Abstract
Deadenylation is the first and rate-limiting step during turnover of mRNAs in eukaryotes. In the yeast, Saccharomyces cerevisiae, two distinct 3′–5′ exonucleases, Pop2p and Ccr4p, have been identified within the Ccr4-NOT deadenylase complex, belonging to the DEDD and Exonuclease–Endonuclease–Phosphatase (EEP) families, respectively. Ngl3p has been identified as a new member of the EEP family of exonucleases based on sequence homology, but its activity and biological roles are presently unknown. Here, we show using in vitro deadenylation assays on defined RNA species mimicking poly-A containing mRNAs that yeast Ngl3p is a functional 3′–5′ exonuclease most active at slightly acidic conditions. We further show that the enzyme depends on divalent metal ions for activity and possesses specificity towards poly-A RNA similar to what has been observed for cellular deadenylases. The results suggest that Ngl3p is naturally involved in processing of poly-adenylated RNA and provide insights into the mechanistic variations observed among the redundant set of EEP enzymes found in yeast and higher eukaryotes.
INTRODUCTION
The poly-A tail is an important structural element affecting both stability and translational efficiency of eukaryotic mRNAs [for a review, see (1)]. For most cytoplasmic mRNAs, turnover is promoted through progressive shortening of the poly-A tail (deadenylation) leading to removal of the 5′ cap (decapping) and subsequent degradation of the mRNA body in either the 5′–3′ (main pathway) or 3′–5′ direction [reviewed in (2–4)]. Deadenylation is thus the first and rate-limiting step of mRNA turnover, making it an important control point during gene expression. In yeast, Saccharomyces cerevisiae, two major deadenylase complexes have been found; the mega-Dalton Ccr4-NOT complex (5–8), which contains two active 3′–5′ exonucleases, Ccr4p and Pop2p, and the heterodimeric Pan2-Pan3 complex (9–11). In addition, higher eukaryotes contain another deadenylation enzyme, poly(A)-specific ribonuclease (PARN) belonging to the RNase D family (12). Based on conserved domain structures, a number of additional nucleases putatively involved in deadenylation or RNA processing have been identified, but in many cases the enzymatic properties and biological roles of these enzymes have not yet been established (1).
Human CCR4 (called CNOT6L) and PARN are highly poly-A specific in vitro but this is not the case for all deadenylases (13,14). Yeast Pop2p, for example, shows a preference for polyadenosine in competition assays but is able to degrade other sequences as well (15–18). Based on sequence and structural comparison of nuclease domains, most deadenylases can be classified as either DEDD or Exonuclease–Endonuclease–Phosphatase (EEP) type exonucleases. DEDD-type nucleases, such as PARN, PAN2 and Pop2p, are RNA-specific and contain a number of conserved catalytic aspartate and glutamate residues in the active site required for catalysis, while EEP nucleases, such as Ccr4p, belong to a much broader class of enzymes capable of degrading both RNA and DNA via both exonucleolytic and endonucleolytic mechanisms (1). Consistent with this, human CNOT6L was shown to have trace activity on polyA-DNA (14).
The EEP domain was first identified in Mg2+-dependent DNA endonucleases such as DNase I and DNA repair enzymes specific for abasic sites, including Escherichia coli Exonuclease III (ExoIII) and the major human AP endonuclease, hAPE1 (19,20). Subsequently, sequence analysis has been instrumental in identifying similar domains in a wide range of proteins such as Ccr4p but also enzymes active on sugars and lipids, such as the inositol polyphosphate-5-phosphatases and sphingomyelinases (21–23). Yeast and higher eukaryotes contain at least 19 enzymes related to Ccr4p having the EEP fold and containing the conserved residues required for them to function as deadenylases (1,24). Of these, S. cerevisiae Ngl1p (yol042), Ngl2p (ymr285) and Ngl3p (yml118) were named by their homology to the human ANGEL proteins and found to be non-essential but containing all the features of known active enzymes (24,25). The best characterized of the three, Ngl2p, has been shown to take part in the final step during 3′-end processing of 5.8S rRNA (26–28). Ngl1p has been detected in highly purified mitochondria in a high-throughput study (29) but the localization, biochemical activities and biological roles of Ngl1p and Ngl3p are still open questions.
In order to expand our knowledge about the activity and specificity of the diverse EEP enzymes, we have undertaken a detailed biochemical analysis of S. cerevisiae Ngl3p. In this article, we demonstrate that the enzyme is an active 3′–5′ exoribonuclease with a relatively low pH optimum, which might suggest a mechanism in which the protonated form of the active site His 485 is directly involved in the RNA hydrolysis reaction. We further show that Ngl3p possesses a preference for poly-A stretches but is able to degrade polypyrimidine tracts as well, albeit at a much slower rate. Finally, we show that deletion of ngl3 does not severely affect the growth of a Δccr4 strain, suggesting that the enzyme is not directly involved in deadenylation.
MATERIALS AND METHODS
Cloning, expression and protein purification
The gene encoding full-length Ngl3p (systematic name, yml118) was amplified from S. cerevisiae genomic DNA using primers Ngl3LICtev1-20 (5′-GACGACGACAAGATGGAGAATCTTTATTTTCAGGGCATGGA TAGCCAAGTAGAAGG-3′) and Ngl3LIC1494-1518 (5′-GAGGAGAAGCCCGGTTTATTTTAAGCTATGCACTTTCTC-3′) and inserted into pET30Ek/LIC (Novagen) by the ligation-independent cloning (LIC) method (LIC sites are underlined). The fusion protein contains an N-terminal 6× His tag and a Tobacco Mosaic Virus (Tev) protease site (bold). The active site Ngl3p mutant (Ngl3pH485A) was constructed by site-directed mutagenesis of pET30 Ek/LIC-Ngl3wt using primers 5′-GAGAATATGCTAGTGATGCCATTTCCCTAATG-3′ and 5′-CATTAGGGAAATGGCATCACTAGCATATTCTC-3′, for which the site of mutation is underlined.
Protein expression was carried out in E. coli BL21 (DE3) Rosetta cells grown at 37°C in 2×TY with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol until a OD600 of 0.6–0.8, cooled on ice for 30 min, then induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated at 20°C overnight. After harvesting, the cells were resuspended in buffer A (50 mM Tris–HCl, pH 8.0, 300 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol (BME), 20 mM imidazole, 10% glycerol and Complete Protease Inhibitor Cocktail tablets (Roche) and lysed by high-pressure homogenization at 15 000 psi. The crude extract was ultra-centrifuged at 50 000 r.p.m. at 4°C before loading on a 5 ml His-trap chelating nickel-column (Qiagen) pre-equilibrated in buffer A. The column was then washed with buffer B (50 mM Tris–HCl, pH 8.0, 1 M KCl, 5 mM MgCl2 and 5 mM BME) and eluted using 500 mM imidazole in buffer A. The eluate was dialysed overnight at 4°C into buffer A in the presence of His-tagged Tev protease at a ratio of 1:200 (w/w) Tev to Ngl3p. The sample was then reapplied to the Ni-column to remove the Tev protease, uncleaved Ngl3, and any proteins non-specifically binding the column. The run-through was concentrated to about 8 mg/ml and loaded on a 24 ml Superdex 200 10/300 GL column (GE healthcare) pre-equilibrated in buffer C (10 mM Hepes, pH 7, 100 mM KCl, and 5 mM BME) at 4°C. Finally, the protein was concentrated to 20 µM using VivaSpin spin filters (10.000 MWCO PES, VivaScience) at 4°C and stored at −80°C in 50% glycerol. Protein concentration was measured using the Nanodrop ND-1000 spectrophotometer and an extinction coefficient of 1.18 OD280/mg (ExPaSy ProtParam). Rrp6p12–536 was purified essentially as described previously (30).
In vitro degradation assays
RNA substrates were synthesized with either a 5′-carboxyflourescein (FAM) or 5′-carboxytetramethylrhodamine (TAMRA) label (RNA-polyC10), except RNA-endoA10, which had both 5′-FAM and 3′-TAMRA labels (DNA Technology, Invitrogen, Microsynth and Dharmacon), gel purified and dissolved in water to 100 nM. The RNA oligo used for creating the circular RNA had the sequence, 5′-CAGCUCCGCAUTCCCUUUCCCAAAAAAAAAA-3′, where UT denotes a modified uracil base to which a TAMRA fluorophore is attached (Microsynth). Circular RNA was produced by incubating the labelled, 5′-phosphorylated, linear RNA with T4 RNA ligase 1 (New England Biolabs) and 1 mM ATP in the supplied buffer for 15 min at 37°C. The circular form was then isolated by gel purification on a 18% denaturing acrylamide gel, eluted into 10 mM Mes, pH 6.5, precipitated and dissolved in water to 140 nM. In vitro reactions with Ngl3p contained at least 50 times molar excess of enzyme (0.5 µM) over substrate (1–10 nM) and reactions at standard conditions were carried out at 30°C in reaction buffer containing 10 mM Mes, pH 5.5, 10 mM KCl, 5 mM MgCl2 and 1 mM DTT. Reactions with Rrp6p contained only 5 nM protein and were carried out in 10 mM Tris, pH 8.0, 50 mM KCl, 1 mM MgCl2 and 2 mM BME at 30°C. After incubation, reactions were quenched by addition of an equal volume of RNA loading buffer (8 M Urea, 50 mM Tris–HCl, pH 7.5, and trace bromophenol blue) and the resulting RNA fragments separated by denaturing gel electrophoresis in 20 cm 18–20% denaturing polyacrylamide gels (8 M urea in 1× TBE) running ∼3–4 h at 9 W in 1×TBE at room temperature. RNA ladders were made by limited alkaline hydrolysis of the individual RNA substrates in 50 mM NaHCO3/Na2CO3 at pH 8.9 for 14 min at 96°C. A Typhoon Trio Imager (GE healthcare) was used to visualize the gels and data quantification was done using TotalLab Quant.
Microscale thermophoresis
Equilibrium binding experiments were carried out at a constant low, fluorescent RNA concentration of 10 nM, while titrating in protein in 16 steps from 0.14 nM to 4.6 μM. The protein titration series was prepared by serial dilution of a 9.22 μM Ngl3p stock solution in protein storage buffer (5 mM Hepes, pH 7, 50 mM KCl, 2.5 mM BME and 50% glycerol) and 1:1 mixing of protein and RNA in binding buffer (30 mM Mes, pH 5.5, 10 mM KCl, 1 mM DTT and 0.05% Tween-20). Experiments were carried out using Ngl3wt in the absence of divalent metal ions, where it remains inactive. The raw fluorescent thermophoresis signal was recorded in triplicate from a Monolith NT.115 microscale thermophoresis system (NanoTemper) and normalized by scaling to 0–100%. Fitting was done using a sigmoidal dose-response model with variable slope and the EC50 value of the curve used as a measure for the KD.
Yeast growth assay
Yeast strains were constructed by back-crossing of the ccr4Δ::KAN and ngl3Δ::KAN strains from BY background (Open Biosystems) with a BY trp1Δ strain. Double deletion mutants were obtained from crossing of the ccr4Δ::KAN and ngl3Δ::KAN single deletions and individual deletions were confirmed by gene-specific PCR. For yeast growth assays, BY trp1Δ derivatives were diluted to A600 = 0.2, spotted in 10-fold dilutions onto YPAD plates, and incubated at 18°C for 5 days and 30 or 37°C for 2 days.
RESULTS
S. cerevisiae Ngl3p is an active 3′–5′ exoribonuclease in vitro
For the biochemical analysis of S. cerevisiae Ngl3p, the full-length enzyme was expressed in E. coli as a His-tagged fusion protein cleavable by Tev protease. The fusion protein (63.3 kDa) was purified by tandem Ni-NTA chromatography separated by Tev cleavage, and subsequent gel filtration yielded highly pure protein in three steps (Supplementary Figure S1A). The cleaved and purified wild type enzyme, denoted Ngl3pwt, elutes corresponding to a molecular weight of 60 kDa during gel filtration, very near the theoretical molecular weight of 58 kDa, suggesting that Ngl3p is a globular, monomeric protein in solution (Supplementary Figure S1B). Gel filtration also served to change the enzyme into a minimal buffer allowing later addition of specific divalent metal ions during the functional analysis.
To analyse the enzymatic activity of Ngl3p in vitro, we carried out a series of RNA degradation experiments in which the enzyme was incubated with defined, chemically synthesized and 5′ fluorescently labelled RNA and DNA substrates. Following incubation, samples were separated by denaturing polyacrylamide gelelectrophoresis and the resulting 5′ fragments visualized with a resolution down to one nucleotide by scanning fluorometry. As deadenylases are relatively slow enzymes, their activity is best studied under pre-steady state conditions, where the protein is in molar excess over RNA (31). Consequently, due to the risk of cross-contamination with other nucleases co-purified from the expression host, inclusion of an active site mutant is critical in such experiments to ensure that the correct activity is being studied. We therefore constructed an H485A active site mutant, Ngl3pH485A, in which an essential histidine in the EEP active site motif was changed into alanine (Supplementary Figure S2, marked with a red asterisk). The mutant was purified essentially as the wild type enzyme.
First, Ngl3wt was incubated with a 30 nt RNA oligo consisting of 20 nt of generic sequence with little or no secondary structure followed by 10 adenosine residues mimicking a poly-adenylated mRNA (RNA-polyA10, Figure 1A). During initial assays carried out at standard conditions at pH 7–8 with 100 mM KCl and 5 mM Mg2+, the enzyme showed only trace activity, and a more thorough screening for the reaction optimum was therefore carried out at pH values ranging from 3.5 to 9, salt concentrations from 5 to 1000 mM, and temperatures from 10–50°C all in the presence of 5 mM Mg2+ (Figure 1B and Supplementary Figure S3). In these experiments, the rate of degradation was quantified as the relative amount of partly or fully degraded RNA relative to full length RNA (30 nt) by integration of the gel bands. Optimal activity was observed at 30°C, low ionic strength (5–10 mM KCl), and a relatively low pH value of 5.5–6.5 with very little activity below pH 5.5 and above 7. All subsequent reactions were carried out in 10 mM KCl at pH 5.5 and 30°C at which conditions the enzyme displayed robust activity.
Figure 1.
Ngl3p is a 3′–5′ exonuclease active at low pH. (A) Fluorescently labelled, mRNA-like substrates with either 10 or 21 adenosine residues at the 3′-end, where (F) represents the 5′-carboxyfluorescein label. (B) pH optimum of Ngl3p shown as the percentage of degraded RNA at different pH values. The pH optimum is pH 6.2 (dashed line). (C) Metal ion requirements. Gel showing degradation of RNA-polyA10 in the presence of 5 mM Mg2+, 5 mM Mn2+, 1 mM Zn2+ and combinations of these. In lane 9, the reaction contained 7.1 mM Mg2+, 75 µM Mn2+ and 220 µM Zn2+(33), and in lane 10, 25 mM EDTA was added. The transiently stabilized fragment (21 nt) is indicated. (D) Degradation of RNA-polyA10 by Ngl3pwt over time. L is an RNA ladder made by partial alkaline hydrolysis. The RNA ladder sizes (30–20 nt) and the stabilized fragment (21 nt) are indicated and for the lane ‘H485A’, RNA-polyA10 was incubated with the active site mutant, Ngl3pH485A. (E) Degradation of RNA-polyA21. The points of pausing at 10 and 8 nt are indicated.
EEP-type nucleases are dependent upon one or more divalent cations for binding RNA, activating water for hydrolysis and stabilizing the negatively charged transition state during the cleavage reaction (32). To investigate the metal ion requirements of Ngl3p, we studied degradation of RNA-polyA10 in the presence of combinations of milimolar quantities of Mg2+, Mn2+ and/or Zn2+ (Figure 1C). Incubation of 10 nM RNA for 1 h at 30°C in the reaction buffer including either 5 mM Mg2+ (lane 1) or 50 times molar excess of Ngl3wt (lane 2) but not both, resulted in little or no RNA degradation illustrating that the enzyme is inactive in the absence of divalent metal ions. Including either 5 mM Mg2+ (lane 3) or 5 mM Mn2+ (lane 4), on the other hand, resulted in degradation of the substrate as visible by the shortening of the 5′-labelled RNA substrate, while a much slower rate of reaction was observed in the presence of Zn2+ (lane 5), similar to what was observed for the cellular deadenylase, Pop2p (31). Mixing two of the ions showed intermediate reaction rates, compatible with Zn2+ having a dominant negative effect on activity (lanes 6–8). Interestingly, a mix of all three ions mimicking as closely as possible their intracellular concentrations [7.1 mM Mg2+, 75 μM Mn2+, 220 μM Zn2+, (33)] resulted in an activity whereby a fully deadenylated species of ∼20 nt was temporarily stabilized (lane 9). Finally, as expected, inclusion of EDTA abolished any trace activity in the purified enzyme (lane 10). We conclude that Ngl3p is a divalent metal ion-dependent nuclease that most likely binds Mg2+ in vivo due to its higher cellular concentration.
Ngl3p transiently stabilizes a fully deadenylated RNA species
To analyse the mechanism by which Ngl3p degrades the poly A-containing RNA, we studied the reaction over time at lower RNA concentration (1 nM) and in the presence of 5 mM Mg2 (Figure 1D). Under these conditions, we observe progressive shortening of the substrate indicating exonucleolytic degradation in the 3′–5′ direction. Degradation continues until all substrates have reached a length of exactly 21 nt (0–30 min) after which the enzyme continues into non-poly A RNA (30–180 min). Ultimately, after overnight incubation, Ngl3p would degrade the substrate completely (data not shown). The RNA species transiently stabilized in this manner corresponds to the 20 nt of generic sequence and the first of the 10 adenosine residues in the 3′-end, which shows that poly-A discrimination by Ngl3p is not confined to the ultimate 3′ base, but probably involves recognition of several 3′ adenosine residues. The active site mutant Ngl3pH485A showed no degradation of the substrate even after 60 min incubation (lane ‘H485A’). To test whether the observed pausing at the beginning of the poly-A tail is indeed due to the sequence of the 3′-end and not the result of complex RNA structure, we repeated the experiment with a substrate of identical length but with 21 adenosine residues at the 3′-end (RNA-polyA21, Figure 1A and E). Consistently, the wild type enzyme degraded this substrate down to about 10 nt length compatible with removal of all but the last adenosine. After 30 min, degradation has continued into non-poly A sequence and appears to stop at the first G encountered (8 nt). We conclude that Ngl3p is active as a 3′–5′ riboexonuclease and tends to pause at the end of poly-A stretches, reminiscent of what has been observed for other known cellular deadenylases, such as Schizosaccharomyces pombe Pop2p (16,17).
Ngl3p does not degrade RNA secondary structure
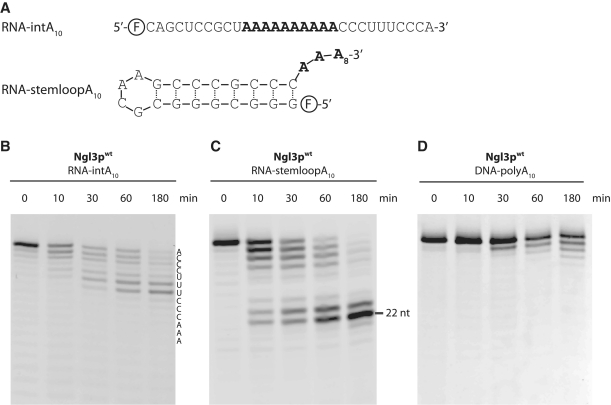
Next, we asked if the poly-A tract has to be located at the very 3′-end to promote degradation by Ngl3p. To analyse this, we used a similar substrate in which the poly-A tract was ‘internal’ to the sequence, i.e. surrounded on both 5′ and 3′ sides by non-poly A sequence (Figure 2A). The 3′ extension to the poly-A stretch was pyrimidine-rich and ended with a single A. In this case, the enzyme does not recognize the poly-A tract and degrades the last seven nucleotides slowly with a complicated degradation pattern (Figure 2B). This shows that the poly-A tract has to be present in the very 3′-end to be recognized correctly by the enzyme. We then asked if Ngl3p could degrade an RNA containing a strong secondary structure element followed by a poly-A sequence (Figure 2A and C). Interestingly, the poly-A tail following the strong secondary structure element is not degraded as fast as the completely single-stranded RNA. This might indicate that the structured RNA interferes with degradation, perhaps restricting entry of the 3′-end into the active site. Degradation eventually halts after removal of eight residues (length = 22 nt) and the enzyme is not able to proceed through the RNA secondary structure (Figure 2C). Finally, as many members of the EEP family are also active on DNA, we tested if Ngl3p would be able to degrade a deoxy nucleotide substrate with a sequence identical to RNA-polyA10 (Figure 2D). In this case, DNA is degraded in a 3′–5′ exonucleolytic fashion similar to RNA, but the reaction is much slower than for poly-A RNA, suggesting that this may not be the natural substrate. We conclude that although Ngl3p is able to degrade both DNA and single-stranded regions of structured RNAs as well as non-poly A RNA, optimal activity is achieved with fully single-stranded RNA bearing a poly-A sequence at the very 3′-end.
Figure 2.
Ngl3p requires a 3′ poly-A tail and does not degrade dsRNA. (A) Oligos used in this figure, where (F) represents the 5′-carboxyfluorescein (FAM) label. (B) Degradation of RNA-intA10, a substrate with an internal oligo-A tract. The sequence of the oligo is shown along the side. (C) Degradation of an RNA containing a strong secondary structure element followed by a 10 nt poly-A tail (RNA-stemloopA10). The shortest product is 22 nt, corresponding to the stem structure plus two overhanging 3′ residues. (D) Degradation of a deoxynucleotide substrate having the same sequence as RNA-polyA10.
Ngl3p is inactive on poly-G RNA
Given the observed poly-A specificity, we asked if Ngl3p would be able to initiate degradation of other monotonous 3′ sequences, poly-uridine, poly-cytidine and poly-guanosine. Four oligos containing the 20 nt generic sequence followed by 10 identical residues (A, U, C or G) were incubated at 10 nM with Ngl3p in parallel reactions, and aliquots removed at different time points. As shown in Figure 3A, Ngl3wt is able to degrade both poly-pyrimidine sequences (RNA-polyC10 and RNA-polyU10) reasonably efficiently and at similar rates, although much slower than the corresponding poly-adenosine sequence. Additionally, the pausing at the end of the monotonous sequence region is not observed. In contrast, the poly-G substrate was left essentially untouched illustrating that Ngl3p cannot initiate degradation of this type of 3′ sequence. A plot of the total amount of NMP produced over time as function of the type of 3′ sequence allows direct comparison of the observed differences in activity (Figure 3B). This reveals approximately constant rates of reaction for the poly-C and poly-U substrates of 3.78 × 10−5 s−1 and 2.20 × 10−5 s−1, while poly-A is degraded about 10 times faster at 2.64 × 10−4 s−1 until reaching a plateau approaching 100 nM NMP corresponding to an average removal of 10 adenosines from each substrate molecule.
Figure 3.
Ngl3p prefers poly-A RNA. (A) Parallel time-course degradation experiments with 10 nM RNA-polyA10, RNA-polyU10, RNA-polyC10 and RNA-poly-G10. The constant diffuse bands seen for RNA-polyC10 around 12, 15 and 20 nt arise from absorption of the bromophenol blue dye at the TAMRA excitation wavelength. (B) Quantification of the degradation experiments in A converted to nanomolar NMP produced at each time point. Error bars represent the standard error of the mean (SEM) of three independent experiments. Errors are relatively large at longer time points due to weakening of the fluorescent label. (C) Measured affinities towards RNA and DNA substrates with different 3′ end sequences. The KD values measured are 226 ± 63 nM (RNA-polyA10), 195 nM ± 95 (RNA-polyU10), 96 ± 22 nM (RNA-polyG10) and 161 ± 37 nM (DNA-polyA10). The error bars represent 95% confidence intervals.
The inability of Ngl3p to degrade poly-G RNA could in theory arise either from lack of binding these RNAs (i.e. low affinity) or an intrinsic lack of activity on such substrates. In order to resolve this ambiguity, we measured the affinity of Ngl3pwt towards RNA-polyA10, RNA-polyG10, and RNA-polyU10 as well as DNA-polyA10 by microscale thermophoresis (MST), a novel technique that measures changes in fluorescent mobility upon heating the sample with an infrared laser (34). Since binding of protein to the fluorescently labelled RNA significantly alters mobility in solution, a signal can be recorded when titrating a fixed, low amount of RNA (10 nM) with increasing protein concentrations at otherwise identical conditions. Binding affinities measured as the dissociation constant, KD, can then be calculated by fitting the normalized MST fluorescence using a dose-response type sigmoidal model (Supplementary Figure S4). The RNA affinities were measured using the wild type enzyme in the absence of Mg2+ to avoid degradation of the substrates during the experiment. Using this approach, we measured dissociation constants of ∼200 nM for RNA-polyA10 and RNA-polyU10, around 100 nM RNA-polyG10 and about 150 nM for DNA-polyA10 (Figure 3C). The measured affinities are all in the 100–200 nM range and thus within the same order of magnitude. Remarkably, however, the affinity towards RNA-polyG10 is significantly higher than those for the other substrates. As the conditions used for the biochemical experiments (10 nM RNA and 500 nM protein) are well above all binding constants we can thus conclude that the inability of Ngl3p to degrade RNA-polyG10 and the slower reaction rate on DNA-polyA10 are not due to lower affinity, on the contrary as RNA-polyG10 was bound strongest, rather that the enzyme is inherently less active on these substrates.
Ngl3p approaches the 3′-end in a different way than DEDD nucleases
As several members of the EEP family are known endonucleases, we wanted to investigate whether Ngl3p, in addition to functioning as a 3′–5′ exonuclease, could also cleave RNA by endonucleolysis. To test this, we constructed an RNA substrate containing two different fluorophores, a 5′-fluoresceine and a 3′-TAMRA attached to the 3′OH group (RNA-endoA10) with the rationale that the presence of a bulky 3′ fluorophore should inhibit 3′–5′ exonuclease activity prior to any endonucleolytic cleavage event. Moreover, this substrate allows tracking the fate of both 5′- and 3′-ends after cleavage by scanning the gels separately for each fluorophore. Apart from this modification, the substrate is identical to RNA-polyA10. To test the assumption that 3′–5′ exonucleases cannot generally degrade this 3′-blocked RNA, we first incubated RNA-endoA10 with the catalytic fragment of the S. cerevisiae nuclear exosome component, Rrp6p (residues 12–536), which is of the DEDD type and shows robust 3′–5′ exonuclease activity in vitro (30). In contrast to RNA-polyA10, which is quickly degraded by Rrp6p (Figure 4A, lane marked ‘polyA’), no degradation is visible for RNA-endoA10 (Figure 4A), showing that DEDD-type exonucleases cannot initiate degradation due to the presence of the 3′ fluorophore. In contrast, incubation of RNA-endoA10 with Ngl3p shows a progressive degradation pattern similar to RNA-polyA10 ending at the end of the poly-A tract. However, as the initial cleavage in this case could have occurred either by endo- or exo-nucleolysis, we repeated the experiment using a circular RNA, in which a TAMRA group had been attached to an internal uracil base prior to circularizing the substrate with RNA Ligase 1 (Figure 4B). We observed no degradation of the circular substrate, thus suggesting that Ngl3p cannot cleave RNA in a classical, endonucleolytic fashion. However, since it is still able to cleave the 3′ blocked substrate, this indicates that it uses a different, and more open, way to approach the 3′-end than DEDD-type nucleases. In summary, our data provide evidence that Ngl3p primarily functions as an exonuclease, but unlike other 3′–5′ exonucleases is able to degrade substrates with a blocked 3′-end. Future structural investigations will reveal the molecular basis for this subtle difference.
Figure 4.
Ngl3p degrades a 3′ blocked but not a circular RNA substrate. (A) Degradation of RNA-endoA10 by Ngl3pwt and Rrp6p12-536 (5′-end fragments). The lane marked ‘polyA’ is Rrp6p12–536 incubated with RNA-polyA10. (B) Incubation of Ngl3pwt with a circular RNA substrate labelled internally with a TAMRA label. The lane marked ‘linear RNA’ is the phosphorylated RNA oligo prior to circularization.
Deletion of Ngl3p does not affect growth of a ccr4Δ strain
Since the poly-A specificity of Ngl3p suggests an involvement of the enzyme in cellular deadenylation, we finally investigated if deletion of ngl3 in S. cerevisiae affected the growth phenotype in a ccr4Δ background, which is already deadenylation compromized (Figure 5). ccr4Δ exhibits a strong cold- and mild temperature-sensitive growth phenotype, while deletion of the gene coding for Ngl3p does not have any measureable effect on the growth of the yeast cells consistent with its non-essential role. In comparison, double mutant, ccr4Δngl3Δ, shows a growth phenotype very similar to ccr4Δ, showing that the deadenylation-compromized cells are not further challenged by the concomitant deletion of Ngl3p. This suggests that Ngl3p is not part of the main Ccr4-NOT mediated deadenylation pathway or another RNA degradation pathway involving the majority of cellular RNAs, but is more likely required for processing of specific poly-adenylated substrates.
Figure 5.
Deletion of ngl3 does not affect the growth of a ccr4Δ yeast strain. Growth phenotypes of the S. cerevisiae wt, ccr4Δ, ngl3Δ and ccr4Δ/ngl3Δ double deletion mutants. Ten-fold serial dilutions of cells were spotted onto agar plates and grown at the indicated temperatures.
DISCUSSION
In this article, we demonstrate that Ngl3p is an active 3′–5′ exonuclease with a preference for poly-A sequences. The in vitro experiments presented here also show that the sequence specificity of Ngl3p can be modulated by the availability of divalent cations. In particular, Mn2+ appears to alter the rate of catalysis and specificity of the enzyme in a subtle way removing some of the specificity of the enzyme (Figure 1C). The molecular basis for this observation is not currently understood, but it is interesting to note that a similar phenomenon was observed for S. pombe Pop2p, for which the presence of Zn2+ alone or in combination with Mg2+ or Mn2+ inhibited the enzyme even at the low physiological level of 220 µM (16). For Pop2p it was shown by a combination of biochemistry and anomalous X-ray diffraction that the enzyme binds one Mg2+ ion and one Mn2+ ion at physiological levels of the two ions. Addition of Zn2+ in micro-molar concentrations, however, causes exchange of Mg2+ for Zn2+ and a concomitant drop in reaction rate and loss of specificity of the in vitro reaction (16,17). Given the structural differences between DEDD and EEP enzymes it is curious that we observe a similar effect in the case of Ngl3p, but it suggests at least that two ions are present in the active site.
Our finding that Ngl3p is only active at low ionic strength and between pH 5.5 and 6.5 also has possible implications for the reaction mechanism. Based on crystal structures and available functional data, several reaction mechanisms have been proposed for EEP type enzymes (20,35,36). In the model for ExoIII from E. coli by Mol et al. (20), a catalytic dyad of D229 (Ngl3p D428, see Supplementary Figure S2) and H259 (Ngl3p H485) work together to activate a water molecule for nucleophilic attack on the phosphate linkage, while E34 (Ngl3p E159) is proposed to coordinate a single Mg2+ ion stabilizing the charged phosphate transition state. In the later model for hAPE1 by the same authors (35), it is D210 (Ngl3p D294) which functions as a general base activating water, while the D283-H300 (Ngl3p D428-H485) dyad instead functions in stabilization of the phosphate group. Finally, in the model for hAPE1 by Beernink et al. (36), several aspartic acid, asparagine, and glutamic acid residues, together with the histidine, combine to coordinate two Mg2+ ions, one of which activates a water molecule for nucleophilic attack. In the structure of human Ccr4 (CNOT6L), the active site configuration most closely resembles the two-ion model proposed for hAPE1 (14). Given the low pH optimum observed for Ngl3p, it is therefore tempting to speculate that catalysis requires a protonated conserved histidine, as this is the only amino acid with a pKa in this range. We therefore suggest that Ngl3p might also employ the same mechanism of cleavage as proposed for hAPE1 by Mol et al. (35) as this is the only model that requires the histidine (H485) to be protonated in order for it to donate hydrogen bonds to both D428 and the phosphate oxygen. Detailed structural studies of enzyme reaction states will be able to address this question in the future.
The observation that yeast Ngl3p is able to cleave RNA with a blocked 3′-end suggests that the enzyme approaches the substrate in a more open way than DEDD nucleases. Intriguingly, when the structure of human CCR4 (CNOT6L) was determined in the presence of a poly-adenosine pentamer (Figure 5A) DNA, two nucleotides were found downstream of the scissile bond (Figure 5B), suggesting that the enzyme approached the substrate in an endonucleolytic fashion. Although the authors of the paper dismiss the relevance of this observation due to DNA being an unnatural substrate, it may in fact show that these enzymes can, under certain conditions (such as when the 3′-end is blocked for example by proteins) function as endonucleases. If endonucleolytic decay of RNA during deadenylation takes place in the cell, i.e. that some deadenylases are able to remove the poly-A tail close to its start by endonucleolysis, it would provide an entirely unknown way of turning over mRNA.
We show that Ngl3p prefers poly-A 3′-ends to other sequences, however, from a yeast growth assay, it does not appear to be directly involved in deadenylation. The localization of Ngl3p is not known, but if it turns out to be nuclear, this opens up the possibility that Ngl3p takes part in degradation of non-canonical nuclear polyadenylated RNAs, such as those produced by the TRAMP complex (37). Our binding experiments demonstrate that at low salt concentrations, the affinity of Ngl3p towards a range of different RNA and DNA substrates is similar; suggesting that the enzyme is able to scan the cellular pool of 3′-ends but only reacting with RNA poly-A 3′-ends. It is also possible that the interactions between Ngl3p and RNA are stronger at the conditions found inside the cell due to binding of co-factors. Deadenylases are often associated with multi-subunit complexes such as the Ccr4-NOT complex (38). Inside these complexes, protein interactions provide additional means of regulation of RNA affinity and the reaction rate. Further in vitro and in vivo studies of Ngl3p are thus needed to determine the exact biological role of Ngl3p and should be combined with structural studies for a full molecular understanding of the enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table, Supplementary Figures S1–S4.
FUNDING
The Lundbeck Foundation (grant number 98/06 to D.E.B.); Novo Nordisk Foundation; Danish National Research Foundation's Centre for mRNP Biogenesis and Metabolism; Danish National Research Council (FNU) (grant number 09-072378 to D.E.B.). Funding for open access charge: The Novo Nordisk Foundation, Denmark.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jesper Godrim Jensen for help with the deletion and yeast growth experiments and Paulina Seweryn for help with purification of the circular RNA substrate.
REFERENCES
- 1.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 2.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 3.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 8.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 9.Boeck R, Tarun S, Jr, Rieger M, Deardorff JA, Muller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 10.Brown CE, Tarun SZ, Jr, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachs AB, Deardorff JA. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 12.Astrom J, Astrom A, Virtanen A. Properties of a HeLa cell 3′ exonuclease specific for degrading poly(A) tails of mammalian mRNA. J. Biol. Chem. 1992;267:18154–18159. [PubMed] [Google Scholar]
- 13.Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Morita M, Yang X, Suzuki T, Yang W, Wang J, Ito K, Wang Q, Zhao C, Bartlam M, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010;29:2566–2576. doi: 10.1038/emboj.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen KR, Jonstrup AT, Van LB, Brodersen DE. The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site. RNA. 2009;15:850–861. doi: 10.1261/rna.1489409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonstrup AT, Andersen KR, Van LB, Brodersen DE. The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 2007;35:3153–3164. doi: 10.1093/nar/gkm178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thore S, Mauxion F, Seraphin B, Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 20.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 21.Dlakic M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo Y, Yamada A, Tsukamoto K, Tamura H, Ikezawa H, Nakamura H, Nishikawa K. A distant evolutionary relationship between bacterial sphingomyelinase and mammalian DNase I. Protein Sci. 1996;5:2459–2467. doi: 10.1002/pro.5560051208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whisstock JC, Romero S, Gurung R, Nandurkar H, Ooms LM, Bottomley SP, Mitchell CA. The inositol polyphosphate 5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J. Biol. Chem. 2000;275:37055–37061. doi: 10.1074/jbc.M006244200. [DOI] [PubMed] [Google Scholar]
- 24.Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 26.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 27.Faber AW, Van Dijk M, Raue HA, Vos JC. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA. 2002;8:1095–1101. doi: 10.1017/s1355838202021027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson E, Tollervey D. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010;30:976–984. doi: 10.1128/MCB.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome. Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 30.Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA. 2008;14:2305–2313. doi: 10.1261/rna.1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen KR, Jensen TH, Brodersen DE. Take the “A” tail–quality control of ribosomal and transfer RNA. Biochim. Biophys. Acta. 2008;1779:532–537. doi: 10.1016/j.bbagrm.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tholey G, Ledig M, Mandel P, Sargentini L, Frivold AH, Leroy M, Grippo AA, Wedler FC. Concentrations of physiologically important metal ions in glial cells cultured from chick cerebral cortex. Neurochem. Res. 1988;13:45–50. doi: 10.1007/BF00971853. [DOI] [PubMed] [Google Scholar]
- 34.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 35.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 36.Beernink PT, Segelke BW, Hadi MZ, Erzberger JP, Wilson DM, 3rd, Rupp B. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J. Mol. Biol. 2001;307:1023–1034. doi: 10.1006/jmbi.2001.4529. [DOI] [PubMed] [Google Scholar]
- 37.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstrohm AC, Hook BA, Wickens M. Regulated deadenylation in vitro. Methods Enzymol. 2008;448:77–106. doi: 10.1016/S0076-6879(08)02605-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.