Abstract
The delivery of Met-tRNAi to the 40S ribosomal subunit is thought to occur by way of a ternary complex (TC) comprising eIF2, GTP and Met-tRNAi. We have generated from purified human proteins a stable multifactor complex (MFC) comprising eIF1, eIF2, eIF3 and eIF5, similar to the MFC reported in yeast and plants. A human MFC free of the ribosome also is detected in HeLa cells and rabbit reticulocytes, indicating that it exists in vivo. In vitro, the MFC-GTP binds Met-tRNAi and delivers the tRNA to the ribosome at the same rate as the TC. However, MFC-GDP shows a greatly reduced affinity to Met-tRNAi compared to that for eIF2-GDP, suggesting that MFC components may play a role in the release of eIF2-GDP from the ribosome following AUG recognition. Since an MFC–Met-tRNAi complex is detected in cell lysates, it may be responsible for Met-tRNAi–40S ribosome binding in vivo, possibly together with the TC. However, the MFC protein components also bind individually to 40S ribosomes, creating the possibility that Met-tRNAi might bind directly to such 40S-factor complexes. Thus, three distinct pathways for Met-tRNAi delivery to the 40S ribosomal subunit are identified, but which one predominates in vivo remains to be elucidated.
INTRODUCTION
Recruitment and positioning of the initiator tRNA on the small ribosomal subunit is required to initiate protein synthesis. In eukaryotes, it is generally believed that the initiator tRNA is recruited to the 40S subunit surface as an eIF2–GTP–Met-tRNAi ternary complex (TC). The TC is further stabilized on the 40S subunit surface by the presence of eIF1, eIF1A, eIF3 and eIF5. The resulting 43S pre-initiation complex then stably associates with an mRNA via interactions with the cap-binding complex located at the 5′ end of the mRNA. During scanning and recognition of the start codon, eIF2–GTP is hydrolyzed in a reaction promoted by the GTPase-activating protein, eIF5. GTP hydrolysis is completed by phosphate release promoted by eIF1 dissociation upon AUG recognition. Then, eIF2 is released as a GDP form, leaving the Met-tRNAi in the P-site of the ribosome. Finally, an 80S initiation complex is formed by joining of a 60S ribosomal subunit and is ready to synthesize the encoded protein. After the release of eIF2–GDP from the ribosome, eIF2 is recycled to its GTP form by the guanine nucleotide exchange factor eIF2B and participates in another round of initiation (1, 2).
Studies in Saccharomyces cerevisiae have shown that eIF1, eIF2, eIF3 and eIF5 can form a stable multi-factor complex (MFC) in the absence of 40S subunits in vitro and in vivo by pull-down and sucrose gradient centrifugation assays (3). The ribosome-free MFC detected in cell lysates also was found to contain Met-tRNAi. Disruption of interactions between MFC components inhibits protein synthesis, implying a functional role for the formation of the MFC during initiation. Extensive studies have determined the precise nature of the interactions between yeast MFC components (3–8). Specifically, affinity tag pull-down assays indicate that eIF1, eIF3c and eIF5 form stable interactions with each other, and eIF5 bridges an interaction between eIF2 and eIF3 (3). An additional contact between eIF3a and eIF2β has been identified (9). Interaction domains within these components have been identified and shown to form a minimal core complex in vitro (3,8). Importantly, eIF5 is recognized as the central component in forming the MFC in S. cerevisiae (8). Attempts to study the MFC in other systems have generated mixed results. The MFC has been identified by pull-down assays using plant proteins (10), while studies using mammalian components have identified only eIF5–eIF2β and eIF1–eIF3c interactions in vitro (11,12).
It has been firmly established that interactions between MFC components are important for protein synthesis initiation in yeast. It is unknown, however, whether the MFC forms prior to 40S subunit binding or if it forms on the surface of the 40S subunit in an ordered or random fashion. It is possible that a pre-formed MFC functions to deliver the initiator tRNA to the 40S subunit, but this has never been tested. Moreover, whether an MFC can form in mammalian cells is currently unknown. Using a native gel electrophoresis assay, we have identified the presence of a stable MFC that exists independently of the 40S subunit in mammalian cell lysates. Using highly purified components, we have been able to successfully reconstitute the human MFC in vitro, confirming that the MFC is conserved throughout evolution. Using our purified system, we have investigated the role of a pre-formed human MFC in delivering the initiator tRNA to the 40S subunit. Interestingly, we show that eIF2 is able to bind to 40S subunits in the absence or presence of other MFC components, creating the possibility that Met-tRNAi delivery to the ribosome is direct rather than through the TC. Thus, the findings suggest three possible pathways for Met-tRNAi–40S binding: TC-mediated; MFC-mediated; and direct.
MATERIALS AND METHODS
Preparation of proteins, ribosomes, tRNA and cell lysates
Endogenous eIF2, eIF3 and 40S ribosomal subunits were purified from HeLa cell extracts as described previously (13,14). Human initiator tRNAMet was in vitro transcribed by T7 RNA polymerase and charged with either non-labeled or 35S-labeled methionine using Escherichia coli methionyl-tRNA synthetase as described previously (15). The [35S]Met-tRNAi (15 Ci/mmol) was stored at −80°C in small aliquots with buffer containing 10 mM K acetate pH 5.2, 2 mM MgCl2 and 1 mM DTT. All recombinant proteins used here (human eIF1, eIF1A and eIF5) were expressed as a fusion with a His-tag, maltose binding protein (MBP) and TEV protease cleavage site on the N-terminus by using modified pET-28c (Novagen) as previously described for eIF1 and eIF1A (15). Recombinant eIF5 was expressed and purified by similar methods, as described in detail in Supplementary Data. Rabbit reticulocyte lysate was from Green Hectares. HeLa cells were grown in 10 ml Dulbecco's modified Eagle's medium (DMEM) media in a 37°C CO2 incubator for ∼60 h (∼70% confluence). Before harvesting, the cells were incubated with fresh media for 20 min to stimulate translation, treated with 100 µg/ml cycloheximide for 3 min and kept on ice. The cells were washed twice with phosphate-buffered saline (PBS), and resuspended in 50 µl buffer containing 20 mM Tris–HCl pH 7.5, 80 mM KCl, 1 mM DTT, 2 mM MgCl2, 5% glycerol, 0.1% Triton-X and protease inhibitor cocktail (Roche), and lysed by vortexing for 10 min at 4°C. The supernatant after centrifugation at 30 000g for 15 min was stored in small aliquots at −80°C.
Native gel electrophoresis
Binary and higher order protein complexes were formed by mixing various combinations of eIF3, eIF2, eIF1 and/or eIF5 at 37°C for 10 min in 5 µl reaction buffer (20 mM Tris–HCl pH 8.1, 80 mM KCl, 2 mM DTT, 2 mM MgCl2 and 5% glycerol). To test the effect of nucleotides or Met-tRNAi, eIF2 was incubated with 1 mM GDPNP or GDP at 37°C for 5 min prior to the addition of magnesium and 15 pmol of Met-tRNAi as indicated in the figure legends. Clarified HeLa cell lysates (10 µl, 0.5 OD260 units) and rabbit reticulocyte lysates (5 µl, 0.4 OD260 unit) were directly used for the analyses without any further treatment. Reaction mixtures were loaded onto a 4% native polyacrylamide gel prepared in 10 cm × 8 cm glass slabs with electrophoresis buffer (50 mM Tris-borate pH 8.1) supplemented with 2 mM MgCl2, and run at 125 V for 1.5–4 h at 4°C. A wide (12 mm) sample comb was used to obtain better resolution. Gels were stained with GelCode Blue stain (Thermo Scientific), or analyzed by SDS–PAGE, western blotting, or reverse transcription-polymerase chain reaction (RT-PCR). For sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis, the native gel was negatively stained with Zinc-imidazole (16). Each band was excised, finely disrupted and shaken overnight at 4°C in 200 µl extraction buffer (25 mM Tris, 250 mM glycine and 0.1% SDS). Each band extract was filtered through a Spin Cups paper filter (Pierce), TCA precipitated and analyzed on an EZ-Run SDS gel (Fisher Scientific). Due to its small size, eIF1 is not detected by Coomassie staining, but its presence in the MFC band was confirmed by western blotting with anti-eIF1 specific to its N-terminus (Santa Cruz Biotechnology). The blots were imaged by using ECL plus/ECL Advance (GE Healthcare) detection reagents and a fluorescent scanner/CCD camera.
RT-PCR
The native gels of HeLa and rabbit reticulocyte lysates were sliced, disrupted, extracted and subjected to RT-PCR as described in detail in Supplementary Data.
Met-tRNAi filter binding assay
The nitrocellulose filter binding assay was done essentially as described previously (17) (see Supplementary Data).
Met-tRNAi–40S ribosomal subunit binding rates
In the TC-mediated Met-tRNAi delivery assay, a TC mixture (20 µl) was prepared by incubating 12 pmol of eIF2 with 1 mM GDPNP at 30°C for 5 min, followed by the addition of 24 pmol of [35S]Met-tRNAi for 10 min at 30°C in reaction buffer (20 mM Tris–HCl pH 7.0, 80 mM KCl, 2 mM MgCl2, 2 mM DTT and 5% glycerol). A 40S ribosome mixture was prepared separately by incubating 12 pmol of eIF1, eIF1A, eIF5, eIF3 and 40S ribosomal subunits in reaction buffer in a total volume of 20 µl. The two mixtures were further incubated separately at 25°C for 10 min, then mixed together at 25°C to initiate the reaction. The reaction was stopped at various time points by loading a 4.5-µl aliquot onto a 2% polyacrylamide/0.5% agarose composite native gel (running at 125 V at 0°C) made with electrophoresis buffer (50 mM Tris-borate pH 8.1) supplemented with 2 mM MgCl2 (18). As judged by the mobility of a xylene cyanol marker dye, which shows a mobility similar to that of the 40S ribosome under these conditions, it takes 30–40 s for the complex to enter the gel after loading, suggesting that the time resolution of this assay is at the level of ∼1 min. Under these conditions, spontaneous deacylation of free Met-tRNAi was measured to be <20% after 40 min. Electrophoresis continued for another 40 min after loading the final time point (40 min). The gel was then fixed by soaking in 10% TCA for 15 min at 4°C, dried on a polyvinylidene difluoride (PVDF) membrane with a vacuum gel dryer at 4°C for 1 h, heat dried for an additional 30 min and exposed to a phosphor screen overnight. Because Met-tRNAi is easily deacylated and diffuses when heating such a soft gel, consistent results were obtained by vacuum drying the gel at 4°C. The amount of Met-tRNAi bound to 40S ribosomes was estimated by the intensity of the radioactive band corresponding to the 43S complex.
In the MFC-mediated delivery assay, the MFC mixture was prepared as in the TC-mediated assay by combining 12 pmol of eIF1, eIF2, eIF3, eIF5, 1 mM GDPNP and 24 pmol [35S]Met-tRNAi, while 40S ribosomal subunits were mixed with eIF1A. The two mixtures were combined, incubated and analyzed as described above.
In the direct binding assay, 40S subunits were mixed with all the initiation factors involved and incubated for 10 min, and then [35S]Met-tRNAi was added to initiate the reaction.
GDP off-rate assay
eIF2 (18 pmol) was incubated with 0.24 µCi [3H]GDP (45 Ci/mmol, Perkin Elmer) in reaction buffer (20 mM HEPES pH 7.5, 80 mM KCl, 1 mM DTT, 1 mg/ml creatine phosphokinase and 5% glycerol) without magnesium at 37°C for 10 min, and then incubated a further 10 min at 37°C with 1 mM MgCl2 and with or without 30 pmol eIF5 in a total volume of 60 µl. The reaction was initiated by adding 6000 pmol unlabeled GDP (>1000 times excess) as a chase, and further incubated at 37°C. At each time point, an aliquot (8 µl) was injected into 300 µl ice-cold stop buffer (reaction buffer supplemented with 5 mM MgCl2), filtered immediately through an HAWP nitrocellulose membrane and washed twice with 1 ml ice-cold stop buffer. The radioactivity on the dried membranes was measured with a liquid scintillation counter. The resulting GDP dissociation curves were fitted to a first order exponential decay to calculate the off rates.
Factor binding to 40S ribosomes
For testing binding to 40S ribosomal subunits, reaction mixtures were prepared essentially as described above by using 2 pmol of 40S, 6 pmol of eIF1A and 6 pmol of the other initiation factors in a total volume of 10 µl. Again, 1 mM GDPNP or GDP, and 12 pmol Met-tRNAi were included, where indicated, in the same manner as above. Reaction mixtures were incubated for 10 min at 37°C, loaded onto a 2% polyacrylamide/0.5% agarose composite native gel prepared with electrophoresis buffer (50 mM Tris-borate pH 8.1) supplemented with 2 mM MgCl2 (18) and run at 125 V for 70 min at 4°C. The resulting gel was stained by ethidium bromide and further analyzed by western blotting.
RESULTS
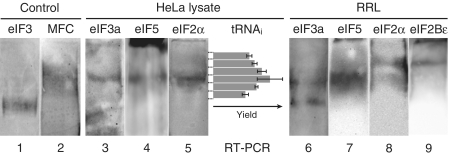
The MFC is present in HeLa cell and rabbit reticulocyte lysates
To test whether an MFC might exist independently of ribosomes in mammalian cells, a HeLa cell lysate was subjected to centrifugation to pellet nuclei and mitochondria, then analyzed by native PAGE and immunoblotting with anti-eIF3a antiserum. Using purified eIF3 as a marker (Figure 1, lane 1), the lysate clearly generates an immunoreactive band whose migration is significantly slower than that of eIF3 (lane 3). Since ribosomes and their complexes do not enter the gel under the conditions used, the retardation of the anti-eIF3a reactive band implies the presence of a ribosome-free complex in the lysate. Unexpectedly, it appears that all of the eIF3 that exists free of ribosomes in the lysate resides in this complex, as no band corresponding to uncomplexed eIF3 is detected. Immunoblotting with eIF5- and eIF2α-specific antisera (lanes 4 and 5) confirms their presence in the complex. To determine if the complex also contains initiator tRNAi, the region of the gel containing the complex was sliced horizontally into pieces, extracted and subjected to RT-PCR to quantitate tRNAi levels as described in ‘Materials and Methods’ section. The yield of the specific product with each gel slice correlates well with the position of the complex in the gel (Figure 1), indicating that the complex contains Met-tRNAi presumably bound to eIF2–GTP. These results suggest that an MFC exists in human cells free of ribosomes. A similar immunoblot analysis of a rabbit reticulocyte lysate with anti-eIF3a, anti-eIF5 and anti-eIF2α also detects a complex migrating with a mobility slower than eIF3 but similar to that seen for the MFC in HeLa cells (lanes 6–8). However, in this case, a substantial amount of eIF3 is also found to migrate as free eIF3; thus, the amount of MFC differs in different cell types and/or in different physiological states. It is interesting to note that in the reticulocyte analyses, a portion of eIF2 migrates more slowly than the MFC at a position corresponding to the band reactive to anti-eIF2Bε (lane 9), suggesting that this complex is eIF2–eIF2B. Elucidation of the level and importance of the eIF2–eIF2B complex and its possible dependence on eIF2 phosphorylation will require further experimentation.
Figure 1.
In vivo presence of the MFC shown by native gel electrophoresis of HeLa cell and rabbit reticulocyte lysates. Purified eIF3 (lane 1), reconstituted MFC with Met-tRNAi (lane 2), HeLa cell lysate (lanes 3–5) and rabbit reticulocyte lysate (lanes 6–9) were subjected to native gel electrophoresis as described in ‘Materials and Methods’ secton, and analyzed by western immunoblotting with anti-eIF3a (lanes 1, 3 and 6), anti-eIF5 (lanes 2, 4 and 7), anti-eIF2α (lanes 5 and 8) or anti-eIF2Bε (lane 9) antiserum. Ribosomes and their complexes do not enter and stay at the top of the gel and thus are clearly separated from ribosome-free factors under the condition used here. For the tRNAi specific RT-PCR, the lane of HeLa cell lysate was horizontally sliced into pieces as shown by dashed lines, and their extracts were analyzed (see ‘Materials and Methods’ section). The yields of the RT-PCR product are the means of two independent experiments, and errors shown are standard errors.
Formation of an MFC with purified human initiation factors
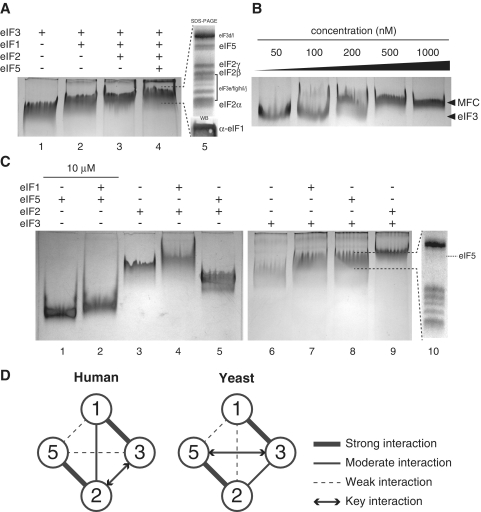
To determine more convincingly if a human MFC comprising eIF1, eIF2, eIF3 and eIF5 might form, we constructed such a complex in vitro using purified human factors. Endogenous eIF2 and eIF3 were purified from HeLa cell cytosol, and recombinant eIF1 and eIF5 were expressed and isolated from bacteria, as described in ‘Materials and Methods’ section. Different combinations of these purified factors were incubated together, followed by analysis using native PAGE to detect their interactions. Since eIF3 is the largest factor among the MFC components (800 kDa), we initially tested whether adding the other factors induces an eIF3 mobility shift, indicative of complex formation. Adding eIF1, eIF2 and eIF5 sequentially to eIF3 results in a clear mobility shift at each step (lanes 1–4 in Figure 2A). The shifted band in lane 4 was excised and subjected to SDS–PAGE and Coomassie blue staining; bands corresponding to eIF2 and eIF5 are seen in addition to those for eIF3, confirming their presence in stoichiometric amounts (lane 5). Western blotting of the shifted band in lane 4 with anti-eIF1 antiserum (lane 5, bottom) further confirms the presence of eIF1 in the shifted complex. The results clearly indicate that a human MFC comprising eIF1, eIF2, eIF3 and eIF5 can stably form in vitro. A dilution analysis indicates that the human MFC is quite stable at concentrations above 300 nM (Figure 2B), suggesting that it can assemble at physiological concentrations. Unexpectedly, the MFC forms in both the presence and absence of GTP/GDP and/or Met-tRNAi (data not shown). This implies that the formation of the MFC is not sensitive to the nucleotide bound state of eIF2.
Figure 2.
In vitro reconstitution of human MFC with purified components. (A) Native gel analysis of protein complexes formed by mixing (in a total of 5 µl) 5, 7.5, 10 and 10 pmol of eIF3, eIF2, eIF1 and eIF5, respectively, as indicated above the gel. Lane 5 shows the Coomassie-stained SDS gel image generated from the excised band in lane 4, where the bands corresponding to each factors are identified. The presence of eIF1 in the band from lane 4 was demonstrated by western immunoblotting of the native gel (lane 5, bottom). (B) Stability of the MFC shown by varying its concentration. MFC components in the amounts defined in (A), lane 4, were mixed in different volumes to generate diluted mixtures, with eIF3 concentrations ranging from 50 to 1000 nM. Following incubation at 25°C for 10 min, reactions were analyzed as in (A). (C) Binary interactions of MFC components shown by native gels. eIF1 (10 pmol), eIF2 (7.5 pmol), eIF3 (5 pmol) and eIF5 (10 pmol) in lanes 3–9 (except 50 pmol each in lanes 1 and 2) were combined in 5 µl of reaction buffer as indicated above the gels. Complex formation was detected by band shifts and/or diminishing of the monomer bands, and confirmed by SDS–PAGE/western immunoblotting. A representative SDS–PAGE analysis of the eIF3–eIF5 complex band is shown (lane 10) (the eIF5 band is identified). Note that a gel with eIF1 alone is not shown, as this non-complexed protein does not enter the gel under the conditions used here. (D) Component interaction models for the human and yeast MFC (this study and ref. 8). Each factor is indicated by circled numbers. Bold lines indicate stable interactions, thin solid lines indicate moderate interactions and dashed lines indicate less stable interactions. The key interaction line proposed to be important for MFC formation in each system is shown by arrows.
Importantly, when the in vitro–generated MFC–Met-tRNAi was analyzed together with cell lysates (Figure 1, lane 2), it exhibited the same mobility as the lysate complex containing eIF2, eIF3, eIF5 and Met-tRNAi. This strongly supports the conclusion that the ribosome-free complex seen in cell lysates is indeed an MFC containing four initiation factors and Met-tRNAi.
To study MFC assembly in greater detail, binary interactions between the four initiation factors were analyzed. eIF2, eIF3, eIF5 and the resulting binary complexes migrate as distinct bands in the gel and thus are easily distinguished from one another (non-complexed eIF1 does not migrate into the gel under these conditions). Accordingly, binary complexes are shown to form in every possible combination (Figure 2C, and summarized in Figure 2D). None of the binary complexes containing eIF2 is affected significantly by the presence of GDP, GDPNP and/or Met-tRNAi (data not shown). It should be noted, however, that formation of the eIF1–eIF5 complex requires higher protein concentrations (10 µM of each protein compared to 1–2 µM for the other complexes). Curiously, the shifted eIF3–eIF5 native gel band contains a substoichiometric amount of eIF5 (Figure 2C, lane 10), even though the shift is almost complete (compare lanes 6 and 8 in Figure 2C). This suggests that this interaction is relatively weak and likely dissociates during analysis but may somehow induce an eIF3 conformation that persists even after dissociation. Since the amount of eIF5 in the eIF2–eIF3–eIF5 trimeric complex is stoichiometric, but not in the eIF1–eIF3–eIF5 and eIF3–eIF5 complexes (data not shown and lane 10), it is likely that eIF5 is anchored to the MFC mostly through its interaction with eIF2. We also find that the eIF2–eIF3 complex starts to dissociate below 200–300 nM for each component (data not shown), thus exhibiting a stability similar to that of the whole MFC (Figure 2B). In contrast, the eIF1–eIF3 and eIF2–eIF5 binary complexes are quite stable even at 50 nM, implying that the association of eIF2–eIF5 with eIF1–eIF3 occurs through the eIF2–eIF3 interaction, which may be the key interaction for MFC formation in humans.
Similar interactions are found between the components of the yeast MFC, except that eIF1 binding to eIF2 appears to be much weaker (8). In addition, yeast eIF5 binding to yeast eIF3 appears to be much stronger than in humans, with yeast eIF5 serving to mediate MFC assembly as the core component (8). Taken together, human and yeast MFC interaction models are similar, though not identical, perhaps in large part due to the difference in eIF3 structures.
The eIF2–Met-tRNAi binding affinity in the presence of GDP is greatly reduced by MFC components
Although the MFC can form in the absence of guanine nucleotides and Met-tRNAi, we wished to determine in vitro if it binds Met-tRNAi in a manner similar to that for eIF2 alone. We first tested Met-tRNAi binding affinities to eIF2–GDPNP with or without MFC components using the standard nitrocellulose filter binding assay (summarized in Table 1; see also Supplementary Figure S1). Under our conditions, eIF2–GDPNP alone binds Met-tRNAi with a dissociation constant of 35 ± 2 nM, which is in good agreement with a previously reported value with the rabbit factor (28–47 nM) (17) and is slightly weaker than that of yeast eIF2 (10 nM) (19). The affinity seen with MFC–GDPNP (Kd = 19 ± 3 nM) is comparable to that for eIF2–GDPNP. Note that saturating amounts of the other MFC components are included throughout this experiment to reduce the possibility of inefficient MFC formation at low eIF2 concentrations (see ‘Materials and Methods’ section). The results indicate that the other three initiation factors in the MFC have little effect on the ability of eIF2 in the GTP-state to bind Met-tRNAi.
Table 1.
Summary of Met-tRNAi Kd values (nM)
| GDPNP | GDPa | |
|---|---|---|
| eIF2 | 35 ± 2 | 4600 ± 170 |
| MFC | 19 ± 3 | N.D. |
| eIF2–eIF1 | – | 6100 ± 140 |
| eIF2–eIF5 | – | 21 000 ± 1100 |
| eIF2–eIF3 | – | N.D. |
Values are means of two independent experiments. The errors shown are standard errors.
N.D., not detected.
aThe values were calculated from the data from 188 to 3000 nM protein concentration range.
In light of this finding with GDPNP, we wished to determine if the MFC and eIF2 are equally responsive to GDP. The affinity of Met-tRNAi to eIF2 is 130-fold decreased in the GDP state compared to the GTP state, with a Kd value of 4600 ± 170 nM. Human eIF2 therefore displays a change in Met-tRNAi affinity between the GTP and GDP states that is much stronger than observed for its yeast counterpart, which shows only an 18-fold lower affinity in the GDP state with a Kd of 180 nM (19). Intriguingly, however, when we determined the Met-tRNAi binding affinity with the MFC in the GDP state, no Met-tRNAi binding is detected within the range tested (Kd >> 6000 nM) (Table 1). To identify the MFC component(s) responsible for this nucleotide-sensing effect, we measured the Met-tRNAi binding affinities for eIF2 binary complexes in the GDP state. Strikingly, we find that the association of either eIF5 or eIF3 with eIF2–GDP dramatically reduces its affinity to Met-tRNAi. Curiously, eIF3 appears to have a much stronger effect, comparable to that of the complete MFC (Supplementary Figure S1). This result suggests that the release of eIF2–GDP from the 40S ribosomal subunit following initiation codon recognition is enhanced by eIF3 and eIF5 (see ‘Discussion’ section).
Because the binding of Met-tRNAi to eIF2 in the GDP state is significantly affected by eIF3 and eIF5, we wondered if human eIF5 might also serve as a GDP-dissociation inhibitor (GDI) of eIF2, as recently reported for yeast eIF5 (20). We therefore tested the human eIF2–GDP dissociation rate in the presence and absence of human eIF5. With eIF2 alone, the GDP off rate at 1 mM MgCl2 and 37°C is 6.6 ± 0.1 × 10−3 min−1, which is in the same range as the previously reported value of 2.0 × 10−3 min−1 with the murine factor at 1 mM MgCl2 and 0°C (21). These values are two orders of magnitude slower than that reported for yeast eIF2 measured at even higher magnesium and lower temperature (1.2 × 10−1 min−1 at 2.9 mM MgCl2 and 10°C) (22,20). When dissociation of GDP from eIF2 is measured in the presence of eIF5, an off rate of 5.9 ± 0.3 × 10−3 min−1 is observed. We therefore detect very little if any inhibition of GDP release by human eIF5. It is important to note, however, that yeast eIF5 decreases GDP release by only 2-fold (20). Considering the high conservation of the consensus motif responsible for GDI activity (20), it is possible that the GDI activity of human eIF5 is stronger under different experimental conditions. However, the evidence to date indicates that human eIF5 does not possess GDI activity, and thus differs from the yeast system in this regard.
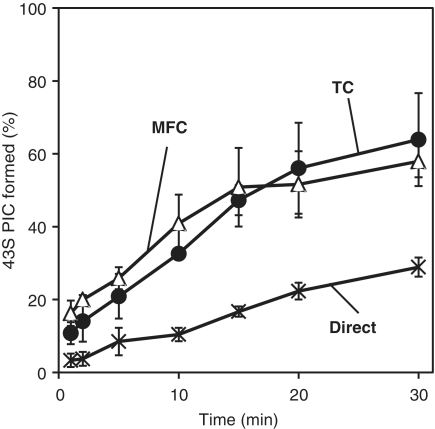
The MFC and TC promote Met-tRNAi binding to the ribosome with equal rates
It is generally accepted that Met-tRNAi is delivered to the 40S ribosomal subunit as a TC. However, since the MFC is present in cells and also binds Met-tRNAi, this raises the possibility that Met-tRNAi binding involves the MFC rather than, or in addition to, the TC. To test this, we set up three different Met-tRNAi–40S binding reactions: (i) a direct binding pathway in which Met-tRNAi binds directly to the 40S subunit already complexed with all additional factors (eIF1, eIF1A, eIF2–GDPNP, eIF3 and eIF5); (ii) a TC-mediated pathway in which the TC is preformed and then added to 40S subunits already carrying the other initiation factors; and (iii) an MFC-mediated pathway in which the MFC is preformed with Met-tRNAi and GDPNP and then is added to 40S subunits containing only eIF1A. Note that the final compositions of the three reaction mixtures are identical and only the orders of addition differ. Moreover, to reduce the extent of possible dissociation of the preformed complexes during the incubations, the concentrations of all the components are kept above 300 nM at near-stoichiometric levels. Met-tRNAi delivery rates to the 40S subunit were monitored at each time point by loading a reaction aliquot to a running native composite gel so that the resulting 43S complex is immediately separated from the other components. Strikingly, almost 60% of input ribosomes bind Met-tRNAi in 30 min in both the TC- and MFC-mediated delivery pathways (Figure 3). In contrast, only 20% binding of Met-tRNAi occurs after 30 min in the direct binding pathway. This indicates that both the MFC and the TC stimulate the rate and degree of Met-tRNAi binding when compared to non-complexed Met-tRNAi. To ensure an efficient rate of delivery in this in vitro assay, Met-tRNAi must therefore bind to eIF2 as part of the TC or MFC prior to delivery to the 40S subunit.
Figure 3.
Rates of Met-tRNAi binding to 40S ribosomal subunits. Met-tRNAi delivery rates involving direct binding (‘×’ marks) and TC- (solid circles) and MFC-mediated pathways (blank triangles) were measured as described in ‘Materials and Methods’ section. Values are the means of two independent experiments, and errors shown are standard errors.
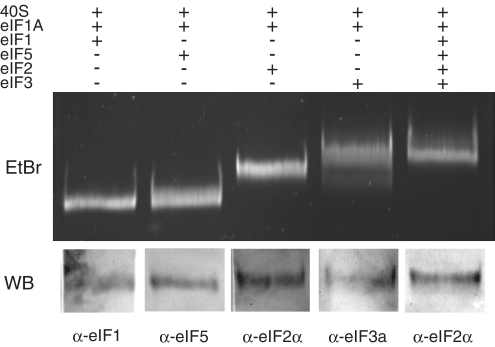
The 40S ribosome binds initiation factors stably
To extend our MFC studies, we further investigated the binding of individual components of the MFC to the 40S subunit. Equilibrium binding of each component is detected by native polyacrylamide–agarose composite gel electrophoresis as described in ‘Materials and Methods’ section. Since eIF1A binds tightly to the 40S subunit (15), it is routinely included in each binding experiment. Gel analysis shows that each initiation factor individually binds stably to the 40S ribosome (Figure 4), as confirmed by band shifts (eIF2 and eIF3) and/or western blotting (eIF1 and eIF5). Interestingly, eIF2 forms a stable 40S complex with or without GDP/GDPNP/Met-tRNAi under the conditions tested (Figure 4 and data not shown). This result is unexpected, as its binding in the absence of Met-tRNAi is very weak when measured by sucrose gradient centrifugation (23). We also find that eIF2 and eIF5 bind stably to 40S ribosomal subunits in the absence of eIF1A (data not shown). The fact that the initiation factors bind stably to 40S ribosomal subunits in the absence of Met-tRNAi raises the possibility that their binding precedes Met-tRNAi binding, as has been reported to occur in bacteria (24) and even more significantly, in archaea (25). This view conflicts with the more generally accepted mechanism that Met-tRNAi delivery is mediated by the TC (see ‘Discussion’ section).
Figure 4.
Individual interactions between each MFC component and 40S-eIF1A are shown by composite native gel electrophoresis. Mixtures containing 40S-eIF1A and the MFC components shown above the gel were generated and analyzed as described in ‘Materials and Methods’ section. Binding was demonstrated by a band shift and/or subsequent western blot analysis of the band with an antibody specific for the initiation factor as shown at the bottom of each lane.
DISCUSSION
The ability to form a human MFC with purified proteins suggests that such a complex may exist in cells free of the ribosome. The MFC is stable at concentrations above 300 nM, but dissociates at more dilute concentrations (Figure 2B). Since initiation factor concentrations in HeLa cells are near 1000 nM (26), their concentrations are sufficiently high to associate into a MFC without involvement of the ribosome. Detection of the MFC–Met-tRNAi complex in the HeLa cell cytoplasm by the gentle native gel electrophoresis procedure (Figure 1) supports the view that the MFC indeed exists in a ribosome-free state in the cell. The finding that all of the ribosome-free eIF3 is found in the MFC further enhances the apparent importance of this complex.
Assuming that the presence of the MFC in the HeLa cell and rabbit reticulocyte cytoplasm is due to its formation off the ribosome rather than to its dissociation from the ribosome during the analysis, a free form of the MFC suggests that it may play an important role in the delivery of Met-tRNAi to the 40S ribosomal subunit. The MFC efficiently binds Met-tRNAi in a reaction requiring GTP (Supplementary Figure S1), and thus is comparable to the TC. This raises the question of which complex, the TC or the MFC, is responsible for bringing the Met-tRNAi to the ribosome. Comparisons of Met-tRNAi delivery rates by the TC and the MFC (Figure 3) show that both complexes are equally efficient, and both stimulate the rate over that obtained with uncomplexed initiator tRNA. Under these conditions, no advantage in forming the MFC is evident, but the results suggest that both the TC and MFC participate in Met-tRNAi delivery to the 40S ribosomal subunit. This issue has been addressed in yeast by measuring the cellular levels of TC and MFC (27). The authors concluded that the TC forms first, followed by the MFC-Met-tRNAi, which then binds to the 40S subunit, but since no kinetic data were reported, the actual pathway remains uncertain.
However, we also find that eIF2 and the other initiation factors as well as the whole MFC bind stably to high salt-washed 40S ribosomal subunits when assayed by native composite gel electrophoresis (Figure 4). Much of our understanding of the initiation pathway has been based on results obtain by sucrose gradient centrifugation analyses. However, detection of binding intermediates requires that such complexes dissociate at very low rates, sufficient to maintain association during the hours of centrifugation, but which may not be physiologically relevant. Thus, the failure to detect binding by such methods may not reflect what is happening in cells. For example, eIF2 in the absence of Met-tRNAi does not bind stably to 40S ribosomes when examined by centrifugation methods, yet forms a stable complex when assayed by native gel electrophoresis. The latter finding suggests that its binding affinity is sufficient to result in eIF2 binding to 40S subunits under physiological conditions.
If the individual initiation factors bind to the ribosome faster than to one another, this would result in the formation of the MFC on the surface of the 40S ribosomal subunit rather than in the ribosome-free state. The stable binding of eIF2 and the other initiation factors to the 40S ribosomal subunit raises the possibility that Met-tRNAi binding occurs to such a 40S-factor complex rather than through the TC- or MFC-mediated pathways described above. Although our finding suggests that direct binding of Met-tRNAi is slower than TC- or MFC-mediated binding, this does not rule out direct binding since the in vitro experimental conditions may not accurately reflect those in intact cells. It is noteworthy that the direct binding pathway is similar to what occurs in bacteria and archaea, where sophisticated fluorescence kinetic assays have shown that IF2/aIF2 binds first to the 30S ribosomal subunit, followed by initiator tRNA binding (24,25). Thus, the situation in eukaryotes remains unclear, as our findings suggest three different ways that the initiator tRNA could be delivered to the 40S ribosome. To resolve this important question in the mammalian initiation pathway, detailed kinetic analyses are required to determine which pathway is most dominant in this regard. Real-time fluorescence assays are needed to determine the association and dissociation rates of eIF2 binding to Met-tRNAi, the 40S ribosome and to other MFC components. In addition, rates must be measured for Met-tRNAi binding to eIF2-GTP in the free state, in the MFC and when bound to the 40S ribosomal subunit. Although some of these binding and dissociation rates have been determined (19), it will be important to elucidate all of these reactions under comparable conditions in order to resolve the pathway of Met-tRNAi delivery to the 40S ribosomal subunit.
Another important finding is that, compared to eIF2, the MFC more severely discriminates Met-tRNAi binding in the GDP state. Although the physiological implication of this finding is not clear, one possibility is that the MFC components or more simply eIF3 and eIF5 promote eIF2–GDP dissociation from ribosome-bound Met-tRNAi after start codon recognition and GTP hydrolysis. A model has been proposed in which eIF3, and possibly other initiation factors, may remain bound to the 80S following initiation (28). This raises the possibility that eIF2–GDP may dissociate as the free factor rather than as part of the MFC. Alternatively, eIF2–GDP may dissociate as a complex with eIF5, as has been proposed to occur in yeast (29). Experiments to measure the rapid association and dissociation rates of individual initiation components with the 40S subunit will therefore ultimately help reveal the true pathway of Met-tRNAi delivery in human cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure S1, Supplementary Methods and Supplementary Reference (30).
FUNDING
Human Frontier Science Program Fellowship (LT00575/2007-L to M.S.); the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (to M.S.); and grants from the United States Public Health Service (GM073732 to J.W.B.H., R01GM092927 to C.S.F.). Funding for open access charge: National Institutes of Health (PO1 GM073732 to J.W.B.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. K. Asano for critical comments on the manuscript.
REFERENCES
- 1.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translational initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 3.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh CR, He H, Ii M, Yamamoto Y, Asano K. Efficient incorporation of eukaryotic initiation factor 1 into the multifactor complex is critical for formation of functional ribosomal preinitiation complexes in vivo. J. Biol. Chem. 2004;279:31910–31920. doi: 10.1074/jbc.M313940200. [DOI] [PubMed] [Google Scholar]
- 5.Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J. Biol. Chem. 2008;283:1094–1103. doi: 10.1074/jbc.M708155200. [DOI] [PubMed] [Google Scholar]
- 6.Asano K, Shalev A, Phan L, Nielsen KH, Clayton J, Valášek L, Donahue TF, Hinnebusch AG. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valášek L, Mathew AA, Shin B-sik, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh CR, Yamamoto Y, Asano K. Physical association of eukaryotic initiation factor (eIF) 5 carboxyl-terminal domain with the lysine-rich eIF2β segment strongly enhances its binding to eIF3. J. Biol. Chem. 2004;279:49644–49655. doi: 10.1074/jbc.M409609200. [DOI] [PubMed] [Google Scholar]
- 9.Valásek L, Nielsen KH, Hinnebusch AG. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis MD, Person MD, Browning KS. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J. Biol. Chem. 2009;284:20615–20628. doi: 10.1074/jbc.M109.007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieniossek C, Schütz P, Bumann M, Limacher A, Uson I, Baumann U. The crystal structure of the carboxy-terminal domain of human translation initiation factor eIF5. J. Mol. Biol. 2006;360:457–465. doi: 10.1016/j.jmb.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher CM, Pestova TV, Hellen CUT, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18:2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JWB, Doudna JA, Robinson CV, Leary JA. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 15.Fraser CS, Berry KE, Hershey JWB, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 16.Castellanos-Serra L, Hardy E. Negative detection of biomolecules separated in polyacrylamide electrophoresis gels. Nat. Protoc. 2006;1:1544–1551. doi: 10.1038/nprot.2006.233. [DOI] [PubMed] [Google Scholar]
- 17.Farruggio D, Chaudhuri J, Maitra U, RajBhandary UL. The A1 x U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol. Cell. Biol. 1996;16:4248–4256. doi: 10.1128/mcb.16.8.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh M-H, Ye P, Datta AB, Zhang M, Fu J. An agarose-acrylamide composite native gel system suitable for separating ultra-large protein complexes. Anal. Biochem. 2005;343:166–175. doi: 10.1016/j.ab.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 20.Jennings MD, Pavitt GD. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature. 2010;465:378–381. doi: 10.1038/nature09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panniers R, Rowlands AG, Henshaw EC. The effect of Mg2+ and guanine nucleotide exchange factor on the binding of guanine nucleotides to eukaryotic initiation factor 2. J. Biol. Chem. 1988;263:5519–5525. [PubMed] [Google Scholar]
- 22.Erickson FL, Hannig EM. Ligand interactions with eukaryotic translation initiation factor 2: role of the gamma-subunit. EMBO J. 1996;15:6311–6320. [PMC free article] [PubMed] [Google Scholar]
- 23.Benne R, Wong C, Luedi M, Hershey JW. Purification and characterization of initiation factor IF-E2 from rabbit reticulocytes. J. Biol. Chem. 1976;251:7675–7681. [PubMed] [Google Scholar]
- 24.Milon P, Carotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010;11:312–316. doi: 10.1038/embor.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasenöhrl D, Fabbretti A, Londei P, Gualerzi CO, Bläsi U. Translation initiation complex formation in the crenarchaeon Sulfolobus solfataricus. RNA. 2009;15:2288–2298. doi: 10.1261/rna.1662609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan R, Hershey JWB. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J. Biol. Chem. 1983;258:7228–7235. [PubMed] [Google Scholar]
- 27.Singh CR, Udagawa T, Lee B, Wassink S, He H, Yamamoto Y, Anderson JT, Pavitt GD, Asano K. Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J. Mol. Biol. 2007;370:315–330. doi: 10.1016/j.jmb.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pöyry T, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh CR, Lee B, Udagawa T, Mohammad-Quereshi SS, Yamamoto Y, Pavitt GD, Asano K. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006;25:4537–4546. doi: 10.1038/sj.emboj.7601339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benne R, Amesz H, Hershey JWB, Voorma HO. The activity of eukaryotic initiation factor eIF-2 in ternary complex formation with GTP and Met-tRNAf. J. Biol. Chem. 1979;254:3201–3205. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.