Abstract
Cellular micro(mi)RNAs are able to recognize viral RNAs through imperfect micro-homologies. Similar to the miRNA-mediated repression of cellular translation, this recognition is thought to tether the RNAi machinery, in particular Argonaute 2 (AGO2) on viral messengers and eventually to modulate virus replication. Here, we unveil another pathway by which AGO2 can interact with retroviral mRNAs. We show that AGO2 interacts with the retroviral Group Specific Antigen (GAG) core proteins and preferentially binds unspliced RNAs through the RNA packaging sequences without affecting RNA stability or eliciting translation repression. Using RNAi experiments, we provide evidences that these interactions, observed with both the human immunodeficiency virus 1 (HIV-1) and the primate foamy virus 1 (PFV-1), are required for retroviral replication. Taken together, our results place AGO2 at the core of the retroviral life cycle and reveal original AGO2 functions that are not related to miRNAs and translation repression.
INTRODUCTION
Viruses are obligatory intracellular parasites that hijack many, if not all, cellular pathways. The RNA interference (RNAi) and micro (mi)RNA pathway is no exception (1–3). The miRNAs control translation and protein production by redirecting the miRNA ribonucleoprotein (miRNP) complex (also called RNA-induced silencing complex, RISC) on mRNAs harboring imperfect micro-homologies (4,5). Though the complete composition of the miRNP is not fully characterized, several key effectors have been identified, such as the Argonaute (AGO) proteins, DCP1 and GW182 proteins (4–6). Mammalian genomes encode four AGOs that play redundant roles in miRNA-mediated repression (7). In contrast, AGO2 is the only AGO that functions in RNA interference because its P-element induced wimpy testis (PIWI) domain permits the cleavage of the mRNA at the center of the siRNA–mRNA duplex (8). The AGO proteins, as well as miRNAs, other component of the miRNP and miRNA targets, are found in Processing (P)-bodies (9,10), cytoplasmic foci that are enriched in mRNA-catabolizing enzymes and translational repressors (9). However, AGO proteins can repress translation in the absence of P-bodies, and P-bodies are formed as a consequence of AGO function (11). In addition, AGO2 is also detected with diffuse cytoplasmic staining (7). Hence, the exact implication of P-bodies in RNA silencing and their importance in AGO function(s) are not yet fully understood.
One aspect of the interplay between viruses and the RNAi pathway is the capacity of host miRNAs to recognize viral mRNAs (12–21). This recognition is detrimental for several viruses (12,15–22) but beneficial for Hepatitis C Virus (HCV) (14,16). Moreover, the replication of certain viruses is not at all affected by cellular miRNAs (15). Hence, the link between viral RNAs and the host miRNA machinery may rely on more complex mechanisms that remain to be clarified (1,2,23).
To this aim, we dissected the interactions between the host miRNA machinery and two unrelated retroviruses: primate foamy virus 1 (PFV-1) (12) and human immunodeficiency virus 1 (HIV-1) (17–19,22). These two viruses were studied because they represent the most distantly related retroviruses and common features are likely to be conserved in the whole Retroviridae family (24–26). We show that AGO2 is also tethered on retroviral RNAs through GAG and the GAG-interacting RNA packaging signals without involving miRNAs and translation repression. Using RNAi experiments, we further revealed that AGO2, as opposed to other AGOs, plays crucial functions in both PFV-1 and HIV-1 replications, a scenario akin to HCV (27,28). Together, our results unveil original AGO2 functions that are unlinked to miRNA and translation regulation, but yet hijacked by both PFV-1 and HIV-1.
MATERIALS AND METHODS
Cells, viruses and transfection
293T cells were maintained in DMEM (Gibco-BRL) supplemented with 2 mM l-glutamine, 100 µg/ml penicillin, 50 µg/ml streptomycin and 10% fetal calf serum and transfected with Lipofectamine 2000 (Invitrogen). Jurkat cells were maintained in RPMI (Gibco-BRL) supplemented with 2 mM l-glutamine, 100 µg/ml penicillin, 50 µg/ml streptomycin and 10% fetal calf serum and transfected using the Amaxa Cell line Nucleofector kit V (Lonza). Cell culture was realized using a Z1 Coulter Particle counter (Beckman Coulter). To produce PFV-1 viruses, 293T were transfected with the pc13 provirus and, 2 days post-transfection, cells and supernatants were collected and lysed by three successive cycles at −80°C/37°C. The virus stock was collected after centrifugation for 15 min at 12 000 rpm and 4°C. To produce HIV-1 virions, 293T cells were transfected with the pNL4.3 provirus and supernatants were collected and cleared using 0.45 µ filters.
Plasmids and mutagenesis
The following vectors were previously described: myc-AGO2 and myc-PAZ9 in (22,29), pc13 in ref. (12), pMH29 and pcgp1 in ref. (30), pFH-AGO2, pFH-AG02-Y529A, pFH-AG02-Y52E, pFH-AG02-Y529F in ref. (31), APOBEC3G-V5 in ref. (32). The pDCP1-flag, pAGO2-EGFP and pGW182-EGFP were provided by W. Filipowicz. The pRFP-p54 was provided by D. Weil. To construct EGFP-GAG vectors, the PFV-1 GAG ORF was PCR-amplified and cloned into the SacII/XmaI restriction sites of pEGFP-C1. The GRI motif was deleted using the QuickChange Mutagenesis kit (Stratagene) and the primers are indicated in Supplementary Data. The PFV-1 encapsidation sequences were amplified from the pMH29 vector (provided by A. Rethwilm) (30). The PCR product was inserted into the XhoI/NotI restriction sites of the psiCHECK2 vector (Promega). The HIV-1 encapsidation sequence was extracted from pLK0-1 using BglII and NotI. This sequence was further cloned in the XhoI site of the psiCHECK2 vector. The sequences of the siRNAs are indicated in Supplementary Data.
RNA-immunoprecipitations
The 293T cells were lysed 48 hpt (hours post-transfection) in 20 mM HEPES pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 250 mM Sucrose, 0.05% NP40, 0.5% Triton X-100, complete EDTA-free protease inhibitor cocktail (Roche). Lysates were pre-cleared on Ig-G/sepharose beads for 1 h at 4°C and incubated overnight with the indicated antibody fixed on Ig-G/sepharose beads. After washes, samples were treated with proteinase-K for 1 h at 37°C and immunoprecipitated RNAs were extracted using Tri-Reagent (Invitrogen).
Quantitative RT–PCR
Total RNA was extracted using Tri-Reagent (Invitrogen). We performed a DNase RQ1 (Sigma) treatment for 30 min at 37°C on 2 µg of RNA to digest residual genomic DNA. RT was performed using the SuperScript II RT (Invitrogen) and oligodT(N). qPCRs were performed using SYBR Green PCR Master Mix (Roche). Primer sequences are indicated in Supplementary Data. Results are means of at least three independent experiments.
Protein immunoprecipitation
Cells were lysed in 50 mM Tris–HCl (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate (SDS), complete EDTA-free protease inhibitor cocktail (Roche) for 15 min at 4°C. Lysates were pre-cleared on Ig-G/sepharose beads and incubated overnight with specific antibody fixed on Ig-G/sepharose beads. Beads directly coupled to anti-GFP antibodies (ChromoTek GmbH) were also used. After washes, beads were heat-disrupted at 100°C for 5 min in Laemmli buffer. When indicated, RNases A+T1 (Ambion) were added to the cell lysates before immunoprecipitation (IP). The IP fractions or ∼50 µg of Laemmli total protein extracts were resolved by SDS–PAGE and transferred onto a Nitrocellulose Membrane (Protran Whatman Schleicher and Schuell). Blots were incubated in 20 mM Tris–HCl (pH 8), 150 mM NaCl, 0.1% Tween-20 and 5% milk and further incubated with specific antibodies diluted in the same buffer. Western blots were revealed using an enhanced chemiluminescence kit (Amersham).
Luciferase assays
The psiCHECK-2 Vector (Promega) contains a reporter gene, firefly luciferase, which allows normalization of the Renilla luciferase expression. This vector was transfected in 293T cells, which were lysed in Passive Lysis Buffer (Promega) 48 hpt. The expression of the Renilla and the firefly luciferases were measured with the Dual-Luciferase Reporter Assay System (Promega). Results are the mean of at least three independent experiments (three biological replicates and for each biological replicate (i.e. same lysate), three technical replicates).
Antibodies
The antibodies used were: anti-myc (clone 9E10, Roche), anti-V5 (Invitrogen), anti EGFP-HRP (MACS), anti-Tubulin (Seotec), anti-Flag M2 (Sigma) and anti-HIV-1 p24 (AIDS reagent). Anti-AGO1 and anti-AGO2 antibodies were provided by G. Meister. The polyclonal anti-PFV-1 antibodies were provided by A. Saïb. The anti-p24 ELISA kit was purchased from Innogenetics.
Virion purification
Retroviral particles were purified from cell culture supernatants, which were cleared with 0.45 µ filter and centrifuged through a 20% sucrose cushion in a solution containing 100 mM NaCl, 10 mM Tris–HCl (pH 7.4), and 1 mM EDTA at 25 000 rpm for 3 h in a SW28 rotor (Beckman) at 4°C.
Electron microscopy
Cells were fixed in 2.5% glutaraldehyde, Sorensen buffer (0.1 M, pH 7.4) overnight at 4°C. Next day, cells were washed in Sorensen's buffer and post-fixed in 1% osmic acid for 1 h at room temperature, then washed twice with Sorensen's buffer, dehydrated in a graded series of ethanol, and embedded in epon resin. Sections were cut with a Leica–Reichert Ultracut E and collected at different levels of each block. The sections were counterstained with uranyl acetate 1.5% in ethanol 70%, and observed using a Hitachi 7100 transmission electron microscope equipped with an AMT digital camera at ‘The Centre de Ressources en Imagerie Cellulaire’ in Montpellier.
Yeast two-hybrid
The HIV-1 GAG and hAGO2 cDNAs were recovered from donor vectors of the Gateway cloning system (Invitrogen) and cloned into two-hybrid plasmids (pACT-II and pAS2ΔΔ). For the two-hybrid assays, plasmids were introduced into the appropriate haploid strains (CG929 or YL455), which were then crossed. Diploids were plated on double or triple selectable media (minus Leu, Trp, or minus Leu, Trp, His), and growth was assessed 3 days later. We used Alix as a positive control for the interaction with HIV-1 GAG (33) and Rsa (a yeast protein) as negative control.
RESULTS
The PFV-1 and HIV-1 mRNAs can interact with the RNAi machinery in a miRNA- and P-body-independent manner
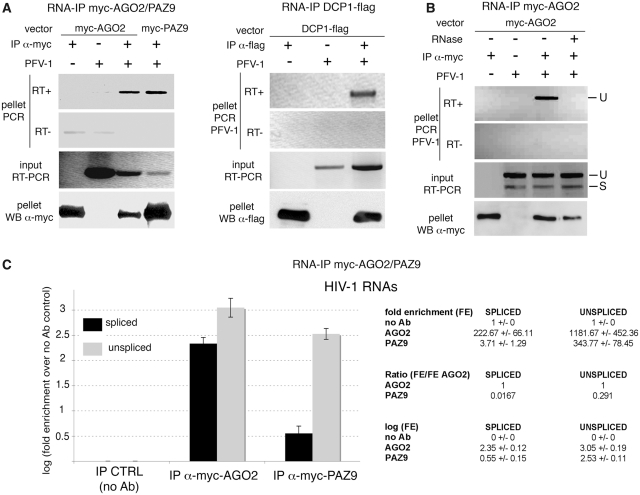
In order to confirm the interactions of PFV-1 RNAs with the RNAi machinery (12), we first performed RNA-immunoprecipitations (RNA-IPs) in 293T cells transfected with the PFV-1 provirus and myc-AGO2 or DCP1-flag vectors. These experiments indeed showed that PFV-1 RNAs co-immunoprecipitated with both AGO2 and DCP1 (Figure 1A). Strikingly, using a different set of primers, we further observed that AGO2 preferentially bound unspliced PFV-1 RNAs (Figure 1B). The spliced RNA form was below PCR detection limit (Figure 1B). Because the interaction between PFV-1 RNAs and miRNAs seemed to differ from that usually observed in the case of cellular mRNAs, we evaluated the role of host miRNAs in the interactions depicted in Figure 1A. We used a mutant of AGO2 that is unable to interact with miRNAs (called PAZ9) (29,31). PAZ9 is not detectable in P-bodies although it is still able to interact with DCP1 [(29) and data not shown] and to induce RNA silencing when artificially tethered to RNA (31). We observed that PFV-1 RNAs also co-immunoprecipitated with PAZ9, suggesting that the interaction between PFV-1 RNAs and AGO2 does not strictly require miRNAs and occurs outside of P-bodies (Figure 1A).
Figure 1.
Retroviral mRNAs interact with the RNAi machinery independently of miRNAs and P-bodies. (A) 293T cells were transfected with the PFV-1 pc13 provirus together with vectors encoding myc-AGO2/myc-PAZ9 (left) or DCP1-flag (right). IPs directed against the myc tag (left) or the flag tag (right) were performed 48 hpt. IP (pellet) and cell-extract (input) RNAs were further analyzed by RT–PCR. IP efficiency was controlled by western blots (WBs). (B) IP RNAs described in (A) were analyzed by RT–PCR using primers discriminating spliced (S) and unspliced (U) RNAs. RNase treatment was performed before IP to assess potential DNA contamination (due to DNA transfection). (C) 293T cells were transfected with the HIV-1 pNL4.3 provirus and the myc-AGO2 or myc-PAZ9 expression vectors. IPs directed against the myc tag were performed 48 hpt. RNAs were extracted from immunoprecipitated fractions and assayed for the presence of spliced (black) and unspliced (gray) viral RNAs by RT–qPCR. The log(fold enrichment, FE) = {log[2∧(Ct noAb – Ct IP)]} was calculated. The log representation was chosen in order to underscore the FE obtained with spliced RNAs bound to PAZ9. The details of our calculation are shown.
We then repeated these experiments with HIV-1 for which key interactions with miRNAs have also been documented (17–19). We performed anti-myc RNA-IPs in 293T cells transfected with the HIV-1 pNL4.3 provirus and myc-AGO2 or myc-PAZ9 expressing vectors. We found that both AGO2 and PAZ9 interacted with spliced and unspliced HIV-1 RNAs although to different extents (Figure 1C): 30% of unspliced RNAs interacted with AGO2 independently of miRNAs (Figure 1C, compare IP AGO2 with IP PAZ9), while the vast majority (98%) of the spliced RNAs interacted with AGO2 in a miRNA-dependent manner (Figure 1C). We concluded that host miRNAs and P-bodies are not strictly required for the interaction between unspliced retroviral RNAs and AGO2. Since unspliced RNAs can be specifically packed into virus-derived ribonucleoprotein complexes en route to virion egress, we hypothesized that viral proteins could also interact with core components of the RNAi machinery.
The PFV-1 and HIV-1 GAG proteins interact with AGO2 in a miRNA- and P-body-independent manner
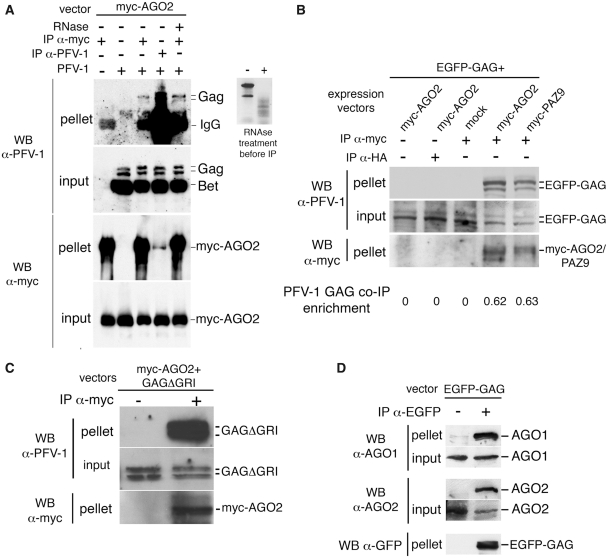
To test this idea, 293T cells were transfected with the PFV-1 provirus and myc-AGO2 expressing vector. Using immunoprecipitation experiments, we observed that PFV-1 GAG co-immunoprecipitated with myc-AGO2 and that, inversely, myc-AGO2 co-immunoprecipitated with PFV-1 GAG (Figure 2A). Interestingly, the GAG:AGO2 interaction was insensitive to RNase treatment (Figure 2A), suggesting that RNAs, in particular viral RNAs, were not required. In fact, when expressed alone in 293T cells, in the absence of a viral genome, PFV-1 GAG co-immunoprecipitated with myc-AGO2 (Figure 2B). We also observed that PAZ9 co-immunoprecipitated with PFV-1 GAG (Figure 2B) indicating that the GAG:AGO2 interaction did not necessitate miRNAs or P-bodies. To confirm that RNAs were not required for the GAG:AGO2 interaction, we mutated the nucleic acid binding domain of PFV-1 GAG (34) and verified that this mutant was still able to co-immunoprecipitate with myc-AGO2 (Figure 2C). We also verified that PFV-1 GAG interacted with endogenous AGO1 and AGO2 (Figure 2D).
Figure 2.
PFV-1 GAG interacts with AGO2 independently of miRNAs and P-bodies. (A) 293T cells were transfected with pc13 and myc-AGO2 vectors. IPs anti-PFV-1 proteins or anti-myc were performed 48 hpt. IP (pellet) and cell lysates (input) were analyzed by WB as indicated. RNase treatment was carried out before IP. (B) 293T cells were transfected with PFV-1 EGFP-GAG and myc-AGO2, myc-PAZ9 or pcDNA3 (mock) vectors. Anti-myc or anti-HA (used as a negative control), IPs were performed 48 hpt. WBs were also performed as indicated. The WB signals were further quantified and the enrichment of co-immunoprecipitated PFV-1 GAG was calculated in each case using the following formula: [(GAG signal in pellet /GAG signal in input)/ AGO2/PAZ9 signal in pellet] × 100. (C) PFV-1 GAG mutated in the GRI domain, which is involved in nucleic acid interaction, was constructed in the pcgp1 background (GAGΔGRI). 293T cells were transfected with the mutated PFV-1 GAG and the myc-AGO2 expression vectors. Anti-myc IPs were performed 48 hpt. Anti-PFV-1 WBs were performed on immunoprecipitated fractions (pellet) and cell extracts (input). An anti-myc WB was also performed to control IP efficiency (bottom). (D) 293T cells were transfected with PFV-1 EGFP-GAG. IPs directed against EGFP-GFP were performed 48 hpt. Immunoprecipitated fractions (pellet) and cell lysates (input) were analyzed by WB as indicated.
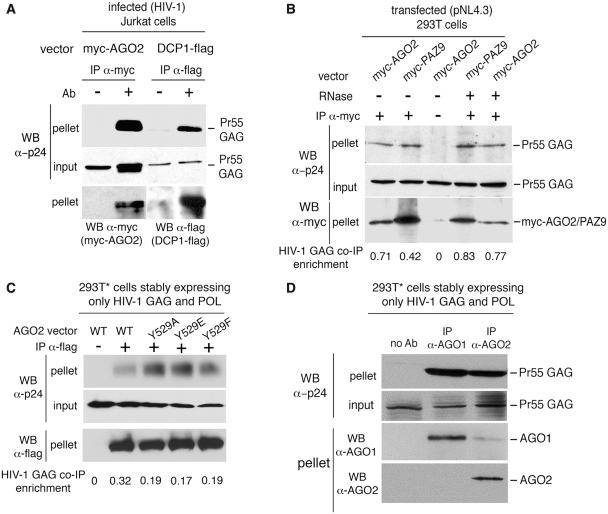
Similarly, we found that the HIV-1 GAG precursor Pr55 interacted with tagged DCP1 and AGO2, independently of RNAs, miRNAs and P-bodies, in infected Jurkat T-cells (Figure 3A) as well as in transfected 293T cells (Figure 3B). In agreement with these observations, we also showed that HIV-1 Pr55 GAG interacted with different AGO2 mutants (Figure 3C), which exhibit distinct miRNA binding capacities and P-body localization (31), in 293T* cells stably expressing only HIV-1 GAG and POL (35,36). The HIV-1 GAG also co-immunoprecipitated with endogenous AGO1 and AGO2 in 293T* cells (Figure 3D). We did not, however, detect any interaction between HIV-1 GAG and AGO2 using yeast two-hybrid assays suggesting that this interaction is indirect and requires additional cellular co-factors (data not shown).
Figure 3.
HIV-1 GAG interacts with AGO2 independently of miRNAs and P-bodies. (A) Jurkat cells were transfected with myc-AGO2 or DCP1-flag vectors and further infected with HIV-1. Anti-myc or anti-flag IPs were performed 48 hpt. IP fractions (pellet) or cell lysates (input) were analyzed by WBs. (B) Similar experiments were conducted in 293T cells transfected with pNL4.3 and myc-AGO2 or myc-PAZ9 vectors. RNase treatment was performed as described in Figure 2A. The WB signals were further quantified and the enrichment of co-immunoprecipitated HIV-1 GAG was calculated in each case using the following formula: [(GAG signal in pellet/GAG signal in input)/ AGO2/PAZ9 signal in pellet] × 100. (C) 293T* cells were transfected with Flag/HA-tagged versions of AGO2 wild-type (FH-AGO-WT) or mutated at position 529 (Y529A, Y529E and Y529F). Anti-flag IPs were performed 48 hpt. WBs were executed as indicated on IP fractions (pellet) or cell lysates (input). (D) IPs directed against endogenous AGO1 and AGO2 were performed in 293T* cells. WBs were executed as indicated on IP fractions (pellet) or cell lysates (input).
Retroviral GAG proteins elicit the recognition of viral mRNAs by AGO2 but do not trigger RNA silencing
Since GAG proteins bind viral RNAs through the encapsidation signals spanning intronic sequences (30,37), we hypothesized that GAG could recruit the RNAi machinery (notably DCP1 and AGO2) to viral RNAs through RNA packaging sequences. We indeed observed that the amount of PFV-1 RNAs interacting with DCP1 paralleled the expression of the cognate GAG protein (Supplementary Figure S1). Thus, to directly test the potential involvement of the RNA packaging signals, we constructed a Renilla luciferase reporter containing the PFV-1 encapsidation sequences (30) (Figure 4A). This reporter was transfected into 293T cells together with the myc-AGO2 expression vector. Anti-myc immunoprecipitations followed by RT–PCR amplifying the Renilla mRNA showed that AGO2 alone was not able to interact with the reporter RNA (Figure 4A). In contrast, in the presence of PFV-1 GAG, AGO2 bound the Renilla reporter containing the PFV-1 packaging signals (Figure 4A). As controls, we repeated these experiments with a Renilla reporter devoid of encapsidation sequences or a reporter containing the HIV-1 packaging signal. We observed that AGO2 did not bind these reporter RNAs, even in the presence of PFV-1 GAG (Figure 4A). Similarly, we did not detect a significant enrichment of GAPDH mRNA in the IP fractions (Figure 4A). These results indicated that PFV-1 GAG specifically elicited the recognition of PFV-1 mRNAs by AGO2 through packaging sequences. We then repeated these experiments with HIV-1 packaging signals. Similarly, we observed that AGO2 bound only the Renilla reporter containing the HIV-1 packaging signals in the presence of HIV-1 GAG (Figure 4D). We verified that HIV-1 GAG was not able to recruit AGO2 to the Renilla reporter containing the PFV-1 encapsidation sequences (Figure 4D). Together with the results shown in Figure 4A, this result indicated that the recruitment of AGO2 through GAG requires the cognate RNA packaging sequences.
Figure 4.
The retrovirus core GAG protein recruits AGO2 on viral RNAs without eliciting RNA silencing. (A) 293T cells were transfected with the empty psiCHECK2 vector or a psiCHECK2 containing the encapsidation sequences of PFV-1 (PFV-1 ψ), as well as myc-AGO2 and PFV-1 GAG expressing vectors as indicated. IPs directed against the myc tag were performed 48 hpt. RNAs extracted from IP fractions were assayed for the presence of the Renilla RNA by RT–qPCR. Fold enrichment was calculated using the following formula: 2^(Ct input-Ct IP) – (Ct input obtained in the no Ab condition – Ct IP obtained in the no Ab condition). As control, a reporter containing the HIV-1 encapsidation sequences (psiCHECK2 HIV-1 ψ) was also tested. The RNAs extracted from myc-AGO2 IPs in the presence or absence of PFV-1 GAG were also assayed for the presence of the GAPDH mRNA by RT–qPCR. Surprisingly, although several miRNAs are predicted to target the GAPDH mRNA (according to miRBase, http://www.mirbase.org/), no RNA enrichment was observed in the myc-AGO2 IP fraction. Anti-myc WBs were also performed to control IP efficiency (bottom, one representative example is shown). (B) 293T cells were transfected with the psiCHECK2 vector or PFV-1 ψ and increasing amount of PFV-1 GAG. Dual-luciferase assays were performed 48 hpt. The results were normalized with the values obtained with each construct in the absence of GAG (Renilla/Firefly ratio or Renilla/Firefly ratio obtained in the absence of GAG). (C) 293T cells were transfected as in (B) and RT–qPCRs directed against Renilla and Firefly luciferases were performed 48 hpt. The ratio Renilla/Firefly was calculated and the values obtained in the absence of GAG were used as reference. (D) 293T cells (devoid of HIV-1 GAG) and 293T* cells (stably expressing HIV-1 GAG-POL) were transfected with the empty psiCHECK2 vector or a psiCHECK2 containing the encapsidation sequences of HIV-1 (HIV-1 ψ) as well as myc-AGO2 as indicated. As control, the reporter described in (A) was also tested. IPs directed against the myc tag were realized 48 hpt. RNAs extracted from immunoprecipitated fractions were assayed for the presence of the renilla RNA by RT–qPCR. Anti-myc WBs were also performed to control IP efficiency (bottom, one representative example is shown). (E) 293T and 293T* cells were transfected with the psiCHECK2 vector or the HIV-1 ψ and Dual-luciferase assays were performed 48 hpt. The results were normalized with the values obtained with each construct in the absence of GAG (Renilla/Firefly ratio or Renilla/Firefly ratio obtained in the absence of GAG). (F) 293T and 293T* cells were transfected with the psiCHECK2 vector or the HIV-1 ψ and RT–qPCRs were performed 48 hpt as in (C).
We then assessed the effect of the GAG-recruited AGO2 on RNA stability and translation using the reporters described in Figure 4A and D. We observed that tethering AGO2 to these reporters via GAG did not inhibit translation (Figure 4B and E) nor affect RNA stability (Figure 4C and F). Accordingly, the expression of PFV-1 GAG did not affect siRNA-triggered RNA interference directed against the luciferase (Supplementary Figure S2). Hence, we concluded that retroviral GAG hijacked AGO2 functions that are not related to RNA silencing and miRNAs.
AGO2 is encapsidated in retroviral particles
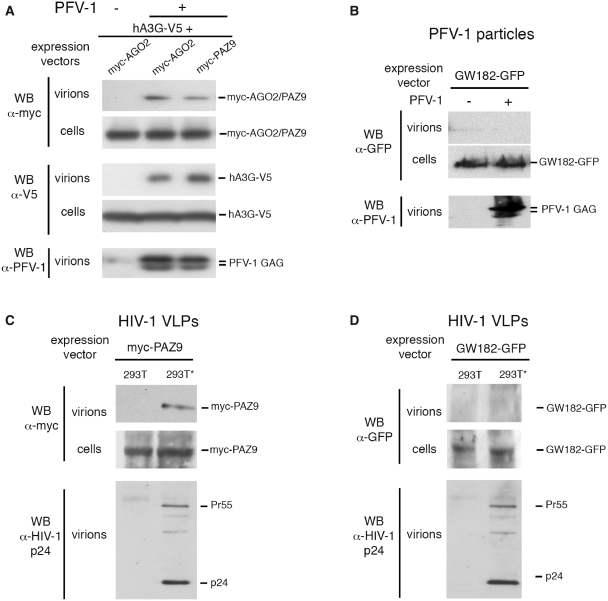
We showed PFV-1 and HIV-1 GAGs specifically elicited the recognition of viral mRNAs by AGO2 through RNA packaging sequences (Figure 4A and D). We thus presumed that AGO2 was encapsidated in retroviral particles. To test this idea, we purified PFV-1 virions produced from 293T cells transfected with the PFV-1 provirus and vectors expressing AGO2-myc or APOBEC3G-V5 (hA3G-V5), used as a positive control (38,39). Both AGO2-myc and APOBEC3G-V5 proteins were detected in PFV-1 particles, whereas no protein was detectable in the supernatant of uninfected cells (Figure 5A). The AGO2 mutant PAZ9 was similarly encapsidated (Figure 5A), suggesting that the encapsidation of AGO2 occurs in a manner independent of P-bodies and miRNAs. In similar settings, we failed to detect GW182-EGFP in PFV-1 particles (Figure 5B). We also tested whether HIV-1 particles likewise encapsidated AGO2. For this purpose, 293T* cells expressing HIV-1 GAG-POL (35,36) and unmodified 293T cells (used as negative control) were transfected with the PAZ9-myc vector (Figure 5C). The PAZ9-myc protein was only detected in the supernatant of 293T* (Figure 5C), indicating that HIV-1 virus-like particles (VLPs) are able to encapsidate AGO2 independently of miRNAs and P-bodies. These results were consistent with the interaction existing between AGO2 and the HIV-1 Pr55 GAG precursor (Figure 3), whose maturation occurs after viral budding (40). As observed in the case of PFV-1, the GW182-EGFP protein was not detected in HIV-1 VLPs (Figure 5D), indicating that not all miRNA-related proteins are packed in retroviral particles.
Figure 5.
AGO2 is encapsidated in retroviral particles. (A) 293T cells were transfected with the PFV-1 provirus and hA3G-V5, myc-AGO2 or myc-PAZ9 vector. hA3G-V5 was used as a positive control (38,39). PFV-1 virions were purified on a 20% sucrose cushion 48 hpt and assayed for the presence of myc-AGO2/PAZ9 and hA3G-V5 by WBs. (B) The experiments described in (A) were repeated except that 293T cells were transfected with the PFV-1 provirus and a GW182-EGFP vector. (C) 293T* and 293T cells were transfected with the myc-PAZ9 expression vector. The culture supernatants were purified on a sucrose cushion 48 hpt and assayed for the presence of myc-PAZ9 by WB. (D) The experiments described in (C) were repeated using a GW182-EGFP vector.
AGO2 is required for retroviral replication
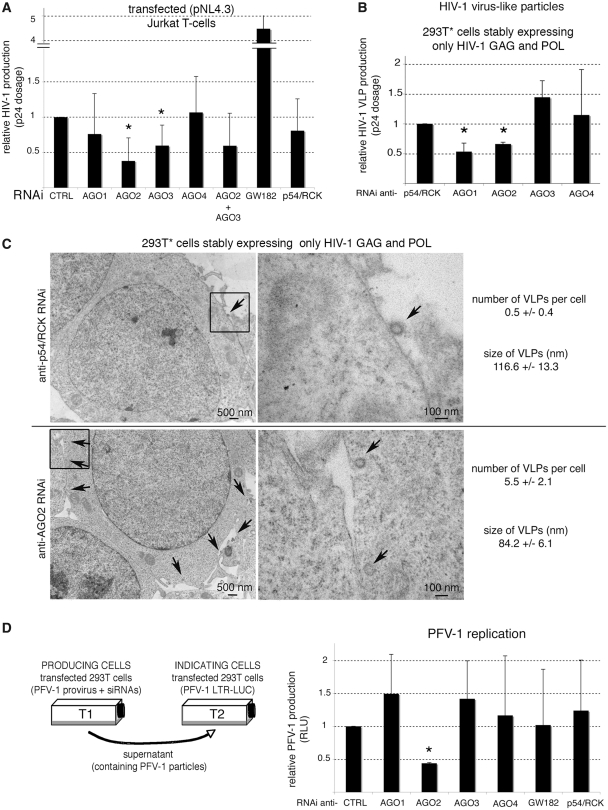
We next questioned the functional consequences of GAG-mediated AGO2 recruitment. We silenced AGO2, as well as AGO1, AGO3, AGO4, GW182 and p54/RCK (also known as DEAD (Asp-Glu-Ala-Asp) box polypeptide 6, DDX6) (Supplementary Figure S3) and measured HIV-1 particle production in Jurkat T-cells (Figure 6A). We observed that siRNAs directed against AGO2 and AGO3 significantly decreased HIV-1 particle production (Figure 6A). Conversely, RNAi directed against GW182 increased viral replication (Figure 6A), as previously reported (19,22). It is noteworthy that siRNAs anti-AGO2 and anti-AGO3 did not synergize to limit HIV-1 replication (Figure 6A). These experiments suggested that AGO2, and possibly AGO3 (41), as opposed to AGO1 and AGO4, play positive functions in HIV-1 replication. Next, we asked whether AGO2 could directly impact retroviral capsid formation and measured HIV-1 VLP formation in 293T* cells (35,36) transfected with siRNAs directed against several RNAi components (Figure 6B). We observed that anti-AGO2 siRNAs significantly decreased VLP formation compared with other siRNAs (Figure 6B), revealing positive functions of AGO2 in HIV-1 capsid assembly. Of note, AGO3 siRNAs, on the other hand, decreased HIV-1 replication [Figure 5A and (41)] but did not affect VLP formation (Figure 6B). Conversely, AGO1 was not significantly implicated in HIV-1 virion production (Figure 6A) but anti-AGO1 RNAi affected VLP production (Figure 6B). These results might be indicative of AGO3 and AGO1 cellular functions that might indirectly impact replication and capsid formation respectively. Using electron microscopy, we further examined HIV-1 VLPs in 293T* cells transfected with siRNAs directed against AGO2 or, as a control, p54/RCK (Figure 6C). These analyses showed that, upon AGO2 RNAi, HIV-1 VLPs were retained in the cytoplasm and were smaller and less dense, compared to VLPs observed upon p54/RCK RNAi (Figure 6C). Together these observations confirmed that AGO2 is required for HIV-1 replication, in particular for the assembly of viral particles.
Figure 6.
AGO2 is required for HIV-1 and PFV-1 replication. (A) Jurkat cells were transfected with the HIV-1 provirus and indicated siRNAs. Viral production was measured 48 hpt using anti-p24 ELISA assays. Results are the mean of at least three independent experiments. *P < 0.01 (Student's test). (B) HIV-1 VLP production from 293T* cells was measured 48 hpt of the indicated siRNAs using anti-p24 ELISA assays. Results are the mean of three independent experiments. *P < 0.005 (Student's test). (C) Electron microscopy of HIV-1 VLPs produced in 293T* transfected with siRNA directed against AGO2 or p54/RCK, as a control. The size and the number of VLPs per cell are indicated in each condition. Arrows show VLPs. Right panels are enlargements of squares indicated in left panels. (D) 293T cells were transfected with the PFV-1 provirus and indicated siRNAs (T1). Separate cells were transfected with Firefly luciferase driven by the PFV-1 Long Terminal Repeat (LTR), activated by the transactivator Tas (T2). Supernatants of T1 were used to infect cells from T2. Luciferase expression was quantified in T2 cells. *P < 0.005 (Student's test).
Similar experiments were also performed in the case of PFV-1. In contrast to orthoretroviruses, PFV-1 GAG proteins expressed on their own do not form VLPs (42). The replication of PFV-1 was measured as described in ref. (12). While RNAi anti-AGO1, -AGO3, -AGO4, -GW182 or -p54/RCK did not significantly impair PFV-1 replication in 293T cells, siRNAs directed against AGO2 consistently diminished virus production (Figure 6D). Hence, in accordance with our initial hypothesis, AGO2, as opposed to other AGOs, seems to play positive functions in both PFV-1 and HIV-1 replications, a scenario akin to HCV (27,28).
DISCUSSION
Here, we show that retroviral GAG and the GAG-interacting RNA packaging signals can recruit AGO2 onto RNA without eliciting translation repression. However, our results also confirm an important contribution of host miRNAs in the interaction of AGO2 with retroviral RNAs (12,17–19,22). In fact, the majority of HIV-1 RNAs requires host miRNAs to interact with AGO2: 70% of unspliced and 98% of spliced HIV-1 RNAs interact with AGO2 in a miRNA-dependent manner (Figure 1C). Thus, we can now distinguish at least two ways to recruit AGO2 on retroviral mRNAs: one elicited by host miRNAs (12,17–19,22) and a second, mediated by GAG and the RNA packaging sequences. These two types of interaction are not exclusive and are probably implicated in distinct steps of the retroviral life cycle. As viruses have co-evolved with the miRNA repertoire of their hosts (14,17,43,44), the first mode, that is dependent on host miRNAs, could have an impact on retroviral replication: for instance, at a particular time point [e.g. latent infection (17)], in specific cells [e.g. resting cells (17)] and/or with certain RNAs (e.g. spliced RNAs, Figure 1C). On the other hand, mRNA recognition by miRNAs [Figure 1 and (12,18,19,22,45)], interaction with other RNAi proteins [such as DCP1 (Figures 1A and 3A) or AGO1 (Figures 2D and 3C)] and sequestration in P-bodies (19,22) could also represent deleterious consequences of the recruitment of AGO2 or other miRNA-related components on viral RNAs. PFV-1 and HIV-1 could have therefore developed protein- or RNA-based strategies to limit the negative effects of cellular miRNAs (12,46–49). Interestingly, host miRNAs also play both beneficial and detrimental roles in HCV replication (13,14,16) and AGO2 was recently shown to be required for efficient HCV replication (27,28). Similar to the retroviral GAG protein, the miR-122 recruits an AGO2-containing complex onto viral mRNAs (27,28) with unclear consequences: miR-122 is able to stimulate HCV translation (50,51) but this effect is not sufficient to fully explain its actions on HCV replication (14,52). Recently, and in accordance with our results, the replication of HCV RNA was shown to depend on recruitment of AGO2 and miR-122 to lipid droplets, not P-bodies, while suppression of HCV RNA by siRNA and AGO2 involves interaction with P-bodies (53). It has been postulated that the miR-122 interaction with HCV RNA changes during the viral life cycle (28), as hypothesized here in the case of the interactions between retroviral RNAs and AGO2. Hence, it is possible that HCV and retroviruses similarly hijack some AGO2 functions that are not related to translation regulation. Provided, the considerable differences existing between Retroviridae and Flaviviridae, it is tempting to speculate that these AGO2 functions are also implicated in the replication of other viruses. The experiments showing that vaccinia, influenza A or encephalomyocarditis viruses are not affected by the blockade of miRNA biogenesis (15) do not exclude an authentic contribution of some RNAi-related proteins, in particular AGO2, in their replication.
Taken together, our results shed a new light on the cellular functions of AGO2 as it can be recruited onto messenger RNAs without eliciting RNA silencing. Our results support the idea that AGO2 has original functions that are not related to miRNAs and RNA silencing (54). In fact, AGO2 has previously been found in specific protein complexes that are not linked to miRNA biogenesis or RNA interference (54). We anticipate that these functions will be deciphered by further studies on the AGO2-containing complex(es) implicated in viral replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS, National Agency for AIDS Research (ANRS #2007/290 and #2008/061); Sidaction (#AI18-3-01372); M.B. is recipient of a PhD fellowship from ANRS (to M.B.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M.-C. Robert for technical help and T. Vasselon, R. Bordonné, I. Robbins and M. Sitbon for critical reading of the article. We are grateful to M. Benkirane, W. Filipowicz, G. Meister, A. Rethwilm, A. Saïb, O. Schwartz, D. Weil and the NIH AIDS Reference and Reagent Program for reagents.
REFERENCES
- 1.Saumet A, Lecellier CH. Anti-viral RNA silencing: do we look like plants? Retrovirology. 2006;3:3. doi: 10.1186/1742-4690-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strebel K, Luban J, Jeang KT. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 11.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 13.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 15.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 18.Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song L, Liu H, Gao S, Jiang W, Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010;84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly EJ, Hadac EM, Cullen BR, Russell SJ. MicroRNA antagonism of the picornaviral life cycle: alternative mechanisms of interference. PLoS Pathog. 2010;6:e1000820. doi: 10.1371/journal.ppat.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, Jacquet JM, Reynes J, Levy Y, Saib A, et al. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffin J, Hughes S, Varmus H. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 25.Llorens C, Fares MA, Moya A. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol. Biol. 2008;8:276. doi: 10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecellier CH, Saib A. Foamy viruses: between retroviruses and pararetroviruses. Virology. 2000;271:1–8. doi: 10.1006/viro.2000.0216. [DOI] [PubMed] [Google Scholar]
- 27.Wilson JA, Zhang C, Huys A, Richardson CD. Human Ago2 is required for efficient miR-122 regulation of HCV RNA accumulation and translation. J. Virol. 2011;85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the Pol region required to transfer human foamy virus vectors. J. Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 2005;280:27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 34.Stenbak CR, Linial ML. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 2004;78:9423–9430. doi: 10.1128/JVI.78.17.9423-9430.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda Y, Takeuchi Y, Martin F, Cosset FL, Mitrophanous K, Collins M. Continuous high-titer HIV-1 vector production. Nat. Biotechnol. 2003;21:569–572. doi: 10.1038/nbt815. [DOI] [PubMed] [Google Scholar]
- 36.Molle D, Segura-Morales C, Camus G, Berlioz-Torrent C, Kjems J, Basyuk E, Bertrand E. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J. Biol. Chem. 2009;284:19727–19743. doi: 10.1074/jbc.M109.019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Souza V, Summers MF. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 38.Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 42.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Muller JG, Rethwilm A. Foamy virus particle formation. J. Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y, Kishi A, Yachie N, Kanai A, Tomita M. Computational analysis of microRNA-mediated antiviral defense in humans. FEBS Lett. 2007;581:4603–4610. doi: 10.1016/j.febslet.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Quintero AL, Neme R, Zapata A, Lopez C. Plant microRNAs and their role in defense against viruses: a bioinformatics approach. BMC Plant Biol. 2010;10:138. doi: 10.1186/1471-2229-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007;35:D381–D385. doi: 10.1093/nar/gkl1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 48.Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J. Biol. Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 49.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diaz-Toledano R, Ariza-Mateos A, Birk A, Martinez-Garcia B, Gomez J. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5′ region of hepatitis C virus RNA. Nucleic Acids Res. 2009;37:5498–5510. doi: 10.1093/nar/gkp553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berezhna SY, Supekova L, Sever MJ, Schultz PG, Deniz AA. Dual regulation of hepatitis C viral RNA by cellular RNAi requires partitioning of Ago2 to lipid droplets and P-bodies. RNA. 2011 doi: 10.1261/rna.2523911. first published online August 25 (doi:10.1261/rna.2523911; Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.