′ elements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Natural Science Foundation of China (90919027); Ministry of Science and Technology (2011CB966300 to J.Q.Z.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Yasumasa Tsukamoto for providing the plasmids.
REFERENCES
- 1.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 2.Louis EJ, Haber JE. The structure and evolution of subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan CS, Tye BK. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- 4.Zakian VA, Blanton HM. Distribution of telomere-associated sequences on natural chromosomes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:2257–2260. doi: 10.1128/mcb.8.5.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 6.Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl Acad. Sci. USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 8.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl Acad. Sci. USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng FL, Hu Y, Shen N, Tong XJ, Wang J, Ding J, Zhou JQ. Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J. 2009;28:1466–1478. doi: 10.1038/emboj.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 12.Bjorklund S, Kim YJ. Mediator of transcriptional regulation. Trends Biochem. Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 13.Flanagan PM, Kelleher RJ, III, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 14.Kelleher RJ, III, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 15.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 17.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu. Rev. Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 18.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 20.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc. Natl Acad. Sci. USA. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 22.Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han SJ, Lee YC, Gim BS, Ryu GH, Park SJ, Lane WS, Kim YJ. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YC, Park JM, Min S, Han SJ, Kim YJ. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 27.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 28.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 29.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 30.Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 33.Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl Acad. Sci. USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 35.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 36.Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 38.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 40.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 41.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki Y, Nishizawa M. The yeast GAL11 protein is involved in regulation of the structure and the position effect of telomeres. Mol. Cell. Biol. 1994;14:3791–3799. doi: 10.1128/mcb.14.6.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piruat JI, Chavez S, Aguilera A. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997;147:1585–1594. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Liao XH, Zhang ML, Yang CP, Xu LX, Zhou JQ. Characterization of recombinant Saccharomyces cerevisiae telomerase core enzyme purified from yeast. Biochem. J. 2005;390:169–176. doi: 10.1042/BJ20050208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 49.van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, Holstege FC. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 50.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 51.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell. Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 52.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 53.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 54.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol. Cell. Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell. 42:127–136. doi: 10.1016/j.molcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 58.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 59.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 60.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 61.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 62.Jacobson S, Pillus L. The SAGA subunit Ada2 functions in transcriptional silencing. Mol. Cell. Biol. 2009;29:6033–6045. doi: 10.1128/MCB.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, Jaspersen SL, Kobor MS, Shilatifard A. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell. 42:118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mondoux MA, Zakian VA. Subtelomeric elements influence but do not determine silencing levels at Saccharomyces cerevisiae telomeres. Genetics. 2007;177:2541–2546. doi: 10.1534/genetics.107.079806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou BO, Wang SS, Xu LX, Meng FL, Xuan YJ, Duan YM, Wang JY, Hu H, Dong X, Ding J, et al. SWR1 complex poises heterochromatin boundaries for antisilencing activity propagation. Mol. Cell. Biol. 2010;30:2391–2400. doi: 10.1128/MCB.01106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchman AR, Lue NF, Kornberg RD. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol. Cell. Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 68.Kurtz S, Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991;5:616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- 69.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 70.Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J. Mol. Biol. 1984;176:307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- 71.Feldman JB, Hicks JB, Broach JR. Identification of sites required for repression of a silent mating type locus in yeast. J. Mol. Biol. 1984;178:815–834. doi: 10.1016/0022-2836(84)90313-9. [DOI] [PubMed] [Google Scholar]
- 72.Lynch PJ, Rusche LN. An auxiliary silencer and a boundary element maintain high levels of silencing proteins at HMR in Saccharomyces cerevisiae. Genetics. 185:113–127. doi: 10.1534/genetics.109.113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30: 235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 76.Zhou BO, Wang SS, Zhang Y, Fu XH, Dang W, Lenzmeier BA, Zhou JQ. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 7:e1001272. doi: 10.1371/journal.pgen.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu X, Zhang Y, Bjornsdottir G, Liu Z, Quan A, Costanzo M, Davila Lopez M, Westholm JO, Ronne H, Boone C, et al. Histone modifications influence mediator interactions with chromatin. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkr551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu X, Liu B, Carlsten JO, Beve J, Nystrom T, Myers LC, Gustafsson CM. Mediator influences telomeric silencing and cellular lifespan. Mol. Cell. Biol. 2011;31:2413–2421. doi: 10.1128/MCB.05242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.