Abstract
Mitochondrial transcription factor A (mtTFA/TFAM) is a nucleus-encoded, high-mobility-group-box (HMG-box) protein that regulates transcription of the mitochondrial genome by specifically recognizing light-strand and heavy-strand promoters (LSP, HSP1). TFAM also binds mitochondrial DNA in a non-sequence specific (NSS) fashion and facilitates its packaging into nucleoid structures. However, the requirement and contribution of DNA-bending for these two different binding modes has not been addressed in detail, which prompted this comparison of binding and bending properties of TFAM on promoter and non-promoter DNA. Promoter DNA increased the stability of TFAM to a greater degree than non-promoter DNA. However, the thermodynamic properties of DNA binding for TFAM with promoter and non-specific (NS) DNA were similar to each other and to other NSS HMG-box proteins. Fluorescence resonance energy transfer assays showed that TFAM bends promoter DNA to a greater degree than NS DNA. In contrast, TFAM lacking the C-terminal tail distorted both promoter and non-promoter DNA to a significantly reduced degree, corresponding with markedly decreased transcriptional activation capacity at LSP and HSP1 in vitro. Thus, the enhanced bending of promoter DNA imparted by the C-terminal tail is a critical component of the ability of TFAM to activate promoter-specific initiation by the core mitochondrial transcription machinery.
INTRODUCTION
Mitochondria are the sites of ATP production via the process of oxidative phosphorylation (OXPHOS), disruption of which causes or is associated with numerous pathogenic states, including neuromuscular and neurodegenerative diseases, cardiomyopathy, diabetes and aging (1–3). Critical to mitochondrial function is the mitochondrial genome (mtDNA), which encodes 13 protein components of the OXPHOX complexes. Therefore, maintenance and transcription of the mtDNA are essential processes needed for efficient ATP production and to sustain normal function of most eukaryotic cells.
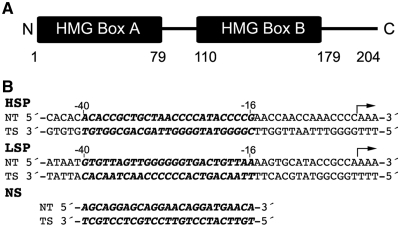
Mitochondrial transcription factor A (mtTFA or TFAM) is a transcription factor and a mtDNA-packaging protein (2,4,5). It regulates transcription, in part, by binding to mitochondrial transcription factor B2 (mtTFB2 or TFBM2) (6) to enhance initiation by mitochondrial RNA polymerase (POLRMT) (2,7,8). TFAM is a member of the high-mobility-group-box (HMG-box) family of DNA binding proteins. TFAM, after mitochondrial import and cleavage of the mitochondrial localization signal, comprises two ∼75-residue DNA binding domains (HMG boxes A and B), a 30-residue linker and a 25-residue basic C-terminal tail (Figure 1A). Box A is the dominant DNA binding domain of TFAM, as box B lacks the ability to bind DNA on its own (9). In addition to its interaction with mtTFB2, the C-terminal tail is required for efficient transcriptional activation by imparting sequence-specificity in binding to promoters (9–11). Through largely non-specific (NS) DNA-binding interactions, TFAM bends mtDNA to facilitate formation of nucleoid structures. These structures are the functional units of mtDNA with regard to transcription and replication and are also thought to provide protection from DNA damage (7,12–14). TFAM bends and unwinds DNA like other HMG-box proteins, and this feature is thought to be integral for both of its ascribed functions (1,4,5). However, the requirement and relative contribution of DNA-bending for these two different binding modes has not been addressed in detail.
Figure 1.
Sequence of TFAM and promoter DNA sequences. (A) Schematic diagram of TFAM. Numbers below the diagram delineate the residues that form HMG box A (1–79), the linker region (80–110), HMG box B (111–179) and the C-terminal tail (180–204). (B) Sequences of the HSP1 and LSP promoter DNA, as well as a protein-coding region of the mitochondrial genome (NS DNA). The 25 bp regions of the promoters that were used here and where TFAM binds (−40 through −16 upstream of the start site) are depicted in bold italics. The NT and TS indicate the non-template, and template strand, respectively. The arrows indicate the POLRMT start site and direction of transcription. For FRET experiments, LSP, HSP1 or NS DNA was 3′-labeled with TAMRA as the acceptor for the template strand, and 3′-labeled with FAM as the donor for the non-template strand.
HMG-box proteins comprise two families based on the specificity of their DNA-binding interactions [see (15–17)]. Sequence-specific (SS) DNA binding HMG-box proteins include transcription factor lymphoid enhancer factor -1 (LEF1), sex determining factor (SRY) and members of the SRY-like box (Sox) family of proteins. The non-sequence-specific (NSS) DNA binding HMG-box family includes vertebrate HMGB1 and HMGB2, Drosophila HMG-D, and the yeast proteins NHP6A and ABF2. Thermodynamic characterization of both modes of DNA interaction found that the SS HMG-box proteins bind to DNA with an enthalpy between 0 and 30 kJ mol−1, and NSS HMG-box proteins bind with an enthalpy of greater than 40 kJ mol−1, which indicates a completely entropy driven mode of DNA binding (18–21). Both modes of binding incur dramatic DNA bending. TFAM is unique in its ability to recognize mitochondrial promoters with sequence-specificity and non-promoter DNA in a NSS manner (11–13). It is not clear how TFAM accomplishes this, whether there is a unique thermodynamic signature for binding, or to what extent DNA bending occurs in these modes of DNA recognition.
Here we examined the ability of TFAM and a mutant lacking the C-terminal tail (TFAM 1–179) to bind and bend various types of DNA. We investigated the interaction of TFAM with 25 bp fragments of DNA comprising the human promoter sequences for heavy-strand promoter 1 (HSP1), the light-strand promoter (LSP) and a NS DNA fragment (Figure 1B). The binding and bending properties were correlated with the transcriptional activation capacity of TFAM in vitro.
MATERIALS AND METHODS
Preparation of DNA and proteins
The LSP, HSP1 and NS DNA sequences are shown in (Figure 1A) (22,23). Forward and reverse single-stranded DNA oligomers of 25 nt corresponding to the LSP DNaseI LSP footprint of TFAM (22), HSP1 TFAM binding region (22,23) and NS protein coding region of the mitochondrial genome were purchased from Eurofins MWG Operon and purified on C18 Sep-Pak cartridges (Waters). The DNA was then annealed in 150 mM NaCl and 10 mM Tris–HCl, pH 7.0, ethanol precipitated, and purified using reversed-phase HPLC [(S.C. Roemer, C.S. Malarkey, C.L. Wysoczynski, M.E.A. Churchill, manuscript in preparation]. Appropriate fractions were collected, ethanol precipitated, and dried in a Savant Speed Vac and resuspended in water. The purity of DNA fractions was assessed using 8% non-denaturing polyacrylamide gel electrophoresis (PAGE) (Supplementary Figure S1A), and 10% acrylamide 7 M urea denaturing PAGE (Supplementary Figure S1B). Fractions of DNA were then pooled and used for isothermal titration calorimetry (ITC) and circular dichroism (CD) experiments. Fluorophore labeled DNA was purified as described above, except DEAE anion exchange chromatography was used for the final purification, with a gradient from 0 to 1 M NaCl in standard TE buffer. Fractions were then ethanol precipitated and resuspended in 50 mM HEPES–Na, pH 7.4, 150 mM NaCl and 1 mM DTT.
Human TFAM and TFAM 1-179 were expressed and purified as previously described (9) (Supplementary Figure S1C). Human POLRMT and TFB2Δ30 (TFB2M lacking the mitochondrial localization sequence) were expressed and purified as previously described (8,25).
Circular Dichroism
CD experiments were performed as described previously (26). CD experiments used a Jasco J-815 CD spectrometer equipped with a Lauda Brinkman ecoline RE106 temperature bath and a 1 mm path-length quartz cell. Samples comprised 10 µM protein or 10 µM protein/DNA complex in 10 mM Na-phosphate buffer, pH 7.4 and 150 mM NaCl. For spectra containing the proteins, the buffer spectrum was subtracted. Although the contribution of DNA to the CD signal was small (Supplementary Figure S2B), spectra of the free DNA in buffer were subtracted from the spectra of the protein–DNA complexes. Spectra were collected from 250 to 195 nm at 20°C (average of six scans for one data set). Thermal denaturation experiments used 10 µM protein or 10 µM protein/DNA in 50 mM HEPES–Na, pH 7.4, 150 and 1 mM DTT. The ellipticity was monitored at 222 nm as the temperature was raised from 4 to 85°C and cooled from 85 to 4°C at a rate of 1°/min and corrected for the elipticity at 222 nm of DNA alone. Sigmoidal best-fit curves were applied to the measured values of ellipticity at 222 nm versus temperature, and the maxima of the derivatives of the best-fit curves were used to estimate the melting temperature of TFAM and TFAM 1-179 in the absence and presence of DNA.
Isothermal Titration Calorimetry
ITC experiments were conducted with a MicroCal VP-ITC. Protein and DNA samples were extensively dialyzed against 4 l of the same 50 mM HEPES–Na, pH 7.4, 150 mM NaCl buffer and degassed before the experiment. DNA at 40 µM was titrated in 10 µl injections to the cell containing the protein at a concentration of 10 µM. The raw heats of injection were measured while the cell was stirred at 300 r.p.m. at 20°C. As a control, DNA was titrated into buffer, and the heat evolved was subtracted from the heats for the protein/DNA injections. The heat of injection measured from the first titration point was discarded. Raw heats of injection were integrated with respect to time, and fit to a binding isotherm to calculate the ΔH of the reaction.
Fluorescence resonance energy transfer assays
Fluorescence resonance energy transfer (FRET) experiments were conducted using a Horiba Fluorolog 3 fluorescence spectrometer equipped with a temperature controller that was thermostated to 20°C. Buffers contained 50 mM HEPES–Na, pH 7.4, 150 mM NaCl and 1 mM DTT. The data were fitted using ligand depletion and cooperative binding models. Further details can be found in the Supplementary Data.
In vitro transcription assays
Run-off transcription assays (Figure 6) using a linearized DNA template containing HSP1 and LSP promoters were performed as described previously (8,27) in a total volume of 25 µl containing 10 mM Tris–Cl, pH 8.0, 20 mM MgCl2, 100 µM DTT, 100 µg/ml BSA, 400 µM ATP, 150 µM CTP, 150 µM GTP, 10 µM UTP, 0.2 µM [α-32P]UTP (3000 Ci/mmol) template DNA (3.4 nM) and 4 U of RNAseOut. Recombinant human transcription proteins were present at the following concentrations: POLRMT and TFBM2 (16 nM each) and TFAM or TFAM 1-179 (10 nM). The reactions were carried out at 32°C for 30 min, and then stopped by the addition of 22.5 µg Proteinase K in 10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 0.5% SDS, 100 ng/µl yeast tRNA and incubated at 42°C for 1 h. The radiolabeled transcription products were then precipitated in ethanol, dried, resuspended in gel-loading buffer and separated on 5% polyacrylamide/7 M urea gels. An end-labeled 10-bp ladder (Invitrogen) was used to estimate transcript sizes.
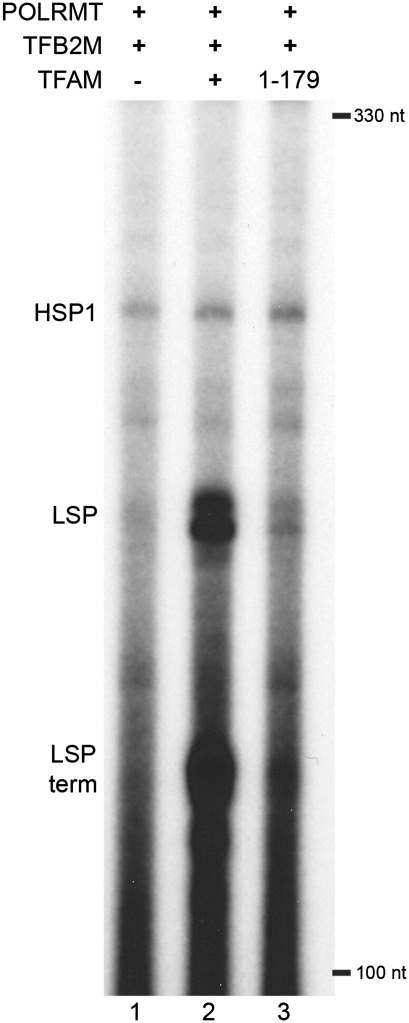
Figure 6.
In vitro run-off transcription reactions with TFAM and TFAM 1-179. Autoradiogram of in vitro run-off transcription products from the LSP3 template (8) using the proteins produced for this study. Transcription products from the mitochondrial promoters are indicated as HSP1, LSP and LSP-term. Each reaction used 85 fmol template (3.4 nM), 400 fmol POLRMT (16 nM), 400 fmol TFB2 (16 nM) and either no TFAM (lane 1), 250 fmol full-length TFAM (10 nM) (lane 2) or 250 fmol TFAM 1-179 (10 nM) (lane 3).
RESULTS
DNA stabilizes TFAM and TFAM 1-179 to thermal denaturation
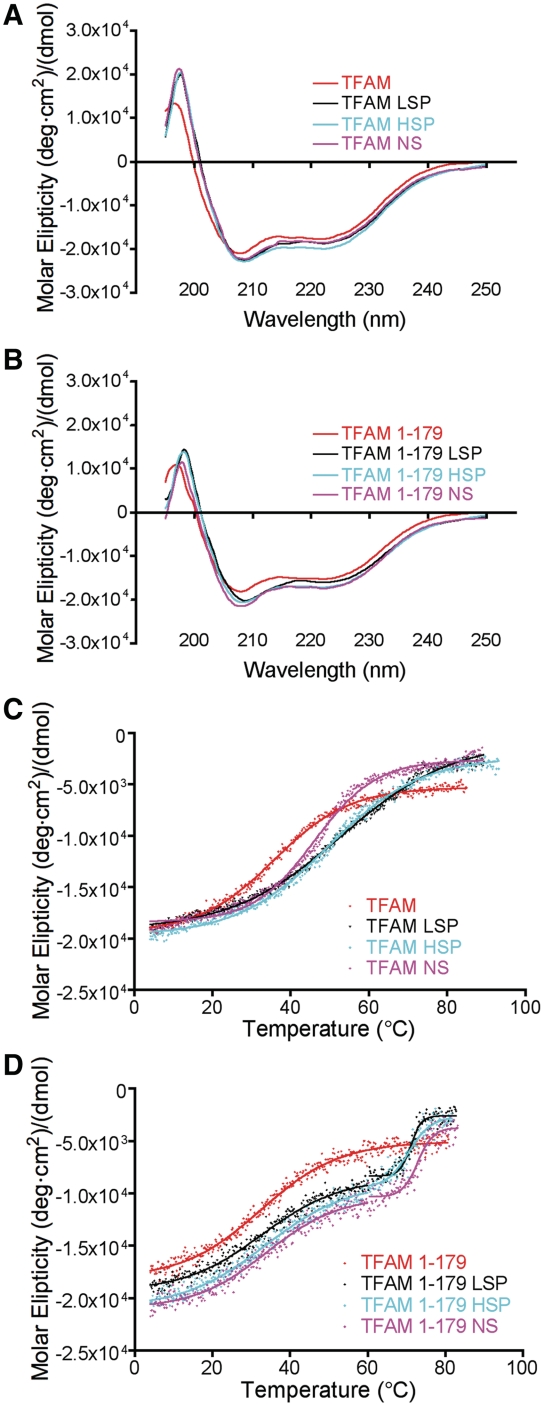
In the absence of DNA, human TFAM is partially unfolded at 37°C and becomes more structured upon binding to HSP1 promoter DNA (28). In order to compare the specific versus NSS binding properties of TFAM, we performed CD studies with 25 bp DNA fragments of the LSP, HSP1 and NS DNA (Figure 1B). TFAM and TFAM 1-179 have a high degree of α-helical content, which was slightly increased in the presence of LSP, HSP1 and NS DNA (Figure 2A and B). The percent change in α-helical content was similar for the LSP (4.4%) and NS (4.1%) DNA fragments and slightly greater for the HSP DNA (11.9%). Similar comparisons for TFAM 1-179, showed increases in α-helical content in the presence of LSP, HSP and NS DNA of 5.1, 12.7 and 14.3%, respectively. Thus, the secondary structure of TFAM appears to be similar in SS and NSS complexes and it is not altered by the presence of the TFAM C-terminal tail.
Figure 2.
CD analysis. (A) CD scans in units of molar protein ellipticity of 10 µM TFAM and (B) TFAM 1-179 in the absence (red) and presence of equimolar concentrations of 25 bp LSP (black), HSP1 (cyan), and NS (magenta) DNA sequences. Spectra represent an average of three independent experiments. The experiental variation at 222 nm was <1% for TFAM, TFAM–LSP, TFAM–HSP and TFAM–NS complexes and <2% for TFAM 1–179, TFAM 1-179–LSP, TFAM 1-179–HSP and TFAM 1-179–NS complexes. (C) CD thermal denaturing curves of TFAM and (D) TFAM 1-179 in the absence and presence of DNA colored as in panels A and B. Curves represent an average of three independent data sets for TFAM and two for TFAM 1-179.
The effect of DNA binding on the thermal stability of TFAM was measured using CD. Thermal denaturation of TFAM is a reversible process (Supplementary Figure S2A) with a melting temperature of 35.8°C, which is consistent with previous results (28). However, thermal denaturation of both TFAM and TFAM 1-179 in the presence of DNA was not reversible. Therefore, only the midpoints of the denaturation curves were available for the comparison. A single transition was observed for TFAM in the presence of DNA (Figure 2C), suggesting that either HMG-box A and HMG-box B unfold at similar temperatures, or unfold in a cooperative manner. All of the DNA fragments stabilized TFAM, with temperatures at each transition midpoint of 53.1°C for LSP DNA, 49.7°C for HSP1 DNA and 46.1°C for NS DNA (Figure 2C and Table 1). In the absence of DNA, TFAM 1-179 denatures with a melting temperature of 32.5 ± 0.3°C (Figure 2D). This is 3.3°C lower than TFAM, which suggests that the C-terminal tail stabilizes TFAM. Interestingly, thermal denaturation of TFAM 1-179 in the presence of DNA shows two distinct transitions (Figure 2D). The temperatures at the midpoints of these transitions were estimated to be 32.7 ± 0.4°C and 71.1 ± 0.2°C for LSP DNA, 33.7 ± 0.5°C and 70.7 ± 0.4°C for HSP DNA and 33.9 ± 0.5°C and 72.9 ± 0.3°C for NS DNA (Figure 2D and Table 2). The temperature of the lower melting transition was similar to the melting temperature of TFAM 1-179 in the absence of DNA. These results suggest that in TFAM 1-179, HMG-box A and HMG-box B may unfold independently of one another in the presence of DNA (Figure 2D). This is in contrast to the single melting transition observed for TFAM in the presence of DNA, and is consistent with the idea that the C-terminal tail of TFAM may interact directy with the DNA, HMG-boxes or both.
Table 1.
TFAM binding to DNA
| DNA | KDapp (M)a | ΔH (kJ/mol) | ΔG (kJ/mol) | TΔS (kJ/mol) | Tmb TFAM (°C) | Δ End to end distance (Å) | Bend angle (°) |
|---|---|---|---|---|---|---|---|
| LSP | 4.4 ± 0.3 × 10−9 | 77.0 ± 3.0 | −46.9 ± 2.8 | 124 ± 7.6 | 53.1 ± 0.3 | 22.7 ± 1.0 | 87 ± 2 |
| HSP1 | 7.4 ± 0.9 × 10−9 | 42.2 ± 4.0 | −45.6 ± 2.4 | 87.8 ± 11 | 49.7 ± 0.2 | 21.9 ± 1.9 | 87 ± 3 |
| NS | 7.4 ± 0.6 × 10−9 | 80.5 ± 3.2 | −45.6 ± 1.9 | 126 ± 9.5 | 46.1 ± 0.3 | 15.2 ± 1.6 | 72 ± 4 |
Table 2.
TFAM 1-179 binding to DNA
| DNA | KDapp (M)a | Tm1b TFAM 1-179 (°C) | Tm2c TFAM 1-179 (°C) | Δ End to end distance (Å) | Bend angle (°) |
|---|---|---|---|---|---|
| LSP | 9.8 ± 0.8 × 10−9 | 32.7 ± 0.4 | 71.1 ± 0.2 | 11.5 ± 0.5 | 62 ± 1 |
| HSP1 | 7.9 ± 0.8 × 10−9 | 33.7 ± 0.5 | 70.7 ± 0.4 | 12.1 ± 0.6 | 63 ± 1 |
| NS | 8.1 ± 1.7 × 10−9 | 33.9 ± 0.5 | 72.9 ± 0.3 | 10.4 ± 0.3 | 61 ± 1 |
aKDapp values were obtained by fitting the data in Figure 5B to a cooperative binding model.
bMelting temperature refers to the midpoint of the first non-reversible melting transition observed between 4 and 60°C (Figure 2D).
cMelting temperature refers to the midpoint of the second non-reversible melting transition observed between 60 and 85°C (Figure 2D).
TFAM binding to DNA is entropically driven with a large unfavorable enthalpy
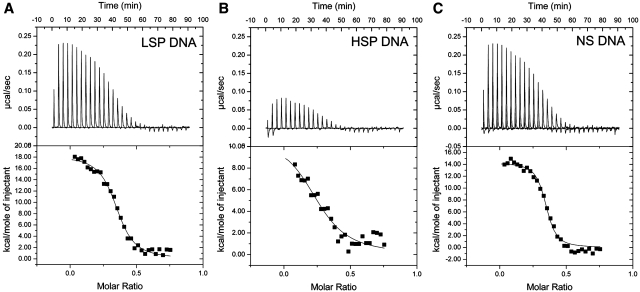
Previous studies of the thermodynamics of DNA binding for SS (19) and NSS HMG-box proteins (18,20) revealed unique thermodynamic signatures for SS versus NSS modes of DNA recognition. However, none of these proteins has the dual functions of TFAM. Since TFAM binds LSP and HSP1 DNA in a SS fashion and NS DNA in a NSS manner, we hypothesized that TFAM might bind LSP and HSP1 DNA with an enthalpy (ΔH) in the range of 0 to 30 kJ mol−1 consistent with SS HMG-box proteins, and bind NS DNA with a large positive enthalpy similar to NSS HMG-box proteins. The enthalpy for the association of TFAM with LSP, HSP1 and NS DNA was measured using ITC. As DNA was titrated into TFAM, the raw heats of injection and the ΔH values for the TFAM–DNA interactions at 20°C indicated a highly endothermic interaction for all three DNA fragments (Figure 3 and Table 1). Analysis of the integrated heats suggested a 2:1 TFAM to DNA binding stoichiometry consistent with previous studies on TFAM binding to LSP DNA (9,28) and NS DNA (28), and our findings below. These highly positive enthalpies are consistent with the positive enthalpies measured previously for NSS HMG-box proteins (18,20), and suggest that the specificity conferred by the C-terminal tail does not alter the underlying DNA binding properties of the HMG boxes.
Figure 3.
Thermodynamic analysis of TFAM binding to DNA. ITC analyses of TFAM binding to (A) LSP DNA, (B) HSP DNA and (C) NS DNA. The top panels show the raw heats absorbed from injecting a 10 µl aliquot of 40 µM DNA into a 10 µM solution of TFAM. The bottom panels show the integrated absorbed heats with respect to time with the heat of mixing subtracted. Molar ratio is DNA:TFAM.
TFAM binds to promoter and NS DNA with nanomolar affinity
A key feature of the function of TFAM in mitochondrial nucleoid formation is thought to be related to its ability to bend DNA (5,12). However, there is little known about the extent to which TFAM bends promoter and genomic mtDNA, or how important the C-terminal tail is for the process of DNA bending. Therefore, we measured the DNA binding and bending abilities of TFAM and TFAM 1-179 using FRET assays, which have previously been used to evaluate the DNA binding and bending abilities of SS and NSS HMG-box proteins (18,19). LSP, HSP1 or NS DNA was labeled with tetramethylrhodamine (TAMRA) as the acceptor and fluorescein (FAM) as the donor (Figure 1B). To measure FRET induced by protein binding to fluorophore labeled DNA, we employed the enhanced sensitization of the acceptor method described by Clegg (29). Titration of TFAM or TFAM 1-179 into each DNA sample produced a decrease in the intensity of the FAM donor signal at 520 nm and an increase in the TAMRA acceptor signal at 580 nm (Figure 4). The increase in FRET is due to a decrease in the distance between the fluorophores, which increases the efficiency of energy transfer through a non-radiative resonance mechanism. The increase in FRET is most likely due to protein-induced distortion of the DNA, which brings the fluorophores at the ends of the DNA closer together.
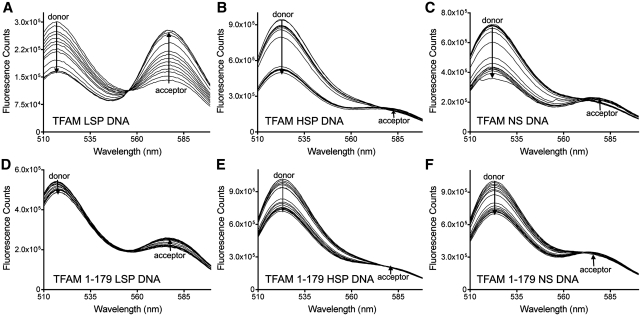
Figure 4.
FRET Measurements for LSP, HSP and NS DNA. Representative fluorescence emission spectra of 3.4 nM FAM/TAMRA labeled DNA with TFAM or TFAM 1-179 titrated as indicated, (A) LSP DNA with TFAM (0–23.8 nM), (B) HSP1 DNA with TFAM (0–40.8 nM), (C) NS DNA with TFAM (0–40.8 nM), (D) LSP DNA with TFAM 1-179 (0–40.8 nM), (E) HSP1 DNA with TFAM 1-179 (0–40.8 nM), (F) NS DNA with TFAM 1-179 (0–40.8 nM).
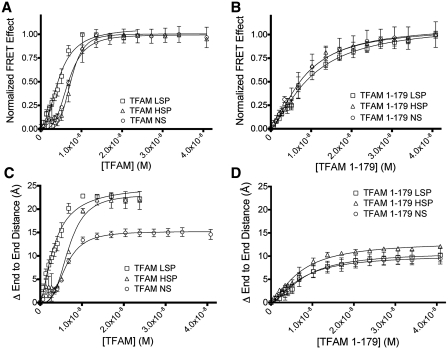
The binding of TFAM and TFAM 1-179 to the different DNA fragments was compared. As TFAM was titrated into the DNA, the relative change in the intensity of donor and acceptor emission was much greater for TFAM binding to LSP DNA (Figure 4A) compared to the TFAM 1-179 LSP interaction (Figure 4D), indicating that TFAM causes a greater decrease in the DNA end-to-end distance than TFAM 1-179. Similar results were found for HSP1 DNA (Figure 4B and E) and NS DNA (Figure 4C and F). However, only qualitative comparisons can be made between TFAM and TFAM 1-179 on the same DNA using the raw data (Figure 4), because the emission spectra of the FAM donor and TAMRA acceptor can differ depending on the DNA to which they are tethered (29). Accordingly, FRET effect (FE) values were calculated by dividing the intensity of the extracted acceptor signal at 580 nm (Supplementary Figure S3A and C–E) by the intensity of the acceptor at 580 nm when TAMRA is excited at 560 nm (Supplementary Figure S3B) (30). The plot of the normalized FE as a function of the concentration of protein gave binding curves and values for the apparent binding affinity (KDapp) of TFAM for LSP, HSP1 and NS DNA of 4.4, 7.4, and 7.4 nM, respectively (Figure 5A and Table 1). A Hill coefficient of ∼2 was measured for TFAM binding to all three DNA sequences. This highly cooperative binding is in agreement with previous studies (28). The ΔG values were calculated from the KDapp values (Table 1). The binding curves of the normalized FRET effect (Figure 5B) gave KDapp values for TFAM 1-179 binding to the LSP, HSP1 and NS DNA of 9.8, 7.9 and 8.1 nM, respectively (Table 2). The Hill coefficient for TFAM 1-179 binding to LSP, HSP1 and NS DNA was 1.5. These KDapp values are similar to previous measurements in this laboratory obtained using electrophoretic mobility shift assays, which gave KDapp values of ∼5 nM for TFAM and ∼10 nM for TFAM 1-179 binding to 30 bp LSP DNA (9). Together these results show that TFAM binds with slightly higher affinity to the LSP compared to the HSP1 and NS DNA, but TFAM lacking the C-terminal tail binds to all of the tested DNA fragments with an equivalent binding affinity. In addition, the C-terminal tail increases the cooperativity of TFAM binding to both SS and NS DNA.
Figure 5.
DNA binding and bending of TFAM and TFAM 1-179. Binding curves of FRET effect for 3.4 nM FAM/TAMRA labeled DNA titrated with (A) TFAM, and (B) TFAM 1-179. Binding isotherms were fitted to a cooperative binding model. The change in end-to-end distance for FAM/TAMRA labeled DNA titrated with (C) TFAM and (D) TFAM 1-179. End-to-end distances were calculated using Equation 4 in the Supplementary Data. The change in end-to-end distance is relative to the distance between the FAM and TAMRA in the absence of protein. The data are an average of three independent experiments, except for five for TFAM with NS and six for TFAM with LSP, with error bars showing the standard deviation.
The C-terminal tail of TFAM confers enhanced DNA bending to promoter, but not NS DNA
Although it has been shown that TFAM bends NS DNA (5,10,12) and promoter DNA (5), the degree of the distortion has been difficult to quantify. We further analyzed the FRET data to assess the change in the end-to-end distance between the fluorophores, which provides information on the degree to which TFAM bends DNA. TFAM binding to LSP and HSP1 DNA caused a decrease in the end-to-end distance of ∼22 Å and a smaller decrease of ∼15 Å for the NS DNA (Figure 5C and Table 1).
Lack of the C-terminal tail decreased the affinity of TFAM for LSP, and as reported previously (11), its transcriptional activation capacity at LSP and HSP1 in vitro (Figure 6), but it did not alter the binding affinity for NS DNA. This prompted us to determine the contribution of the C-terminal tail to the DNA bending ability of TFAM. TFAM 1-179 induced a change in the end-to-end distance of the DNA of ∼11 Å for the LSP, HSP1 and NS DNA (Figure 5D). This is a 2-fold decrease for the LSP and HSP1 DNA and a 50% decrease for NS DNA compared to the bending caused by full-length TFAM binding to the same DNA fragments (Figure 5C). Remarkably, TFAM 1-179 bends all three DNA fragments to a similar extent (Figure 5D and Table 2), which suggests that the C-terminal tail of TFAM facilitates DNA bending with all of the types of DNA that were tested. The larger differences in the change of end-to-end distance for the promoter DNA fragments (∼22 Å) versus the NS DNA (∼15 Å) (Figure 5C) suggests that the C-terminal tail plays an important role in specifically configuring the promoter DNA.
DISCUSSION
This investigation of the thermodynamic and biophysical properties of various TFAM–DNA interactions provides novel insights into the binding and bending properties of TFAM on both promoter and non-promoter DNA, as well as the role of the C-terminal tail of TFAM in facilitating transcriptional activation. Our interpretations of these findings relate to the relationship of TFAM to other HMG-box proteins and the mechanism of activation of human mitochondrial transcription.
TFAM DNA recognition in relation to other HMG-box and DNA binding proteins
Studies here and from the Fersht laboratory (28) indicated that in the absence of DNA, TFAM is largely unfolded at the human body temperature, with a Tm of near 37°C. Increased conformational flexibility allows for proteins to interact with a greater number of ligands than conformationally constrained proteins (31). This idea is consistent with the observations that TFAM binds to multiple promoter regions (11) and at apparently random sites along the mtDNA (12). Given that TFAM binds to multiple DNA sequences to function properly, existing in a partially unfolded state could be advantageous for the different modes of DNA recognition. In the presence of DNA however, TFAM adopts a slightly more ordered α-helical structure (Figure 2A). Whereas TFAM was stabilized substantially and nearly equivalently by the presence of LSP and HSP1 promoter DNA, it was stabilized less by the presence of the NS DNA (Figure 2C). Such an increase in thermal stability in the presence of DNA has been observed for other HMG-box proteins and has previously been correlated to the strength of the protein interaction with DNA (18,19). However, here, the degree of stabilization appears to be better correlated to the degree of DNA bending induced by TFAM than to the affinity of the TFAM–DNA interaction.
Interstingly, the CD studies with TFAM lacking the C-terminal tail provided unexpected insights into the importance of this region of the protein. First, the C-terminal tail does not contribute to the increased α-helical content that was observed in the presence of DNA (Figure 2). As, two regions of TFAM, the linker and the C-terminal tail (Figure 1A), are predicted to be unstructured in the absence of DNA, these results indicate that the increase in α-helical content is either due to increased α-helix formation within the HMG-boxes or in the linker region between the HMG boxes. In addition, the two melting transitions observed for DNA complexes of TFAM 1-179 suggest that the C-terminal tail aids in the formation of a cooperatively folded TFAM–DNA complex. The idea that the C-terminus is important for cooperativity is further supported by the FRET results (Figure 5A and B), where deletion of the C-terminal tail decreases the Hill coefficient from 2 to 1.5 for the SS and NSS DNA fragments. As a transcription factor and nucleoid protein, TFAM exhibits a high and rather narrow range of DNA binding affinities, compared to other DNA binding proteins. Previous work by the Fersht laboratory (28) using fluorescence anisotropy showed that TFAM binds 28 bp HSP1 DNA with moderate affinity (Kd = 68 nM) and cooperativity (Hill = 1.61) (28). However, our data, fitted with a Hill equation as in (28), gave a calculated KDapp value of 7.4 nM and a Hill coefficient of 2 for HSP1 DNA. We attribute this difference to the experimental temperatures, buffer conditions, methods, and choice of DNA sequence in the different studies. Despite these differences, TFAM binds well to all of the DNA fragments studied, including the NS DNA, for which there is only a slight decrease in binding affinity relative to the LSP DNA. This is in contrast to other NSS HMG-box proteins that bind to DNA with affinities in the range of 1–2 µM (HMGB1) (20), but is similar to the affinity of HMGD and NHP6A (18,19). The non-HMG NSS DNA binding protein HU binds dsDNA with an affinity of ∼25 µM, however this affinity is drastically increased to ∼8 nM for damaged DNA (32). TFAM has also been shown to bind damaged DNA with a higher affinity than dsDNA (33), however, not to the extent observed for HU (32).
The TFAM–DNA interaction is entropically driven
We originally hypothesized that TFAM would bind LSP and HSP1 DNA with a lower positive enthalpy than NS DNA, but our results showed that TFAM binds to promoter sites and NS DNA in an entropy-driven manner that is consistent with NSS, as opposed to SS HMG-box proteins (21–23). The enthalpy values summarized in Table 1 compare well to values of 77.9, 64 and 42 kJ mol−1, observed for the NSS proteins HMGB1 (20), HMGD (19) and NHP6A (19), respectively. They are not similar to the values of 12, 27 and 6 kJ mol−1, observed for LEF1, SRY and Sox5, binding to their specific DNA sequences, respectively (19,34). This result is not surprising for the TFAM–NS DNA interaction, because it has been shown to bind to protein coding regions of the mitochondrial genome with little sequence specificity (4,5,12).
HMG-box proteins bind to the minor groove of DNA, and the interaction is characterized by positive changes in enthalpy (21). Indeed, structural studies on Sox2 (35,36), SRY (37), HMGD (38) and HMGB1 (39) revealed extensive hydrophobic interactions in a highly bent and distorted minor groove of DNA (15). NS binding to the minor groove has also been observed for HU (40) and IHF (41), two non-HMG-box proteins that preferentially bind to AT-rich segments of DNA. On the other hand, most proteins that recognize the major groove of DNA have a favorable binding enthalpy (21). The high enthalpy value associated with binding to the minor groove is likely due to the displacement of highly ordered water molecules lining the minor groove of AT-rich sites in the DNA, as well as the energy required for DNA bending, which is not compensated by favorable bond formation (21,42). Indeed, HSP, which is the most G/C-rich DNA fragment has the lowest enthalpy. Our results (Figure 3) suggest that TFAM may make the closest interactions with DNA in the minor groove of AT-rich regions of the DNA fragments (21). The results are also consistent with the idea that the specificity imparted by the C-terminal tail does not alter the thermodynamic signature for NSS HMG-box–DNA recognition, and supports the conclusion that the C-terminal tail imparts specificity through another mechanism.
Impact of DNA bending on TFAM binding specificity and mitochondrial transcriptional activation capacity
The C-terminal tail of TFAM was known from previous studies to be important for SS DNA binding and the transcriptional activation capacity of TFAM (9,11), but the mechanism underlying its effect was unclear. Dairaghi et al. (11) showed that TFAM 1-179 had a severely reduced capacity to stimulate transcription from the LSP, and that TFAM mutants with increasing lengths of the C-terminal tail (TFAM: 1-184, 1-189, 1-194 and 1-199) had incrementally increased transcriptional activity. Furthermore, the yeast homolog of TFAM, ABF2, which lacks a C-terminal tail, has only modest transcriptional activation capacity (43), highlighting a unique transcriptional role for the tail in mammalian TFAM (44). Despite lacking a C-terminal tail, ABF2 has the ability to package and maintain yeast mtDNA (45). Interestingly, expression of TFAM 1-179 in HeLa cells maintains mtDNA architecture, but was not able to stimulate transcription (46), which further supports the idea that the C-terminal tail of TFAM is essential for activating mtDNA transcription.
Here, we confirmed that TFAM enhances transcription dramatically from the LSP and the C-terminal tail contributes significantly to this effect (Figure 6). Our finding that the C-terminal tail confers increased affinity and bending to the LSP promoter DNA suggests that tight binding and DNA bending are important features of the ability of TFAM to activate the LSP. The C-terminal tail increases binding affinity for LSP DNA by ∼2-fold compared to TFAM 1-179, but it does not significantly alter the binding affinity of TFAM for HSP1 or NS DNA. Surprisingly, HSP1, despite being bent to an equivalent degree, requires a higher concentration of TFAM to achieve significant activation by TFAM in vitro [Figure 6 and ref. (8)]. We conclude that activation of LSP by TFAM is much more dependent on DNA bending and potentially other distortions of the promoter than HSP1 and/or that the structural requirements at HSP1 are much different than LSP and perhaps involve other cis-acting sequences such as the inter-promoter region (8).
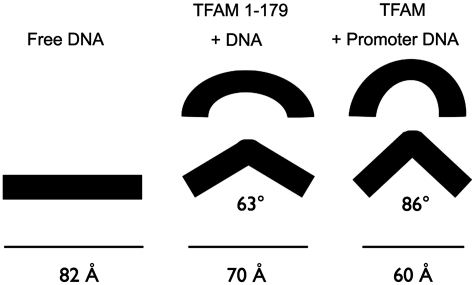
Given the importance of the C-terminal tail in transcriptional activation from the LSP, it was interesting to observe its important role in bending or distorting the DNA. Binding of TFAM had a distinctly greater effect on the distortion of the LSP and HSP1 sequences than on NS DNA, and loss of the tail resulted in a reduced and similar degree of DNA bending for all of the DNA fragments (Figures 4 and 5). Therefore, the C-terminal tail contributes to DNA bending of both the promoter and NS DNA. The degree of bending is difficult to estimate, but Figure 7 shows models for how the DNA either by kinking or smooth bending could achieve the end-to-end distances reported here. HMG-box proteins generally do not kink the DNA at a single site, but instead induce smooth bends and underwinding in the DNA (15), which could easily result in substantially greater overall bend angles than reported in Tables 1 and 2.
Figure 7.
DNA bending models. The estimated impact of TFAM-induced DNA bending visualized for the end-to-end distances for TFAM and TFAM 1-179 binding to LSP, HSP1 and NS DNA. Limiting FRET effect values were used for the estimation of bend angles. The calculated end-to-end distance for the LSP 25 bp DNA fragment is 82 Å. Assuming that TFAM induces a DNA kink, the bend angles corresponding to the end-to-end distances of 70 and 60 Å are shown. TFAM induces a greater bend in the DNA than TFAM 1-179. HMG-box proteins more typically create smooth bends in the DNA, and as illustrated, the apparent overall bend can be greater than the bend angles calculated from the kink-only model.
Other SS and NSS HMG-box proteins have N- or C-terminal tails, which are important for DNA bending and binding affinity. Structural analysis of the LEF1–DNA complex (47) and HMGD–DNA complex (48) show that each C-terminal tail, and in the case of NHP6A, the N-terminal tail (49), make contacts in the DNA major groove on the opposite side of the DNA from where the HMG-box docks in the minor groove (15). This configuration has been proposed to stabilize the highly bent form of the DNA, which in some cases has been bent by 110° (18,19,47,48). In the case of TFAM, we suggest that the interactions of the C-terminal tail with the bent DNA confers increased distortion of the DNA and potentially SS contacts with the promoter sequences, which allows TFAM to distinguish between SS promoter DNA and NS genomic DNA. Knowledge of the structure of TFAM–DNA complexes will be important for a complete understanding of the ability of TFAM to discriminate between promoter and NS DNA. We expect that differences in interactions of unstructured regions of TFAM with various aspects of the DNA structure (50), and possibly also subtle differences in DNA intercalation (26,51) will likely be important for DNA bending and specificity. No matter which detailed interactions lead to these differences, this study suggests that the specialized conformation of the DNA in the TFAM-promoter complexes facilitates the recruitment of the core mitochondrial transcription machinery and/or stimulation of other critical steps in transcription initiation at mitochondrial promoters.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3, Supplementary references [18,29,30].
FUNDING
Muscular Dystrophy Association #68859 and American Heart Association 11GRNT7380064 grants (to M.E.A.C.), NIH grant HL-059665 (to G.S.S.); James Hudson Brown-Alexander Brown Coxe Fellowship and NIH-NIDDK postdoctoral fellowship, F32DK091042 (to M.B.). Funding for open access charge: American Heart Association.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Wallace Liu, Catherine Musselman, Sarah Roemer and Jean Scorgie for critical reading of the manuscript. The authors also thank the Scott Pagan laboratory at the University of Denver for access to their ITC instrument.
REFERENCES
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scarpula RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 3.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am. J. Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher RP, Parisi MA, Clayton DA. Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes Dev. 1989;3:2202–2217. doi: 10.1101/gad.3.12b.2202. [DOI] [PubMed] [Google Scholar]
- 5.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 6.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill MEA. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohgaki K, Kanki T, Fukuoh A, Kurisaki H, Aoki Y, Ikeuchi M, Kim SH, Hamasaki N, Kang D. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J. Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- 11.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 12.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Murphy FVIV, Churchill MEA. Nonsequence-specific DNA recognition: a structural perspective. Structure. 2000;8:R83–R89. doi: 10.1016/s0969-2126(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 16.Teo SH, Grasser KD, Thomas JO. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex determining factor SRY. Eur. J. Biochem. 1995;230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 18.Dragan AI, Klass J, Read C, Churchill MEA, Crane-Robinson C, Privalov PL. DNA binding of a non-sequence-specific HMG-D protein is entropy driven with a substantial non-electrostatic contribution. J. Mol. Biol. 2003;331:795–813. doi: 10.1016/s0022-2836(03)00785-x. [DOI] [PubMed] [Google Scholar]
- 19.Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill MEA, Crane-Robinson C, Privalov PL. DNA binding and bending by HMG boxes: energetic determinants of specificity. J. Mol. Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Muller S, Bianchi ME, Knapp S. Thermodynamics of HMGB1 interaction with duplex DNA. Biochemistry. 2001;40:10254–10261. doi: 10.1021/bi0100900. [DOI] [PubMed] [Google Scholar]
- 21.Privalov PL, Dragan AI, Crane-Robinson C. The cost of DNA bending. Trends Biochem. Sci. 2009;34:464–470. doi: 10.1016/j.tibs.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 23.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy SM, Gilar M, Gebler J. Reversed-phase ion-pair liquid chromatography analysis and purification of small interfering RNA. Anal. Biochem. 2009;390:181–188. doi: 10.1016/j.ab.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 25.Lodeiro MF, Uchida AU, Arnold JJ, Reynolds SL, Moustafa IM, Cameron CE. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J. Biol. Chem. 2010;285:16387–16402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klass J, Murphy FVIV, Fouts S, Serenil M, Changela A, Siple J, Churchill MEA. The Role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong TS, Rajagopalan S, Freund SM, Rutherford TJ, Andreeva A, Townsley FM, Petrovich M, Fersht AR. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009;37:6765–6783. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 30.Stuhmeier F, Hillisch A, Clegg RM, Diekman S. Fluorescence energy transfer analysis of DNA structures containing several bulges and their interaction with CAP. J. Mol. Biol. 2000;302:1081–1100. doi: 10.1006/jmbi.2000.4089. [DOI] [PubMed] [Google Scholar]
- 31.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 32.Pinson V, Takahashi M, Rouviere-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, et al. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem. Biophys. Res. Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 34.Jelesarov I, Crane-Robinson C, Privalov PL. The energetics of HMG box interactions with DNA: thermodynamic description of the target DNA duplexes. J. Mol. Biol. 1999;294:981–995. doi: 10.1006/jmbi.1999.3284. [DOI] [PubMed] [Google Scholar]
- 35.Williams DC, Jr, Cai M, Clore GM. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 2004;279:1449–1457. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 36.Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 38.Murphy FVIV, Sweet RM, Churchill MEA. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stott K, Tang GS, Lee KB, Thomas JO. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 2006;360:90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 40.Serban D, Benevides JM, Thomas GJ., Jr HU protein employs similar mechanisms of minor-groove recognition in binding to different B-DNA sites: demonstration by Raman spectroscopy. Biochemistry. 2003;42:7390–7399. doi: 10.1021/bi030050r. [DOI] [PubMed] [Google Scholar]
- 41.Yang CC, Nash HA. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989;57:869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- 42.Privalov PL, Dragan AI, Crane-Robinson C, Breslauer KJ, Remeta DP, Minetti CA. What drives proteins into the major or minor grooves of DNA? J. Mol. Biol. 2007;365:1–9. doi: 10.1016/j.jmb.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diffley JF, Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- 45.Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural Basis for DNA Bending by the Architectural Transcription Factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 48.Dow LK, Jones DN, Wolfe SA, Verdine GL, Churchill MEA. Structural studies of the high mobility group globular domain and basic tail of HMG-D bound to disulfide cross-linked DNA. Biochemistry. 2000;39:9725–9736. doi: 10.1021/bi000723v. [DOI] [PubMed] [Google Scholar]
- 49.Allain FH-T, Yen Y-M, Masse JE, Schultze P, Dieckmann T, Johnson RC, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Churchill MEA, Travers AA. Protein Motifs that Recognize Structural Features of DNA. Trends Biochem. Sci. 1991;16:92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- 51.Churchill MEA, Klass J, Zoetewey DL. Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending. J. Mol. Biol. 2010;403:88–102. doi: 10.1016/j.jmb.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.