Abstract
Human telomerase reverse transcriptase (hTERT) is localized to mitochondria, as well as the nucleus, but details about its biology and function in the organelle remain largely unknown. Here we show, using multiple approaches, that mammalian TERT is mitochondrial, co-purifying with mitochondrial nucleoids and tRNAs. We demonstrate the canonical nuclear RNA [human telomerase RNA (hTR)] is not present in human mitochondria and not required for the mitochondrial effects of telomerase, which nevertheless rely on reverse transcriptase (RT) activity. Using RNA immunoprecipitations from whole cell and in organello, we show that hTERT binds various mitochondrial RNAs, suggesting that RT activity in the organelle is reconstituted with mitochondrial RNAs. In support of this conclusion, TERT drives first strand cDNA synthesis in vitro in the absence of hTR. Finally, we demonstrate that absence of hTERT specifically in mitochondria with maintenance of its nuclear function negatively impacts the organelle. Our data indicate that mitochondrial hTERT works as a hTR-independent reverse transcriptase, and highlight that nuclear and mitochondrial telomerases have different cellular functions. The implications of these findings to both the mitochondrial and telomerase fields are discussed.
INTRODUCTION
Telomerase is a ribonucleoprotein responsible primarily for telomere maintenance. The enzyme is composed of two distinct subunits, a protein core that mediates DNA catalysis (TERT) and a non-coding RNA template, TERC or TR, used for telomeric DNA synthesis (1–10). TERT also forms a complex in the nucleus with the RNA subunit of the mitochondrial RNA processing endoribonuclease (RMRP), an RNA that is only partially mitochondrial. As such, it works as a RNA-dependent RNA polymerase that regulates gene expression through the production of double stranded RNAs (5). Telomere- and TR-independent functions of TERT had been previously described but limited information was available in regards to its non-canonical roles (6–10).
In addition to the non-telomere-related functions for nuclear telomerase, an increasing body of evidence shows that TERT is also present in mitochondria. Human telomerase reverse transcriptase (hTERT) has a mitochondrial targeting signal (MTS) that is sufficient and required for its mitochondrial localization (1,2). Ectopically expressed hTERT has been found in human mitochondria (1–4,11–14) and telomerase enzymatic activity was detected in purified mitochondrial extracts (1,4). hTERT was also found to bind two regions of mitochondrial DNA (mtDNA), to improve respiratory chain function and to decrease reactive oxygen species (ROS) production (2–4,12). More recently, systemic mitochondrial defects were observed in a TERT knockout model (15), altogether supporting a direct role for TERT in mitochondrial function or regulation.
Despite some descriptive work about mitochondrial TERT (1–4,11–14), fundamental questions about its biology and function in the organelle remain unanswered. For instance, it is unclear whether TERT is present in mitochondria at physiologically meaningful levels and whether its differential subcellular distribution is conserved in other mammalian species. It is also unknown whether TERT uses its associated nuclear RNA in mitochondria and whether its organellar role relies on its reverse transcriptase (RT) activity. Finally, it is yet to be established that the mitochondrial defects associated to the lack of TERT are caused directly by its absence in mitochondria and are not an indirect effect because of its absence in the telomeres. The present work was aimed at addressing these questions. Combining various approaches we show that a fraction of endogenous TERT from human, mouse and rat are mitochondrial. Classical import assays demonstrate that TERT localizes to the mitochondrial matrix, in an import process dependent on the mitochondrial membrane potential. Using iodixanol gradients and chromatin immunoprecipitations we show that TERT co-fractionates with mtDNA and nucleoids proteins, and it also interacts with mitochondrial tRNAs. In contrast the canonical nuclear RNA, hTR, is not detectable in human mitochondria. Nevertheless, the mitochondrial effects of hTERT rely on its RT activity, which we show is reconstituted in vitro in the absence of hTR. Finally, we demonstrate that abolishing the mitochondrial localization of hTERT while maintaining its nuclear function leads to mitochondrial defects, thereby providing direct evidence that its absence specifically in mitochondria negatively impacts the organelle.
Taken together, our results indicate that TERT works in mitochondria as a hTR-independent reverse transcriptase, establishing it as a new player in mtDNA metabolism. Our data also point to fundamentally different roles for nuclear and mitochondrial telomerases.
MATERIALS AND METHODS
Cell culture, plasmids and viral infections
NHF, GM7532 and GM847 fibroblasts along with their wild-type WT hTERT, DNhTERT or nuchTERT derivatives have been previously described (2,12). SQ20B and SCC61 were cultured as in ref. (44). VA13 cells and the lentiviral vector coding full-length hTR were a kind gift from Dr Elizabeth Blackburn (UCSF). Protocols for lentiviral infections were described elsewhere (30). Hek 293 cells were cultured as recently described (45).
Mitochondrial isolations, immunoblots and RT-PCR
Mitochondrial isolations were performed as recently described by us (12,45). Anti-TERT antibody (Rockland Immunochemicals) was used in 1:500 dilution. Information about antibodies against HSP60, TOM20, TIM23 and SF2 can be found in our previous work (45). For RT-PCR, crude extracts were treated with RNase A (50 µg/ml final concentration) prior to mitochondrial isolations. Isolated organelles were treated with 0.3 mg/ml (final concentration) of digitonin to strip the outer mitochondrial membrane. RNA was isolated using the Qiagen mini kit, and submitted to DNase I treatment to remove contaminating DNA. Sequence of primers used to amplify GAPDH, hTR and cytochrome b was described previously (31,34). Reactions were performed with 32–40 cycles.
Mitochondrial protein import
Mitochondrial protein import assays were performed as described by us previously (19).
Nucleoid analysis
Sucrose-gradient purified mitochondria from HEK cells (2 mg/ml) were suspended in 20 mM HEPES pH 7.8, 2 mM EDTA, 210 mM mannitol, 70 mM sucrose and treated with or without 100 µg/ml of trypsin at room temperature for 30 min. After washing and pelleting mitochondria three times, the organelles were lysed with 0.4% DDM and centrifuged for 10 min at 1000 gmax; the supernatant was loaded on a 20–42.5% iodixanol gradient (20 mM HEPES, pH 7.8, 1 mM EDTA, 50 mM NaCl, 2 mM DTT, 0.05% DDM with protease inhibitor) and centrifuged at 100 000 gmax for 14 h. Nucleic acid was extracted from a portion of each fraction of the gradient and after Southern blotting, hybridized to a radiolabeled probe.
Mitochondrial chromatin immunoprecipitations
The Mitochondrial chromatin immunoprecipitations (mIP) was followed as described in ref. (22). Briefly, GM847 fibroblasts stably expressing empty vector (EV) or hTERT-HA were crosslinked with formaldehyde and hTERT immunoprecipitated with an anti-HA antibody (Covance). DNA was isolated and mtDNA was amplified using 27 overlapping primer pairs covering the entire mtDNA (22). Immunoprecipitations were also performed with the IgG control or with an antibody against TFAM (Aviva).
H2O2 treatment and mtDNA integrity analysis
Protocols for H2O2 exposure and QPCR analysis of mtDNA integrity were followed as previously described (31–33). Data were normalized for changes in mtDNA copy number based on amplification of a short mtDNA fragment (221 bp). Statistical analysis was performed using unpaired Student's t-test. In order to obtain basal level of damage, non-transfected, WT and nuchTERT values were compared to a reference control (DNA from HeLa cells), which was included in every run. Results were independently reproduced in two different cellular backgrounds [NHF and GM7532, (12)].
Telomeric repeat amplification protocol
The telomeric repeat amplification protocol (TRAP) was performed as done by us previously (1) using 100 ng of total protein lysates. In the case of rabbit reticulocyte lysate (RRL)-translated TERT, TRAP was performed using 2 µl of 5× diluted RRL-translated protein. A sample of RRL-translated hTERT was heat treated for 10 min at 85°C to inactivate hTERT.
RNA immunoprecipitations
RNA immunoprecipitations (RIP) was followed as previously described (25). The sequence of the primers used can be found in Supplementary Table S1 and in ref. (22).
TERT-driven first strand cDNA synthesis
Total RNA was isolated using Trizol reagent (Invitrogen) and treated with DNase I to remove contaminating DNA. Wild-type and DNhTERT were translated in RRL using the plasmid described by Beattie et al. (46), in which the mutant DNhTERT vector (Addgene) was subcloned. RT reactions were performed using the first strand cDNA synthesis kit from Invitrogen using RRL-translated hTERT or the RT included in the kit. hTR was included in reactions where indicated. The hTR+1 plasmid was described earlier (47). Reactions were run at 50°C and 37°C. Primers used for cDNA amplification were the same as used for the RIP. CystRNA was transcribed in vitro using a commercially available kit (Riboprobe, Promega), and used as template for RT reactions. RNA was treated with DNase I to remove contaminating DNA.
Mitochondrial ROS detection
Levels of superoxide anion generated in mitochondria of non-transfected, WT or mutant hTERT were measured using Mitosox as previously described by us (12). Data were collected both by flow cytometry and confocal microscopy, where a mitochondrial uncoupler was included to assure the specificity of the signals (data not shown). Data obtained by flow cytometry was used for the graphs presented in Figure 5 as it allows for more accurate quantification of signals at the population level.
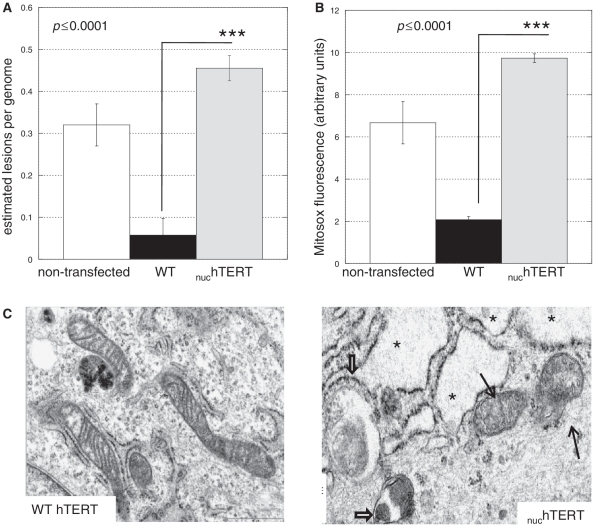
Figure 5.
Lack of hTERT specifically in mitochondria negatively impacts the organelle. (A) MtDNA integrity was measured using QPCR (31–33), and estimated lesion frequency/genome calculated. Results represent the mean of six independent analyses, three in the NHF background and three in the GM7532. Since results were similar the data have been pooled. Error bars represent ± standard error of the mean (SEM). Statistical significance was evaluated using unpaired Student's t-test. (B) Mitochondria superoxide generation was evaluated using the fluorescent probe Mitosox by flow cytometry. Data represent three independent experiments. Statistical significance was evaluated using ANOVA. (C) Non-transfected parental (data not shown), WT- and nuchTERT-expressing cells were evaluated for their subcellular ultrastructures using electron microscopy. At least two independent slides were analyzed per cell type; approximately 20 cells were analyzed per slide. Data shown are representative; criteria to define damaged cellular components are described in the ‘Materials and Methods’ section. Thin black arrows indicate damaged mitochondria and thick black arrows show autophagosomes. The presence of several vacuoles in the cytoplasm is evident in the mutant cells (see asterisks).
Electron microscopy analysis
EM analysis was performed in non-transfected parental, and WT or nuchTERT-overexpressing cells as described by us previously (12). The criteria to define damaged mitochondria included loss or dissolution of ≥25% of cristae, alteration of size and vacuolization. Micrographs were evaluated by a pathologist.
RESULTS
A fraction of mammalian TERT is imported into the mitochondrial matrix in a membrane potential-dependent manner
We previously identified a predicted MTS on TERT of higher eukaryotes, which encompassed the first 20 amino acid residues of the protein (1). Mutations in the MTS of hTERT abolished its mitochondrial localization, and addition of only the MTS of hTERT to green fluorescence protein (GFP) rendered the fluorescent protein completely mitochondrial (1,2), indicating that the identified MTS was necessary and sufficient to target hTERT to the organelle.
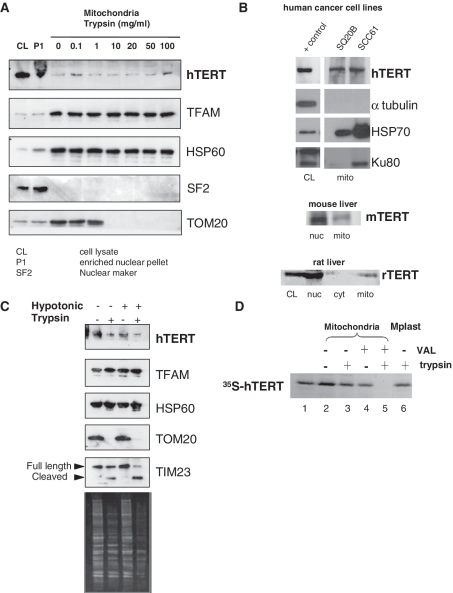
To confirm the mitochondrial localization of endogenous TERT, we performed a series of experiments using antibodies and isolated mitochondria obtained from human cells and rodent tissue. Specificity and suitability of the anti-TERT antibody available from Rockland Immunochemicals has been demonstrated previously (4,16); data obtained by us can be found in Supplementary Data (Supplementary Figure S1A and S1B). In HEK 293 cells, a telomerase-positive human kidney cell line, the majority of hTERT was associated with enriched nuclear pellets (P1). Notwithstanding this, the fraction of hTERT associated with mitochondria, the established mtDNA binding protein TFAM, and the mitochondrial matrix protein HSP60 all proved equally resistant to trypsin treatment, whereas nuclear splicing factor 2 (SF2) and the outer mitochondrial membrane protein TOM20 were susceptible to trypsin (Figure 1A). Similar results were obtained when analyzing highly purified mitochondria (also submitted to protease treatment) from two different human squamous carcinoma cell lines (SQ20B and SCC61, Figure 1B upper panel) and from mouse and rat liver (Figure 1B lower panels). Taken together these data indicate that a fraction of endogenous mammalian TERT is mitochondrial.
Figure 1.
Endogenous TERT is localized to the mitochondrial matrix. (A) Mitochondria from HEK 293 cells were isolated based on differential centrifugation. The first pellet from mitochondrial isolation, consisting on nuclei and non-disrupted cells, was used as enriched nuclear pellet. Purified mitochondria were treated with increasing concentrations of trypsin to assess mitochondrial localization of proteins by western blot analysis. TFAM and HSP60 were used as mitochondrial markers while SF2 controlled for nuclear contamination. TOM20 was included to monitor trypsin efficiency. (B) Western blots showing TERT in mitochondria isolated from two human cancer cells, mouse and rat liver; CL (crude cell lysate) and nucleus: 50 µg/lane, mitochondria: 250 µg/lane. Markers for mitochondrial enrichment (HSP70) or purity (tubulin and Ku80) were included. Ku80 has a truncated version that is mitochondrial (48). Additional analysis of mitochondrial purity can be found in the Supplementary Data (Supplementary Figure S1B–S1D). (C) Mitochondria were purified from HEK 293 cells, resuspended in isotonic or hypotonic buffer and subjected to different treatments with trypsin. TOM20 was used as control for trypsin efficiency. TIM23 is present in the inner membrane with a domain facing the intermembrane space and another facing the matrix. The former is degraded by trypsin when the outer membrane is disrupted. Bottom panel shows Coomassie staining of the gel to confirm trypsin activity. (D) 35S-labeled hTERTN33-PrA was incubated with intact mitochondria in the presence of ATP (4 mM) and GTP (1 mM) at 30°C for 10 min. Valinomycin (5 mg/ml; VAL) was included as indicated. Following import, mitochondria were subjected to osmotic swelling to generate mitoplasts (lane 6). Samples were trypsin-treated and analyzed by SDS–PAGE and followed by autoradiography. Lane 1 shows 50% of 35S-labeled hTERTN33-PrA used per import assay.
Previously subcellular fractionation followed by western blot analysis suggested hTERT is present in the outer and inner mitochondrial membranes, as well as in the matrix (4,14). Using a similar approach we found hTERT in the same fractions as TFAM, HSP60 and TIM23 but not TOM20 (Figure 1C). While these data indicate the presence of hTERT in the matrix and inner mitochondrial membrane, this approach is not ideal to define submitochondrial localization as it largely relies on integrity of the mitochondrial membranes after fractionation and on the susceptibility of marker proteins to protease treatment (17).
To better define the submitochondrial localization of hTERT, we carried out classic in vitro mitochondrial import assays (18–20). Briefly, we generated a chimeric construct (hTERTN33-PrA) in which the first 33 N-terminal amino acids of hTERT (including the MTS) were linked to a non-mitochondrial passenger protein, Protein A. Protein A is not able to enter mitochondria by itself (19). 35S-labeled hTERTN33-PrA was synthesized in RRL, incubated with isolated mitochondria and then treated with trypsin as indicated to distinguish between imported (trypsin-resistant) and unimported (trypsin-sensitive) molecules.
The chimeric protein was efficiently imported into mitochondria as judged by trypsin-resistance (Figure 1D, compare lanes 2 and 3). No change in the molecular mass of the chimeric protein was observed upon import into mitochondria (compare lanes 1 and 3) suggesting that hTERT does not contain a cleavable MTS of more than a few amino acids. Import was strictly dependent on a mitochondrial membrane potential (ΔΨm) since no protease-protected hTERTN33-PrA molecules were observed in the presence of valinomycin, which dissipates the ΔΨm (Figure 1D lane 5). In the absence of ΔΨm, the chimeric protein remained bound to the mitochondrial outer membrane but was not imported (Figure 1D, compare lanes 4 and 5). Imported hTERTN33-PrA remained protected from trypsin digestion even when mitochondria were subjected to hypotonic shock, a condition that disrupt the outer membrane but leave the inner membrane intact (Figure 1D lane 6). Hence, imported hTERTN33-PrA localizes to the matrix side of the mitochondrial inner membrane.
hTERT co-purifies with mtDNA and tRNAs
Mitochondrial DNA is organized in complexes with proteins called nucleoids. Nucleoids are predicted to orchestrate faithful copying, expression and transmission of mtDNA, and have been shown to divide and redistribute in the mitochondrial network (21). Given that hTERT is in the mitochondrial matrix where the mtDNA is located, we next asked whether hTERT co-fractionates with mtDNA and other nucleoid proteins.
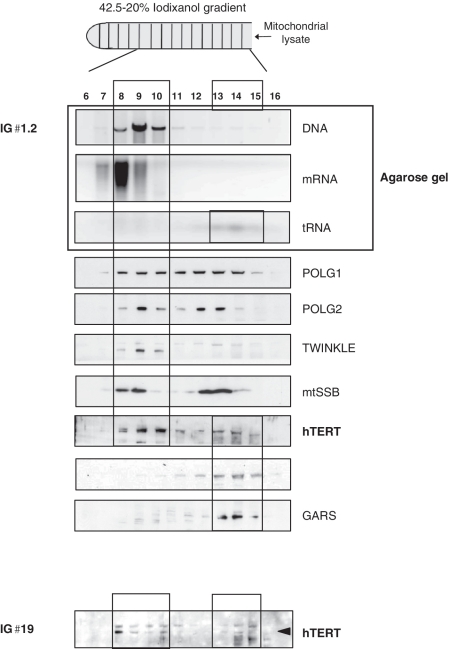
To this end, purified mitochondria from HEK 293 cells were lysed and loaded onto an iodixanol gradient. Gradient fractions were analyzed for the presence of nucleic acids (by agarose gels), and hTERT and nucleoid proteins such as the mitochondrial DNA polymerase γ (POLG 1 and 2), the DNA helicase twinkle and the mitochondrial single stranded binding protein (mtSSB, 21) were evaluated by western blots. Mitochondrial hTERT co-fractionated primarily with mtDNA and established nucleoid proteins (Figure 2A, lanes 8–10) suggesting it is a nucleoid interacting protein. A smaller population of hTERT co-fractionated with mitochondrial tRNAs and amino-acyl tRNA synthetases (lanes 13–15).
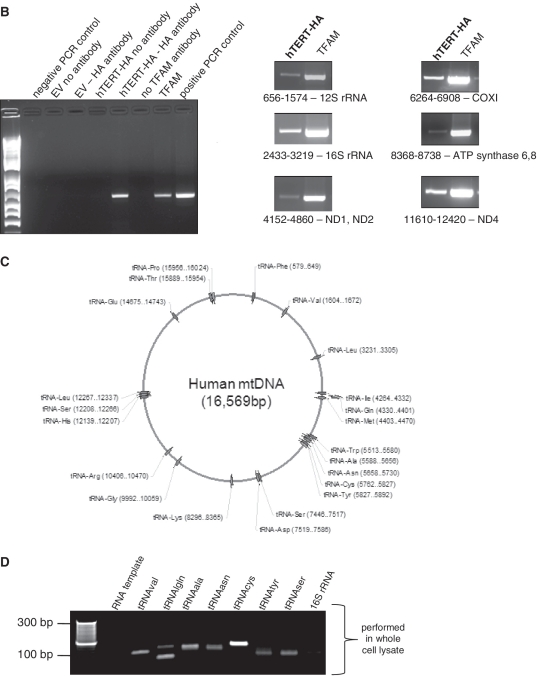
Figure 2.
TERT co-purifies with mitochondrial nucleic acids. (A) Purified mitochondria from HEK 293 cells were lysed and loaded onto an iodixanol gradient. IG♯1.2: no trypsin treatment was applied to mitochondria prior lysis, IG♯19: mitochondria were first treated with trypsin and then lysed. Two mitochondrial tRNA synthetases (GARS and TARS2) were used as controls for the tRNA fraction (B) mIP was performed in cells expressing hTERT-HA or empty vector (EV). Left panel shows controls as indicated in the figure; primers used encompassed region 10712–11249. Right panel shows amplification of mtDNA when using HA or TFAM for the IPs. Location of the primers and the genes present in the amplified region are shown. (C) Schematic representation of human mitochondrial genome with position of tRNA genes indicated. (D) RNA immunoprecipitations were performed in GM847 cells expressing EV or hTERT-HA. Panel shows a sample of tRNAs amplified when immunoprecipitating hTERT. First lane used RNA as template and the tRNAcys primers.
Previously, hTERT was found to bind to regions of the ND1 and ND2 genes of mtDNA only (4). We revisited this issue by performing a modified chromatin immunoprecipitation assay that we developed earlier to identify mtDNA-interacting proteins [mIP, see methods and ref. (22)]. We used cells overexpressing tagged hTERT as, in our hands, the commercially available anti-TERT antibodies failed to immunoprecipitate endogenous hTERT. GM847 cells are immortalized through a telomerase-independent pathway and therefore do not have detectable levels of endogenous hTERT (23). Immunoprecipitations were also performed with the IgG control and with an antibody against TFAM, which binds indiscriminately to the mtDNA (2). We found that hTERT binds to various regions of the mtDNA, including the 12S and 16S rRNAs, ND1, 2, 4 and 5, COX I and III, tRNAs and the subunits 6 and 8 of the ATP synthase (right panel of Figure 2B, lane 1 in all panels). These results suggest that hTERT binds to mtDNA non-specifically (directly or indirectly), or that it is bound to mtDNA via a widely distributed cis-element. Why hTERT was found bound to ND1 and ND2 exclusively in a previous study is not clear but could relate to the use of cells transiently expressing hTERT (4).
The data presented in Figure 2A also show that hTERT co-purifies with mitochondrial tRNAs, consistent with a previous report (5). To determine whether hTERT binds specifically to mitochondrial tRNAs, we performed RIP as previously described (25). In short, GM847 cells expressing empty-vector (EV) or hTERT were crosslinked with formaldehyde and hTERT immunoprecipitated with the anti-HA antibody. Protein–RNA crosslinks were reversed by heat-treatment and RNA purified using Trizol reagent. Given that mtDNA encodes all the tRNAs required for translation of mtDNA-encoded proteins, we designed primers to amplify each of the 22 mitochondrial tRNAs. Location of the tRNA genes and sequence of the primers are shown in Figure 2C and Supplementary Table S1, respectively. We also amplified the two mitochondrial rRNA genes and larger regions of mtDNA, since the RNA transcribed from the mtDNA is polycystronic.
Results depicted in Figure 2D are representative of three independent experiments and show some of the tRNAs detected as bound to hTERT. No RNA was retrieved in the EV control or when DNase I-treated RNA was used as a template for PCR (Supplementary Figure S2B; Figure 2D, lane 1). Of the 22 mtDNA-encoded tRNAs, we detected interaction of hTERT with 14, but were unable to amplify rRNA or other larger mtDNA-encoded fragments (Figure 2D and data not shown), indicating that the interaction is specific to tRNAs.
In their extensive analysis, Maida et al. (5) showed that TERT also co-purified with the RMRP and with the 5.8S rRNA, two RNA species that are nuclear-encoded but known to partially localize to mitochondria (26–28). We also probed for their interaction with mitochondrial hTERT in organello, using the RIP protocol as above. Results presented in Supplementary Figure S2C indicate that hTERT interacts with these RNAs in mitochondria although it is unclear whether their assembly occurs in the nucleus and they are co-imported into mitochondria or whether they are imported separately and associate in the organelle. Of note, a recent study on the mitochondrial transcriptome confirmed the presence of these nuclear-encoded RNAs in mitochondria using ultra deep sequencing (29).
hTR is not present in human mitochondria and is not required for the mitochondrial effects of hTERT
The data presented above suggest that mitochondrial hTERT may function with different RNA templates instead of, or in addition to, the canonical telomeric template hTR. Previously we showed that hTERT gives rise to active telomerase in isolated mitochondria as judged by the telomeric repeat amplification protocol (TRAP; 1). However, this assay indirectly indicates the presence of hTR in mitochondria, and we cannot rule out the possibility that extra-mitochondrial RNA contamination may have contributed to our TRAP-positive results. Therefore to directly test for hTR in mitochondria RT-PCR was carried out on RNA extracted from highly purified organelles isolated from GM847 cells.
To rule out the presence of extra-mitochondrial RNAs, crude extracts were treated with RNAse and mitochondria subsequently isolated. Total mitochondrial RNAs were then purified using a commercially available kit, and samples DNase I-treated. We amplified cytochrome b, which is encoded by the mtDNA, to monitor mitochondrial RNA integrity (Figure 3A, left panel) and GAPDH to identify cytosolic contamination (Figure 3A, right panel). hTR RNA was undetectable in purified mitochondria pre-treated with RNase A (Figure 3A, middle panel), even when PCR were performed for 40 cycles (data not shown). The absence of hTR RNA cannot be explained by degradation of mitochondrial RNAs as cytochrome b was detected in both mitochondrial and crude extracts (Figure 4A). Taken together, these data indicate that hTR is not imported into mitochondria.
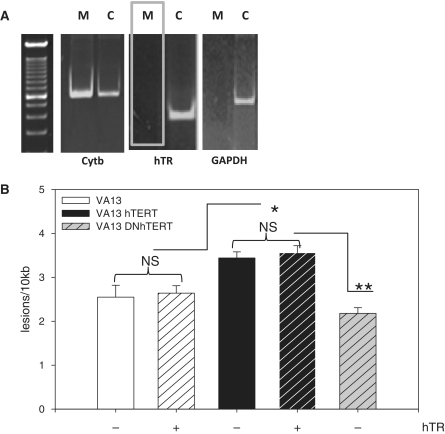
Figure 3.
hTR is not present in human mitochondria and not required for the mitochondrial effects of hTERT. (A) Whole cell lysates were treated with RNase A, washed and mitochondria were isolated. Following RNA extraction and DNase I treatment, RT-PCR was performed using 500 ng of total RNA. M = mitoplast, C = crude extract. (B) VA13 cells were infected with hTR lentivirus and/or WT hTERT or DNhTERT, submitted to treatment with 200 µM H2O2 for 60 min, and mtDNA damage analyzed by QPCR. Data are the mean of three independent experiments ± SEM. NS = not significant. No statistical difference was observed between DNhTERT and VA13 (P = 0.35).
Figure 4.
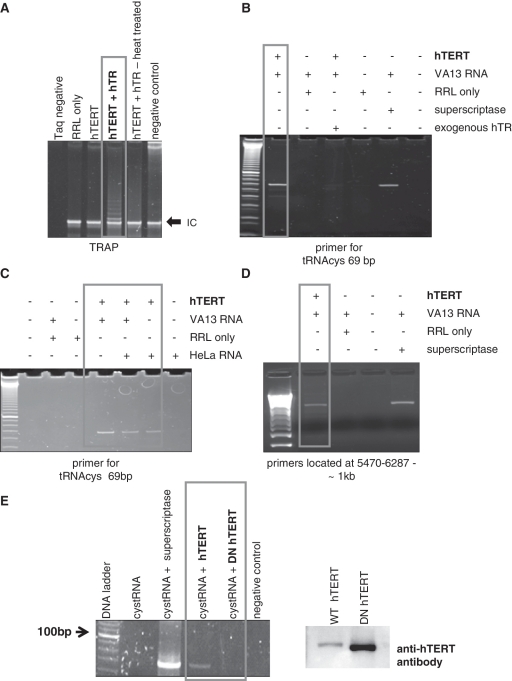
hTERT drives reverse transcription independent of hTR. hTERT translated in RRL was used for first strand cDNA synthesis. (A) TRAP was performed with RRL-translated hTERT to confirm its catalytic activity. Negative control omitting Taq polymerase in the PCR was also included. (B–D) First strand cDNA synthesis reactions were performed using RNA from HeLa (hTR-positive) or VA13 (devoid of hTR) cells and random hexamers. Commercially available RT was used as control. (E) The mitochondrial tRNAcys was transcribed in vitro and the RNA used as template for first strand cDNA synthesis. WT and catalytically inactive (DN) hTERT were translated in RRL and used in the reactions. The commercially available RT was used as control. Western blots (right panel) show RRL-translated proteins; only WT hTERT is FLAG-tagged (46).
Requirement for hTR to the mitochondrial function of telomerase was tested taking advantage of VA13 cells as they are devoid of endogenous hTR (30). Because the function of TERT in mitochondria is currently unknown, devising an assay to detect its mitochondrial activity is not possible. To address the need of hTR for the mitochondrial effects of hTERT, we revisited our original data showing that overexpression of hTERT promotes mtDNA damage induced by H2O2 (1,2). We reasoned that if hTR was required for such effects, then cells expressing hTERT but lacking hTR should have less mtDNA damage when compared to controls. We overexpressed WT hTERT in VA13 cells and re-introduced hTR on a lentivirus (30). As controls, we used VA13 cells and VA13 expressing only hTR. Expression of the genes was confirmed by RT-PCR and reconstitution of catalytic activity was assayed by TRAP (data not shown). Cells were submitted to H2O2 treatment and mtDNA integrity analyzed by gene-specific quantitative PCR (qPCR) as described previously (31–33). Briefly, immediately after exposure to 200 µM H2O2, total genomic DNA was isolated and large (∼9 kb) and small (∼200 bp) fragments of mtDNA were amplified in a quantitative manner. The latter was used to normalize the data to changes in mtDNA copy number. Amplification of treated samples was compared to controls and relative amplifications were calculated. These values were then used to estimate the lesion frequency present on the mtDNA based on a Poisson distribution (31–33).
The presence of hTR itself did not significantly impact levels of mtDNA damage in VA13 cells (Figure 3B compare white bars). Consistent with our previous results (1,2), estimated levels of mtDNA damage were higher in the presence of overexpressed WT hTERT (Figure 3B, compare white and black bars). More importantly, mtDNA damage in the WT background was independent of hTR (Figure 3B, compare black bars), supporting the findings that hTR is not present in mitochondria.
Using a hTERT variant that bears two mutations in its third reverse transcriptase motif (DNhTERT, 34), we previously showed that telomerase RT activity was required for the mitochondrial effects of hTERT (2). The above data indicate that such RT activity is independent of hTR. To directly address this possibility, we introduced DNhTERT in VA13 cells, and submitted them to H2O2 treatment and mtDNA damage analysis as above. If RT activity is required for the mitochondrial effects of hTERT, mtDNA should be damaged to a lesser extent in DNhTERT compared to WT hTERT. Levels of mtDNA damage were significantly decreased in DNhTERT cells relative to WT hTERT (Figure 3B, compare gray to black bars) but were not statistically different than the VA13 controls (Figure 3B, compare gray to white bars). These data indicate that to affect mtDNA hTERT must work as a RT. Given the lack of hTR in this cellular background, these results can only be interpreted as reflecting that telomerase RT activity in mitochondria is reconstituted with mitochondrial RNAs.
hTERT drives first strand synthesis in vitro in the absence of hTR
Reverse transcriptases other than telomerase use cellular tRNAs to prime reverse transcription (35). Thus, it is possible that in mitochondria hTERT behaves as other RTs. However, it is yet to be demonstrated that hTERT can drive a RT reaction in the absence of its canonical RNA (hTR). Thus, we next asked whether hTERT can perform first strand cDNA synthesis in vitro as shown for other RTs.
We translated WT hTERT in RRL in the presence or absence of hTR and first assessed its TRAP-based reverse transcriptase activity. In this assay TERT uses hTR as the template RNA to drive telomeric-DNA synthesis in vitro. Panel A of Figure 4 shows that the RRL-translated hTERT was catalytically active in the TRAP in the presence of hTR. Subsequently, we isolated total cellular RNA from HeLa (hTR-positive) or VA13 cells (hTR-negative) to be used as template for first strand cDNA synthesis. RNA was treated with DNase I to remove contaminating DNA and cDNA synthesis promoted using random hexamers and either RRL-translated hTERT or a commercially available RT. The cDNA was then amplified by PCR using various primers to mtDNA-encoded genes (Supplementary Table S1 and ref. 22). Reactions were performed at 37°C and 50°C and showed similar results (Figure 4 and Supplementary Figure S3). Data presented in Figure 4 are representative of independent experiments, which included independent RNA isolations, RRL translations and RT reactions.
Clearly, hTERT was able to drive cDNA synthesis in the absence of hTR (Figure 4C and D). The reaction was as efficient as with the commercially available RT (Figure 4, compare lanes 1 and 5 of panel B, and lanes 1 and 4 of panel D) and was dependent on hTERT as no products were observed when the enzyme was omitted (Figure 4, lane 2 in all panels). hTERT did not generate any products when pre-incubated with hTR (Figure 4B, lane 3), which was not surprising since the template used for the reactions was not telomeric. When hTR RNA was present in the same sample (HeLa RNA) the level of product generated by hTERT was decreased (Figure 4C), suggesting that hTR and other RNAs compete for binding to hTERT. In support of this conclusion, addition of purified mitochondrial RNAs to the TRAP reaction inhibited the classical telomeric-laddering resulting from telomerase activity (data not shown).
To assure the specificity of the reactions, we used in vitro transcribed RNA of the mitochondrial tRNAcys gene as template for the cDNA synthesis reactions. We also included the catalytically inactive DNhTERT mutant to determine requirement for reverse transcriptase function. As can be seen in Figure 4E, hTERT was able to make the cDNA of the specified RNA template (lane 3) and this activity was clearly dependent on its reverse transcriptase function (lane 4).
Together with data presented in Figures 2 and 3, these results support the hypothesis that in mitochondria TERT works as a hTR-independent RT, using mitochondrial tRNAs as template for reverse transcription.
Lack of hTERT specifically in mitochondria negatively impacts the organelle
The data presented so far indicate that hTERT may be involved in mtDNA metabolism. If this is indeed the case, then lack of hTERT in the organelle should lead to mitochondrial defects. In this regard, it was shown that siRNA for hTERT is associated with increased mitochondrial ROS production (3). Likewise, mouse embryonic fibroblasts (MEFs) derived from a TERT KO mouse showed decreased mitochondrial complex I activity, and targeting of hTERT specifically to mitochondria improved mitochondrial function (4). More recently, decreased mtDNA content accompanied by systemic mitochondrial defects was observed in a TERT KO mouse model (15).
Although together these data support a direct role for TERT in mitochondria, the lack of TERT also in the nucleus makes it difficult to determine whether the mitochondrial defects observed were caused by the lack of the protein in the organelle or resulted from telomere erosion. Given that mitochondrial impairments in the TERT KO mouse were observed already in early generation animals, which essentially had normal telomeres (15), we hypothesized a direct role for TERT in mitochondria.
To tease out the effects of hTERT in mitochondria versus in the nucleus (telomeres), we utilized a unique mutant previously described by us in which two amino acid residues on the MTS of the protein were substituted (R3E/R6EhTERT). This protein is catalytically active in the nucleus, being proficient in cellular immortalization, but is unable to enter mitochondria (2). Herein this mutant is referred to as nuchTERT. Using normal fibroblasts untransfected or stably expressing WT or nuchTERT, we monitored mtDNA integrity, mitochondrial superoxide generation and evaluated mitochondrial ultrastructure using electron microscopy (EM) as surrogate markers of mitochondrial proficiency. If the defects observed in mitochondria were indeed caused by the lack of hTERT in the organelle, increased mtDNA damage and ROS production should be observed in nuchTERT when compared to WT cells. Likewise, we would expect a higher degree of mitochondria with altered ultrastructure in cells expressing the mutant.
Results presented in Figure 5 support the notion that the lack of mitochondrial localized hTERT is responsible for the defects observed. Indeed, significant degree of mtDNA damage (Figure 5A) and increased ROS production (Figure 5B) were observed in cells lacking hTERT in mitochondria. In support of these findings, EM analysis indicated that nuchTERT cells had 50% more mitochondria with altered ultrastructure when compared to controls (Figure 5C, data not shown). Increased vacuolization of the cytoplasm (Figure 5C, see asterisks) along with mitochondria distension (Figure 5C, black arrows) were observed in nuchTERT-expressing cells whereas no significant differences were observed between non-transfected and WT hTERT-expressing cells (data not shown). While approximately 20% of non-transfected cells and 30% of WT hTERT-expressing cells showed signs of autophagy (data not shown) ∼60% of mutant-expressing cells had autophagosomes, some of which contained mitochondria (Figure 5C red arrows). As autophagy has been increasingly recognized as a line of defense to remove damaged organelles (36), our data indicate that in the absence of TERT in mitochondria a pathway to increase clearance of damage organelles is activated. Interestingly, the TERT KO animals also presented with decreased mitochondria density in both heart and liver (15).
Taken together, our data demonstrate that absence of hTERT specifically in mitochondria leads to mitochondrial dysfunction, which likely results from the lack of its reverse transcriptase activity in the organelle.
DISCUSSION
After our initial discovery that hTERT has a MTS that is both necessary and sufficient to target hTERT to mitochondria, several other studies have indicated the presence of ectopically expressed hTERT in the mitochondria of human cells (1–4,11–14). The data presented here give compelling evidence that mammalian TERT is a mitochondrial protein. Protease-protection assays showed that a fraction of endogenous TERT is mitochondrial, and classical mitochondrial protein import assays defined that it translocates into the matrix side of the inner mitochondrial membrane. We also show that TERT co-purifies with mitochondrial nucleic acids along with other nucleoid proteins, and that in cells it is associated with both mtDNA and mitochondrial tRNAs. Collectively, these data give strong evidence that TERT is a dually targeted protein in mammals.
Based on our analysis it is clear that the intracellular distribution of TERT is unequal. In fact, we estimate that ∼10–20% of the total cellular content of the protein is mitochondrial under normal conditions, which is consistent with previous findings (3,4,13). Note that under oxidative stress hTERT accumulates in a time-dependent manner in mitochondria. In such circumstances ∼80% of the protein is present in the organelle (3). The reason for hTERT accumulation in mitochondria under oxidative stress is still unknown. The non-equal intracellular distribution of dually targeted proteins is not a new phenomenon, and has been termed ‘eclipsed distribution’ (37). Such uneven distribution is believed to hamper the detection of the less abundant isoform given its presence is masked by the dominant isoprotein in the other subcellular compartment (37). This phenomenon together with the poor efficiency of the commercially available anti-TERT antibodies likely explain why the mitochondrial localization of TERT was not described earlier.
Indirect evidence had suggested that hTR was also mitochondrial (1,4). However, our data strongly support the conclusion that hTR is not present in human mitochondria (Figure 3) or required for the function of organellar telomerase (Figures 2–4). Indeed, together with recent work from others (5) our data indicate that mitochondrial TERT assembles with other RNAs to reconstitute catalytic activity. The lack of hTR in mitochondria and of its need for telomerase function in the organelle is not surprising since the mtDNA is circular and devoid of telomeres, making it improbable that mitochondrial and nuclear TERT function the same way. While others have reported hTR-independent functions of TERT (5–10), the notion that hTERT can drive reverse transcription independent of hTR is new, and brings to question whether such a function also occurs in the nucleus. We believe this to be a possibility but would rely on TERT binding to cellular tRNAs (or other RNAs) as the mitochondrial ones are mtDNA-encoded and as such are confined to the organelle. The binding of nuclear TERT to cellular RNAs other than hTR or the RMRP has yet to be demonstrated.
Maida et al. (5) identified in their comprehensive analysis that hTERT also co-purifies with the RMRP and the 5.8S rRNA, which we recapitulated in this study using isolated mitochondria (Supplementary Figure S2C). In the nucleus, when assembled with the RMRP TERT works as a RNA-dependent RNA polymerase (5). Whether it attains this same activity in mitochondria has yet to be verified. It also remains to be tested whether TERT binding with the 5.8S rRNA leads to any catalytic activity.
It is interesting that all RNAs found to interact with hTERT [this study and ref. (5)] are thought to be involved in mtDNA replication (26–28, 38). It has also been proposed that priming of the light strand origin, which is still poorly understood, may rely on activity of a reverse transcriptase (38). Thus, it is tempting to speculate that mitochondrial telomerase is involved in organellar DNA replication. Interestingly, mtDNA depletion was observed in the TERT KO animals (15), and targeting hTERT specifically to mitochondria alleviated mtDNA loss caused by low doses of ethidium bromide, a classical means to deplete cells of mtDNA (4). Alternatively, TERT may work in mtDNA repair, specifically, in mtDNA double strand break (DSB) repair. DSB repair has yet to be identified in mitochondria although DSB rejoining activity has been described in the organelle (39). Recent evidence in yeast indicates that DSBs can be repaired using RNA, which is then converted into DNA in a subsequent step. Such activity is predicted to be carried out by a RT (40). A role in mtDNA DSB repair could justify why under oxidative stress hTERT accumulates in mitochondria (3) and also explain the viability of TERT KO animals (15,41): in the absence of this non-essential function non-repairable DNA DSBs would be eliminated (39). Finally, such activity would also be consistent with the improved mitochondrial function observed upon overexpression of TERT in cells otherwise considered telomerase negative (3,4,12). Evidence that hTERT can work in mtDNA replication or repair and its relevance in vivo has yet to be provided.
The findings that TERT has a role in mitochondrial biology have led to the proposal of a dual role for telomerase in aging (2,3,42). Since hTERT is overexpressed in the vast majority of cancers (24,35), which curiously remodel mitochondrial ‘usage’ from oxidative phosphorylation to aerobic glycolysis (43), it is possible that hTERT also has a dual role in carcinogenesis. How exactly hTERT impacts mitochondrial function in cancer is an open area of research.
Finally, our data underscore that manipulation of TERT (ablation or overexpression) is not simply affecting telomere biology, and knocking out the associated RNA is not the same as TERT deletion. Thus, data obtained with such approaches should be carefully interpreted. The recent study that found systemic mitochondrial defects in the TERT KO animals concluded that the phenotypes resulted from telomere dysfunction, despite the fact that early generation animals had essentially normal telomeres (15). Although the authors clearly demonstrated that similar phenotypes were observed when using TERC KO animals, the TERC −/− animals examined had strong telomere dysfunction (15), making it difficult to eliminate confounding effects associated to telomere shortening. Thus, a firm prediction of our results is that comparison of TERC and TERT knockout mice with long telomeres will help reveal the specific contribution of mitochondrial TERT to organellar function and organismal fitness.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Department of Defense, Army Research Office (grant number 56027LS, to J.H.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs Elizabeth Blackburn (University of California San Francisco), Lea Harrington (University of Edinburgh, UK) and Chantal Autexier (McGill University, Canada) for kindly sharing reagents. We thank technical help from Ms Perihan Ulema, Dr Senyene Hunter for optimizing protocol for RNAse digestion of mitochondria, and Dr C. Cummings for pathology reports from the EM micrographs. We thank the members of the Thomas and Santos’ laboratories for fruitful discussions, and Dr Gerald Shadel (Yale School of Medicine) for critical review of the manuscript.
REFERENCES
- 1.Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Mitochondrial hTERT exacerbates free radical-mediated mtDNA damage. Aging Cell. 2004;6:399–411. doi: 10.1111/j.1474-9728.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- 2.Santos JH, Meyer JN, Van Houten B. Mitochondrial localization of hTERT as a determinant for hydrogen peroxide-induced mtDNA damage and apoptosis. Human Mol. Gen. 2006;15:1757–1768. doi: 10.1093/hmg/ddl098. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell. Sci. 2008;121:1046–1053. doi: 10.1242/jcs.019372. [DOI] [PubMed] [Google Scholar]
- 4.Haendeler J, Dröse S, Büchner N, Jakob S, Altschmied J, Goy C, Spyridopoulos I, Zeiher AM, Brandt U, Dimmeler S. Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler. Thromb. Vasc. Biol. 2009;6:929–935. doi: 10.1161/ATVBAHA.109.185546. [DOI] [PubMed] [Google Scholar]
- 5.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lue NF, Bosoy D, Moriarty TJ, Autexier C, Altman B, Leng S. Telomerase can act as a template- and RNA-independent terminal transferase. Proc. Natl Acad. Sci. USA. 2005;102:9778–9783. doi: 10.1073/pnas.0502252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl Acad. Sci. USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W, Cheung P, Jun S, Artandi MK, Shah N, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;1:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Sung YH, Cheong C, Choi YS, Jeon HK, Sun W, Hahn WC, Ishikawa F, Lee HW. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene. 2008;27:3754–3760. doi: 10.1038/sj.onc.1211037. [DOI] [PubMed] [Google Scholar]
- 11.Del Bufalo D, Rizzo A, Trisciuoglio D, Cardinali G, Torrisi MR, Zangemeister-Wittke U, Zupi G, Biroccio A. Involvement of hTERT in apoptosis induced by interference with Bcl-2 expression and function. Cell Death Differ. 2005;11:1429–1438. doi: 10.1038/sj.cdd.4401670. [DOI] [PubMed] [Google Scholar]
- 12.Kovalenko OA, Caron MJ, Ulema P, Medrano C, Thomas AP, Kimura M, Bonini MG, Herbig U, Santos JH. A mutant telomerase defective in nuclear-cytoplasmic shuttling fails to immortalize cells and is associated with mitochondrial dysfunction. Aging Cell. 2010;9:203–219. doi: 10.1111/j.1474-9726.2010.00551.x. [DOI] [PubMed] [Google Scholar]
- 13.Büchner N, Zschauer TC, Lukosz M, Altschmied J, Haendeler J. Downregulation of mitochondrial telomerase reverse transcriptase induced by H2O2 is Src kinase dependent. Exp. Gerontol. 2010;45:558–562. doi: 10.1016/j.exger.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Indran IR, Hande MP, Pervaiz S. hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res. 2011;71:266–276. doi: 10.1158/0008-5472.CAN-10-1588. [DOI] [PubMed] [Google Scholar]
- 15.Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YL, Dudognon C, Nguyen E, Hillion J, Pendino F, Tarkanyi I, Aradi J, Lanotte M, Tong JH, Chen GQ, et al. Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J. Cell Sci. 2006;119:2797–2806. doi: 10.1242/jcs.03001. [DOI] [PubMed] [Google Scholar]
- 17.Sato T, Mihara K. Mammalian Oxa1 protein is useful for assessment of submitochondrial protein localization and mitochondrial membrane integrity. Anal. Biochem. 2010;397:250–252. doi: 10.1016/j.ab.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 18.Stonjanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- 19.Gordon DM, Kogan M, Knight SA, Dancis A, Pain D. Distinct roles for two N-terminal cleaved domains in mitochondrial import of the yeast frataxin homolog, Yfh1p. Hum. Mol. Genet. 2001;10:259–269. doi: 10.1093/hmg/10.3.259. [DOI] [PubMed] [Google Scholar]
- 20.Stuart RA, Koehler CM. In vitro analysis of yeast mitochondrial protein import. Curr. Protoc. Cell Biol. 2007;11:19. doi: 10.1002/0471143030.cb1119s34. [DOI] [PubMed] [Google Scholar]
- 21.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;4:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu B, Yadav S, Shah PG, Liu T, Tian B, Pukszta S, Villaluna N, Kutejová E, Newlon CS, Santos JH, et al. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J. Biol. Chem. 2007;282:17363–17374. doi: 10.1074/jbc.M611540200. [DOI] [PubMed] [Google Scholar]
- 23.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl Acad. Sci. USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;1:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 26.Wong TW, Clayton DA. DNA primase of human mitochondria is associated with structural RNA that is essential for enzymatic activity. Cell. 1986;45:817–825. doi: 10.1016/0092-8674(86)90556-8. [DOI] [PubMed] [Google Scholar]
- 27.Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 28.Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- 29.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Crothers J, Haqq CM, Blackburn EH. Cellular and gene expression responses involved in the rapid growth inhibition of human cancer cells by RNA interference-mediated depletion of telomerase RNA. J. Biol. Chem. 2005;280:23709–23717. doi: 10.1074/jbc.M502782200. [DOI] [PubMed] [Google Scholar]
- 31.Santos JH, Mandavilli BS, Van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol. Biol. 2002;197:159–176. doi: 10.1385/1-59259-284-8:159. [DOI] [PubMed] [Google Scholar]
- 32.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 33.Kovalenko OA, Santos JH. Analysis of oxidative damage by gene-specific quantitative PCR. Curr. Protoc. Hum. Genet. 2009;19:1. doi: 10.1002/0471142905.hg1901s62. [DOI] [PubMed] [Google Scholar]
- 34.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 1999;10:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 35.Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell Mol. Life Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;12:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta. 2011;1808:1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 39.Liu P, Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ. Mol. Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 40.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 42.Saretzki G. Telomerase, mitochondria and oxidative stress. Exp. Gerontol. 2009;44:485–492. doi: 10.1016/j.exger.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 44.Kovalenko OA, Kaplunov J, Herbig U, Detoledo S, Azzam EI, Santos JH. Expression of (NES-)hTERT in cancer cells delays cell cycle progression and increases sensitivity to genotoxic stress. PLoS One. 2010;5:e10812. doi: 10.1371/journal.pone.0010812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39:5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr. Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 47.Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 48.Coffey G, Campbell C. An alternate form of Ku80 is required for DNA end-binding activity in mammalian mitochondria. Nucleic Acids Res. 2000;28:3793–3800. doi: 10.1093/nar/28.19.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.