Abstract
The pathogenic bacterium Group A Streptococcus pyogenes produces several extracellular DNases that have been shown to facilitate invasive infection by evading the human host immune system. DNases degrade the chromatin in neutrophil extracellular traps, enabling the bacterium to evade neutrophil capture. Spd1 is a type I, nonspecific ββα/metal-dependent nuclease from Streptococcus pyogenes, which is encoded by the SF370.1 prophage and is likely to be expressed as a result of prophage induction. We present here the X-ray structure of this DNase in the wild-type and Asn145Ala mutant form. Through structural and sequence alignments as well as mutagenesis studies, we have identified the key residues His121, Asn145 and Glu164, which are crucial for Spd1 nucleolytic activity and shown the active site constellation. Our wild-type structure alludes to the possibility of a catalytically blocked dimeric form of the protein. We have investigated the multimeric nature of Spd1 using size-exclusion chromatography with multi-angle light scattering (SEC-MALLS) in the presence and absence of the divalent metal ion Mg2+, which suggests that Spd1 exists in a monomeric form in solution.
INTRODUCTION
Non-specific nucleases play broad biological roles in various cellular processes, such as scavenging of nucleotides and phosphates for cell growth, DNA repair and recombination, DNA fragmentation during apoptosis, host defence against bacterial invasion and notably establishment of infection (1). The human pathogen Group A Streptococcus pyogenes M1 (GAS) uses extracellular DNases to establish infection by degrading the DNA in neutrophil extracellular traps (NETs) (2). NETs are web-like DNA structures, built from chromatin, proteases and antimicrobial peptides that entrap and kill pathogenic bacteria (3). The DNase activity destroys the NETs and enables the S. pyogenes to adopt a more invasive and aggressive mode of growth, which may ultimately lead to life-threatening manifestations of streptococcal infection, such as necrotizing fasciitis (4).
It was not until the era of genomic sequencing that the degree of phage involvement in the transfer and regulation of streptococcal virulence factors was fully appreciated (5,6). In the S. pyogenes strain SF370, ∼10% of the genome was found to be prophage-encoded, making up four complete or partially integrated sequences (SF370.1, 370.2, 370.3 and 370.4) (7). Spd1 [formerly known as mitogenic factor 2 (MF2), streptodornase and DNase C (8)] is a type I extracellular DNase expressed by an inducible prophage SF370.1 Spd1 is a 28 KDa protein which possesses an N-terminal signal peptide, cleaved during extracellular secretion. It has an interesting genomic location, being situated adjacent to the SpeC toxin gene (Scarlatina toxin) (9). Spd1 and SpeC co-expression is likely to occur as a result of prophage induction. This has been demonstrated in the related CS112 strain (8,10), where after addition of soluble phage inducing factor (SPIF) secreted from pharyngeal cells, several virulence factors were induced (8,10). Another study on 568 emm28 GAS strains isolated from invasive and pharyngitis cases identified 29 distinct phage encoded virulence genes. Eighty-four percent of these strains contained both the Spd1 and SpeC toxin genes, making this pair quantitatively highly important (11). Other bioinformatic and phylogenetic evidence points to a recombination basis for this observation. This may be in part due to the frequent association of a so-called ‘paratox’ open reading frame adjacent to these virulence genes (12).
Structure
The main structural characteristic of this class of DNase is the presence of a ββα-metal (ββα-Me) finger motif (13). Examples for which the three-dimensional structure has been determined include: the site-specific homing endonuclease I-PpoI from Physarum polycephanum (14), the non-specific nuclease Sm from Serratia marcenscens (15), the Escherichia coli colicin E7 (ColE7) (16), the phage T4 endonuclease VII (17), the homing endonuclease I-HmuI (18), the non-specific endonuclease from Anabaena sp. (19), the Vibrio vulnificus endonuclease Vvn (20), an extracellular endonuclease VcEndA from Vibrio cholerae (21), the caspase-activated DNase CAD (22) and the EndA nuclease from Streptococcus pneumoniae (23). These proteins are structurally highly diverse, but all display a ββα-Me motif. The ββα-Me topology is comprised of two anti-parallel β-strands and an α-helix with a centrally located divalent metal ion. This ββα-Me contains the key residues essential for catalysis.
Here, we present the crystal structures of Spd1 DNase wild-type (WT) at 1.7 Å resolution and that of a Spd1 Asn145Ala mutant at 2.5 Å resolution. Structural analysis confirms Spd1 belongs to the ββα-metal finger nucleases, with a conserved RGH active site sequence motif. Mutagenesis studies and structure comparisons of Spd1 with other ββα-metal nucleases identify catalytically important residues and infer a DNA hydrolysis mechanism. The crystal structure of Spd1 Asn145Ala highlights the importance of Asn145 in the active site metal ion coordination and maintenance of activity.
EXPERIMENTAL PROCEDURES
Cloning, expression and purification
The Spd1 gene (NCBI ID: NC_002737.1) was amplified from S. pyogenes M1 SF370 (ATCC 700294) genomic DNA template and annealed into the pET-YSBLIC vector (24). Escherichia coli BL21 (DE3)-RIPL was transformed with the native Spd1 gene and expressed in the autoinduction media at 20°C for 18 h (25). Cultures were harvested and lysed using a high-pressure cell disruptor. Cell debris was separated from soluble fraction by centrifugation at 48 000 g. The Spd1 protein was purified by nickel-affinity chromatography, followed by anion exchange (MonoQ, GE Healthcare) and gel filtration chromatography. Pure protein sample was concentrated to 26 mg ml−1 in 25 mM Tris/HCl pH 7.5 and used in crystallization trials. Selenomethionine-labelled protein was produced in E. coli B834 (DE3) cells, in PASM-5052 Se-Met-labelling auto-induction media (25). SeMet Spd1 was purified following the protocol for the native protein.
Point mutations of the active site residues were introduced using Stratagene QuikChange® site-directed mutagenesis kit. Fourteen mutants (Asp88Ala, Arg90Ala, Lys92Ala, Asn118Ala, Arg119Ala, His121Ala, His121Asp, His121Asn, Gln126Ala, Met140Ala, Asn145Ala, Asn145Gln, Glu164Ala and Asn165Ala) were produced (see Supplementary Table S1 for primer sequences). The constructs were expressed and purified on a small-scale format. Briefly, the plasmids were transformed into E. coli BL21 (DE3) strain and grown in 50 ml of LB media until OD600 reached 0.6–0.8. After that cultures were induced with IPTG and grown for another 3.5 h. Cellular pellets were harvested by centrifugation and resuspended in 1 ml of BugBuster® Protein Extraction Reagent (Merck) and incubated for 30 min on ice. Cell suspensions were spun down and loaded onto disposable Ni-NTA Spin columns (Qiagen). WT Spd1 was included as positive control and cells transformed with empty pET-YSBLIC vector were used as a negative control.
Crystallization and data collection
The Spd1 crystals were grown in 24-well Linbro plates using a hanging drop vapour diffusion method. A total of 0.5 µl of the protein was mixed with 0.5 µl of the mother liquor, containing 0.2 M sodium citrate, 0.1 M bis–Tris propane 7.5 and 20% PEG 3350. Selenomethionine-derivative crystals grew in a 96-well MRC crystallization Plate™ (Molecular Dimensions Ltd) set-up by Mosquito Nanodrop crystallization robot (Molecular Dimensions Ltd). A total of 100 nl of the protein was mixed in 1:1 ratio with the reservoir solution, containing Malic/MES/Tris system pH 6.0 and 25% PEG 1.5 K. The crystals were cryoprotected in the mother liquor solution by the addition of 25% glycerol, vitrified in liquid nitrogen and tested for diffraction. The native crystal diffraction data were collected to 1.7 Å at the ESRF, Grenoble on beamline ID14-1 at a temperature of 100 K at a wavelength of 0.9334 Å and processed using Mosflm (26) and Scala (27). Crystals belong to space group P3221 with cell dimensions: a = b = 76.4 Å, c = 80.4 Å, α = β = 90°, γ = 120°.
Selenomethionine labelled crystals diffracted to 1.9 Å at Diamond, UK, on beamline I03 at a temperature of 100 K and a SAD data set at a wavelength of 0.9764 Å was collected. The data were processed using Mosflm (26) and Scala (27). Spd1 Se-Met crystals are isomorphous with the native crystals, with cell dimensions a = b = 80.4 Å, c = 78.9 Å, α = β = 90°, γ = 120°. The unit cell volume indicates one molecule in the asymmetric unit, with a solvent content of 53%.
Using both the native and SAD data, four selenium sites were found by SHELXD embedded in the AUTOSHARP pipeline (28), which was also used to perform phase calculations and density modification. The resulting phases were of sufficient quality for ARP/wARP (29) to build nearly all of the model. The model was refined by subsequent cycles of model building and refinement in COOT (30) and REFMAC5 (31).
Data for the Spd1 Asn145Ala mutant were collected using a Rigaku Micromax-007HF generator equipped with Osmic multilayer optics and a MAR345 imaging plate detector to 2.5 Å resolution. Data were processed using Mosflm (26) and reduced in Scala (27). Crystals belong to space group P212121 with cell dimensions a = 49.8 Å, b = 101.8 Å, c = 113.5 Å, α = β = γ = 90°. The structure was solved by molecular replacement with Phaser (32) using the coordinates of the WT structure as a search model. The solvent model was built in ARPWARP (29). The model was corrected manually in COOT (30) and refined further in REFMAC5 (31). The structures were validated by PROCHECK (33) and MOLPROBITY (34). The crystal diffraction data and refinement statistics are presented in Table 1. The coordinates and structure factors were deposited at the Protein Data Bank in Europe (http://www.ebi.ac.uk/pdbe/), the structure validation can be seen in Supplementary Table S3. Structure figures were drawn with PyMOL (35) and CCP4mg (36).
Table 1.
Spd1 and Asn145Ala mutant crystallographic data collection and refinement statistics. Values in parentheses are for the outer shell
| Native | SeMet | Asn145Ala | |
|---|---|---|---|
| Data processing | |||
| Space group | P3221 | P3221 | P212121 |
| Unit cell dimensions (Å) | a = 76.4, b = 76.4, c = 80.4 | a = 77.1, b = 77.1, c = 80.9 | a = 49.8, b = 101.8, c = 113.5 |
| Unit cell angles (°) | α = β = 90, γ = 120 | α = β = 90, γ = 120 | α = β = γ = 90 |
| Molecules in AU | 1 | 2 | |
| Wavelength | 0.93400 | 0.97640 | 1.54179 |
| Resolution range (outer shell) | 51.10–1.70 (1.79–1.70) | 66.81–1.88 (1.95–1.88) | 50.92–2.48 (2.61–2.48) |
| Rmerge | 0.090 (0.461) | 0.076 (0.34) | 0.056 (0.300) |
| <I/σI> | 19.8 (5.3) | 37.9 (6.9) | 17.5 (4.5) |
| Completeness | 100 (100) | 99.9 (98.6) | 90.3 (82.5) |
| Redundancy | 10.7 (10.8) | 17.4 (14.0) | 3.9 (3.6) |
| Refinement statistics | |||
| Resolution range (Å) | 38.21–1.70 (1.74–1.70) | 50.92–2.48 (2.55–2.48) | |
| Rcryst | 0.168 | 0.166 | |
| Rfree | 0.208 | 0.244 | |
| No. protein atoms | 1668 | 2993 | |
| RMSD bonds (Å) | 0.018 | 0.019 | |
| RMSD angles (°) | 1.782 | 1.831 | |
| Mean B-value protein atoms (Å2) | 14 | 37 | |
| Mean B-value solvent atoms (Å2) | 29 | 39 | |
| ESU (Å2) | 1.686 | 6.637 | |
| Ramachandran statistics | |||
| Preferred regions (%) | 98.86 | 93.12 | |
| Allowed regions (%) | 1.14 | 5.44 | |
| Outliers (%) | 0 | 1.43 | |
| PDB code | 2XGR | 2XH3 |
DNase activity assays
Protocols for nuclease activity experiments were adapted from Broudy et al. (8) and Aziz et al. (37). DNase activity was assayed by incubating 1 µg of calf thymus DNA, plasmid DNA and RNA in 100 mM Tris/HCl pH 7.0–8.0, 1 mM CaCl2 and 1 mM MgCl2 with 0.35 µg of Spd1 and Spd1 mutants at 37°C for 45 min. The reaction was stopped by addition of 20 mM ethylenediaminetetraacetic acid (EDTA). Duplicates of each reaction with 20 mM EDTA were used as negative controls as was a cell-free extract produced from an ‘empty’ LIC expression construct. DNA degradation was visualized on 1.2% agarose gel.
Sequence alignment
Sequence alignments of available structural homologues were performed using the Multialign program (38).
Size-exclusion chromatography with multi-angle light scattering
About 100 μl of WT Spd1, Spd1 Asn145Ala and Spd1 Glu164Ala at concentration of 0.5 mg ml−1 samples were injected and separated on a BioSep-SEC-S 3000 column (Phenomenex) equilibrated in 25 mM Tris pH 8.0, 150 mM NaCl at room temperature, at 1 ml/min. To determine the effect of Mg2+ ion concentration on the quaternary structure of WT Spd1, gel filtration buffers were prepared containing 150 mM NaCl with the addition of 0, 2 and 50 mM MgCl2. A control buffer where the MgCl2 was replaced by 10 mM EDTA was also included in order to chelate trace metal ions. Light scattering data were recorded on an in-line Dawn Heleos II 18-angle light scattering detector with an in-line Optilab rEX refractive index monitor (both from Wyatt Technology). Data were analysed by ASTRA software and fitted to the Zimm model with an estimated dn/dc value of 0.183 ml/g (39).
Determination of haplotonic or diplotonic mechanism
Analysis of plasmid DNA states during cleavage by Spd1 at a concentration of 0.01 mg ml−1 in 20 mM HEPES at pH 7.4, 5 mM MgCl2 and 1 μg of pUC19 plasmid DNA; 20 μl aliquots were taken from the reaction mixture at the indicated time points, and the reaction stopped by the addition of DNA to the loading buffer. Digested products were analysed by electrophoresis in 1% agarose gel, and stained with SYBR® Safe (Invitrogen) after running. Untreated control (reaction mixture before the addition of enzyme) and EcoRI linearized pUC19 were included.
Continuous assays
DNase activity was assayed by a continuous method based on the differential fluorescence output of a DNA dye called PicoGreen® (Invitrogen) as described by Tolun et al. (40)
Preparation of DNA substrate
The plasmid vector pUC19 was linearized with EcoRI (Fermentas) and used as a homogeneous substrate for the continuous assay. The linearization was verified by agarose gel electrophoresis, and the restriction endonuclease inactivated in accordance with the manufacturer's instructions (heat denaturation at 65°C for 20 min).
Measurement of fluorescence
Fluorometer measurements were taken using a Fluoromax-4 spectrofluorometer set to the following parameters: excitation wavelength, 484 nm; emission wavelength, 522 nm; slit width (all slits), 2 nm; data sampling frequency, 1 s ± 1; assay period, 600 s. Reaction conditions: all reactions were carried out at the temperature inside the spectrofluorometer of 25°C. Reaction mixtures (0.5 ml) contained 20 mM HEPES pH 7.4, 5 mM MgCl2 and linearized pUc19 at a concentration of 400 ng/ml. PicoGreen® (Invitrogen) was diluted in the reaction buffer and added to the mixture at a dilution of 1:2000. The reaction mixture was placed in a Quartz cuvette and incubated in the spectrofluorometer with continuous stirring for 5 min without light. The data collection was started and the reaction indicated by the addition of 10 μl enzyme after 100 s. The Spd1 concentration was adjusted by a series of two dilutions and tested at 2.5, 1.25 and 0.620 mg/ml. DNase I (from bovine pancreas, Sigma cat. no. D56K7680) was used as a positive control at a concentration of 1 mg/ml. As a negative control Spd1 at a concentration of 2.5 mg ml−1 was assayed in the presence of 5 mM EDTA.
Chemical cross linking reactions
Crosslinking reactions were carried out using Dimethyl adipimidate (DMA) and Dimethyl suberimidate (DMS).To determine the possible effect of combinations of Mg2+, NaCl and EDTA on the multimeric nature of WT Spd1, cross-linking experiments were performed. DMA and DMS stock solutions were freshly prepared at a concentration of 6 mg ml−1 and the pH was adjusted to 8.5 using NaOH. The reaction was carried out in a volume of 250 μl in 20 mM HEPES buffer (pH 7.4) with DMA and DMS at a final concentration of 1.5 mg ml−1 with a WT Spd1 protein concentration of 0.5 mg ml−1. Varying combinations of MgCl2, NaCl and EDTA were tested in a matrix over the following concentration ranges: MgCl2 0, 5, 10 and 50 mM, NaCl 0, 50, 150, 500 mM and EDTA 10 mM. The cross-linking reactions were incubated for 3 h at room temperature and then stopped by the addition of SDS denaturing sample buffer. Samples were analysed using SDS–PAGE with a 12% gel.
RESULTS
Gene cloning and expression
The expression of the catalytically active WT Spd1 DNase was achieved in E. coli. Cellular toxicity was not found to be problem as in previous studies (23,41). The expression construct incorporated the streptococcal secretion signal, which was cleaved by E. coli during expression, this was confirmed by electrospray mass spectrometry.
Spd1 structure determination
The WT Spd1 structure was solved by conducting a SAD experiment using the data shown in Table 1. The WT Spd1 sequence was modelled from residue Arg33 to Pro250 out of the 223 residues expected after the cleavage of the signal peptide between Ala29 and Arg30, according to the SignalP server (42). The region from residue Asn94 to Ser114 is disordered and there is no interpretable electron density for these residues. There are eight polyethylene glycol side chains modelled in the structure. The final crystallographic R factors for all reflections are 20% (Rfree) and 16.8% (Rcryst).
The Spd1 Asn145Ala inactive mutant crystallized in a different (P212121) space group from the WT Spd1. The structure was refined to crystallographic R factors of 16% (Rcryst) and 24% (Rfree) and built at a resolution of 2.5 Å with two molecules in the asymmetric unit. A larger proportion of the Spd1 Asn145Ala structure is disordered than of the WT structure. The model was built from Thr48 to Asn252 out of 253 residues. Residues from Lys87 to Lys112 are also missing from the density.
Structure characteristics
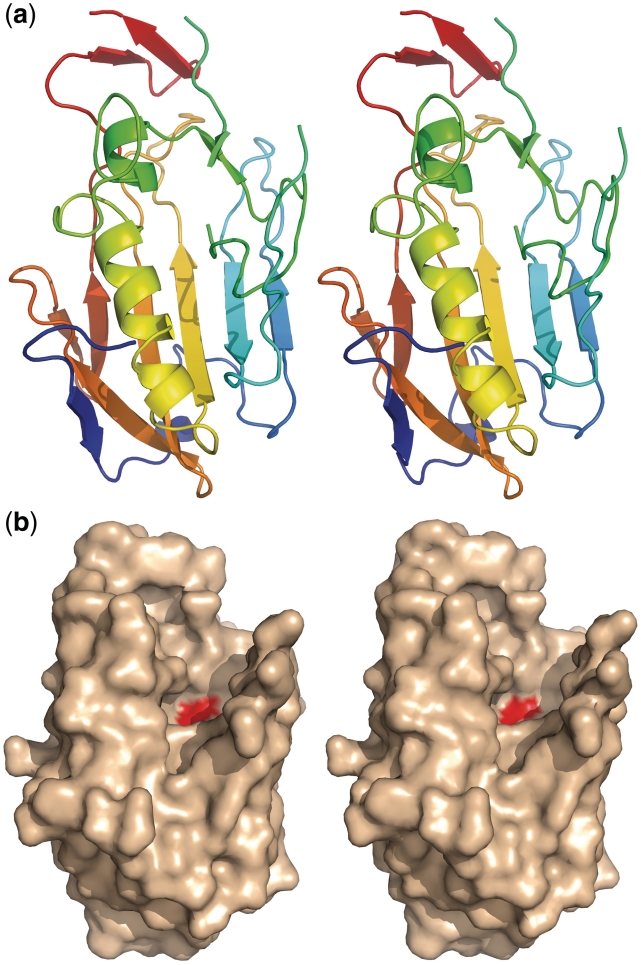
Spd1 displays a mixed α/β-fold, with 12 β-strands and 2 α-helices. Figure 1a shows the constituent secondary structure of the Sdp1 protein. A series of β sheets run anti-parallel around the main α2 helix (green to yellow). These are β2–β3 (light blue to turquoise) β6–β7 (yellow to orange) β10–β9 (orange to light red). The N- terminal β1 and β8 (deep blue and light red) lie toward one end of the main α2 helix. The β-sheets 4 and 5 (green) lie adjacent to the smaller α1 helix; these motifs would harbour residues involved in metal ion coordination and catalysis. Figure 1b shows the position of the active site histidine (red). The DNA binding region can be seen as a major groove sculpted within the structure (Figure 1b.) The dimensions of the cavity are ∼13 × 11 × 8 Å.
Figure 1.
(a) Colour ramped N- to C-terminal (ramped blue to red) cartoon representation showing the secondary structure of Spd1. The following colours correspond to the secondary structure features in the text β1–β2 deep to light blue, β3–β5 turquoise to green, α1–α2 green to yellow, β6–β8 yellow to orange and β9–β12 light red to deep red. (b) Modelled surface of the WT Spd1 structure in the same orientation as (a), showing the position of the catalytic histidine His121 (red).
Nuclease activity
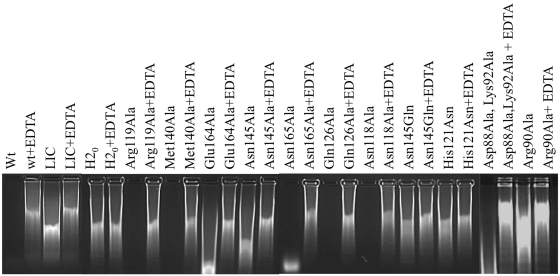
WT Spd1 was shown to degrade both single- and double-stranded DNA as well as RNA. When purified samples of Spd1 were assayed in the presence of calf thymus DNA and RNA and run on agarose gels, a complete degradation of the nucleic acid was observed after 45 min of incubation. The nuclease activity was also assessed by the measurement of absorbance at 260 nm. In this case, the complete degradation of the sample was observed during a time course of 40 min after the addition of enzyme, Figure 2. The digestion experiments were carried out in the presence of magnesium and calcium ions as well as in the presence of EDTA. We observed that the presence of metal is crucial for activity and ion chelation by the EDTA completely inhibited the reaction.
Figure 2.
Spd1 and Spd1 mutants assayed in the presence and absence of EDTA; visualized on 1.2% agarose gel. A LIC control has been included to account for ‘background DNAse activity’ (see ‘Experimental Procedures’ section) and H2O as a negative control.
DNA activity was also measured for Spd1 active site mutants. Arg90Ala, His121Ala, His121Asp, His121Asn, Asn145Ala, Asn145Gln and Glu164Ala substitutions inactivated the Spd1 nuclease. The Asp88Ala, Lys92Ala, Asn118Ala, Arg119Ala, Gln126Ala, Met140Ala and Asn165Ala mutants were found to be catalytically active Figure 2.
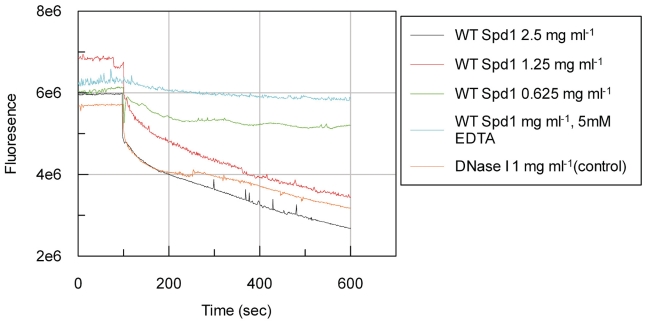
Continuous assay
WT Spd1 was assayed by continuous means (Figure 3) on a linearized pUc19 substrate together with a EDTA and DNase I control. Activity is indicated by a decrease in fluorescence, due to the dissociation of PicoGreen® dye when the DNA is digested. A decrease in fluorescence was observed for all three dilutions of the WT Spd1 and the positive control indicating activity. This activity was inhibited at the highest enzyme concentration of WT Spd1 by the inclusion of 5 mM EDTA in the reaction buffer.
Figure 3.
The activity of Spd1 on 400 ng/ml linearized pUC19 substrate. Activity can be monitored by a decrease in fluorescence due to the dissociation of PicoGreen® dye when the DNA is digested. WT Spd1 was tested over a series of 2-fold dilutions. DNase I was used as a positive control. The inclusion of EDTA at a final concentration of 0.5 mM inhibited 2.5 mg ml−1 WT Spd1.
Size-exclusion chromatography with multi-angle light scattering
To determine the oligomeric state of Spd1 and Spd1 active site mutants in solution (25 mM Tris pH 8.0) size-exclusion chromatography with multi-angle light scattering (SEC-MALLS) experiments were carried out. Molecular weights were calculated by ASTRA software (Wyatt Technologies) by splitting the chromatogram into two ‘slices’. The first slice covers the maximum of the peak (Peak 1) and the second one covers the whole chromatogram peak (Peak 2). Average molecular weights were calculated for each ‘slice’ separately. The average calculated molecular weights for the native Spd1, Spd1 Glu164Ala, Spd1 Asn145Ala, in the presence and absence of MgCl2 and 10 mM EDTA are in the range of 24–28 kDa, suggesting that Spd1 exists as a monomer in the solution, as the estimated mass of Spd1 is 28 kDa (Supplementary Table S2).
Determination of haplotonic or diplotonic mechanism
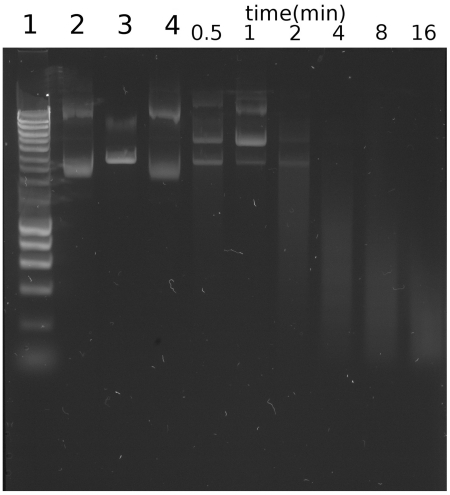
To determine whether the enzyme operates via a haplotonic or diplotonic mechanism (induces a single- or double-strand break in DNA), we performed a timed assay on plasmid DNA (pUc19) using a dilute preparation of Spd1. It can be seen (Figure 4) that the digestion of plasmid DNA proceeds via a single strand nick, mediating relaxation of supercoiled DNA. This can be seen to emerge 30 s to 1 min into the reaction, within the same time frame a linear form of the vector also emerges as both species occur simultaneously. This points to a more complex mechanism as observed in enzymes such as bovine DNase I (43). This digestion has been shown to pass through three phases, (i) relaxation of the plasmid (ii), partial linearization of the relaxed plasmid and (iii) plasmid degradation and the formation of low-molecular weight products (43).
Figure 4.
Observation of the pUC19 plasmid DNA states during cleavage by Spd1. Aliquots were taken from the reaction mixture at the indicated time points, and digested products were analysed by electrophoresis in 1% agarose gel, staining with SYBR® Safe (Invitrogen). Control lanes 1: Hyperladder I (Bioline); Lane 2: Untreated pUC19; Lane 3: EcoRI linearized pUC19; Lane 4: Reaction mixture prior to the addition of Spd1. The digestion of plasmid DNA proceeds via single-strand nick mediated relaxation of supercoiled DNA, this can be seen 30 s to 1 min into the reaction; within the same time frame a linear form can also be seen emerging.
Spd1 cross-linking
The effects of MgCl2, NaCl and EDTA on WT Spd1 were assessed in a combination matrix in the presence of the cross-linking reagents Dimethyl suberimidate and Dimethyl adipimidate. On analysis of the SDS–PAGE gels (results not shown) it was found that no cross-linking was observed in any of the conditions tested, with the protein band corresponding to 25 kDa, consistent with the size of the monomeric Spd1 protein.
DISCUSSION
Quaternary structural characteristics
The family of ββα/metal-dependent nucleases contain enzymes showing both monomeric and dimeric quaternary structures. Using light scattering and structural studies Miller and Krause (44) showed the Serratia nuclease (PDB code 1SMN) to be a homodimer composed of two polypeptide chains linked through the C-terminal sub domain by a series of hydrogen bonds and salt links at the dimer interface. The two active site regions are accessible by substrate and are located on opposite sides of the dimer and function independently. Using a combination of point mutagenesis and chemical cross-linking at the dimer interface Franke et al. (45) were able to produce and assay monomeric and dimeric forms of the Serratia nuclease and these were compared to the monomeric Anabaena nuclease. This study concluded that dimerization is required for the processive degradation of nucleic acids, but not for the diplotonic (double-stranded) cleave of DNA (45).
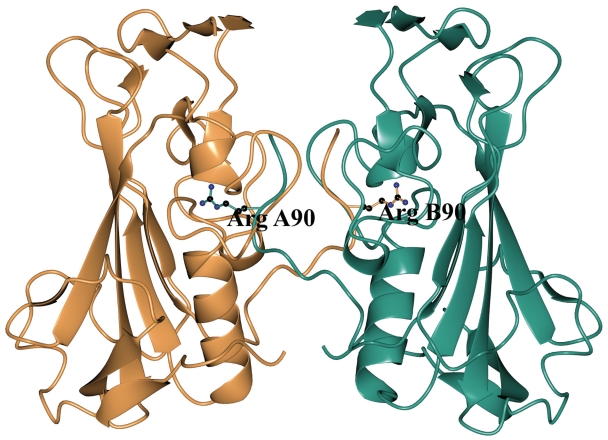
The Spd1 WT protein crystallized in a dimeric form, through a series of interactions made by a loop Lys 87–Lys 92, which protrudes into the active site cleft (Figure 5). This dimerization is unlike the Serratia nuclease, as interactions are made between different areas of the protein. In addition to this the active site is blocked by Arg 90 from an adjacent symmetry related molecule (Figure 5). This residue appears to have occupied the site associated with the active site metal ion. This has resulted in a ‘self inhibiting’ dimer and shows a structural resemblance to the Nuclease A-Inhibitor Complex (46). To investigate this further and rule out the possibility of a naturally occurring self inhibiting dimeric form of the protein we submitted the model to the PISA Interfaces analysis http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html. This resulted in a complexation significance score of 0.977 indicating that this observation is not a crystallographic artefact. To observe the multimeric nature of Spd1 in solution, we performed a series of SEC-MALLS experiments over a range of Mg2+ ion concentrations and in EDTA (see Supplementary Data).
Figure 5.
The Spd1 WT protein shown in dimeric form, a series of interactions made by Lys 87–Lys 92, which protrudes blocking the active site cleft. Arg90 (ball and stick representation) has displaced the active site metal ion. The A molecule is shown in teal blue and the B molecule in salmon pink.
These results indicated that the WT Spd1 protein is monomeric in solution irrespective of the presence of EDTA or Mg2+ ions. We have shown the WT Spd1 to be active in solution (Figure 3), and this activity to be dependent on the presence of Mg2+. Our Spd1 WT dimeric structure is devoid of metal ions and so cannot be the active species we observe in our activity assays. This does not rule out the possibility of Spd1 dimerizing in another way, perhaps on association with DNA. We have observed that Spd1 is able to cleave plasmid DNA through relaxed and linear intermediates (Figure 4) similar to DNaseI (43). Production of the linear form could involve either the sequential presentation of each strand of DNA to a monomeric enzyme or a dimeric enzyme orientated to cleave both DNA strands simultaneously. Our cross-linking studies are consistent with a monomeric enzyme, but proved inconclusive as we were unable to cross-link WT Spd1 under the conditions tested. This does not prove that Spd1 is active as a monomer, given that a dimer that is unable to cross-link would also give this result.
The structure of the Asn145Ala mutant shows an active site free from symmetry-related molecules. The effect of mutation appears to have disrupted metal ion coordination, leaving this structure again devoid of an active site metal ion. This is consistent with the proposed role of the active site Asn in other characterized DNases (47). Our activity assays have indicated this residue is required for the maintenance of activity.
Comparison with other nucleases
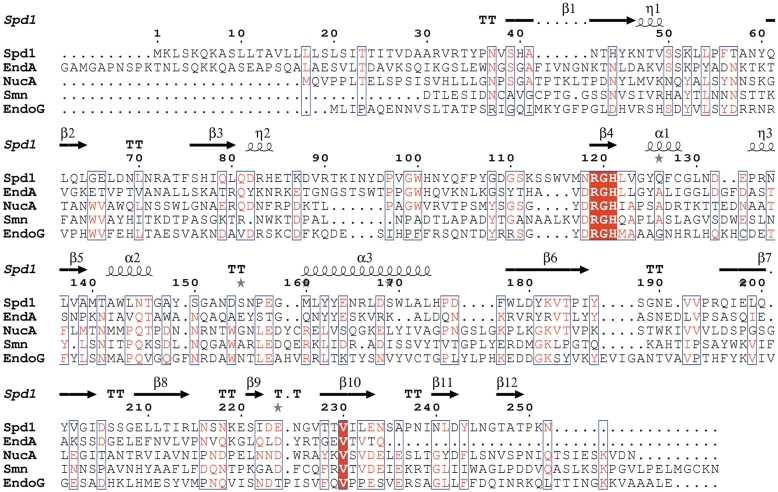
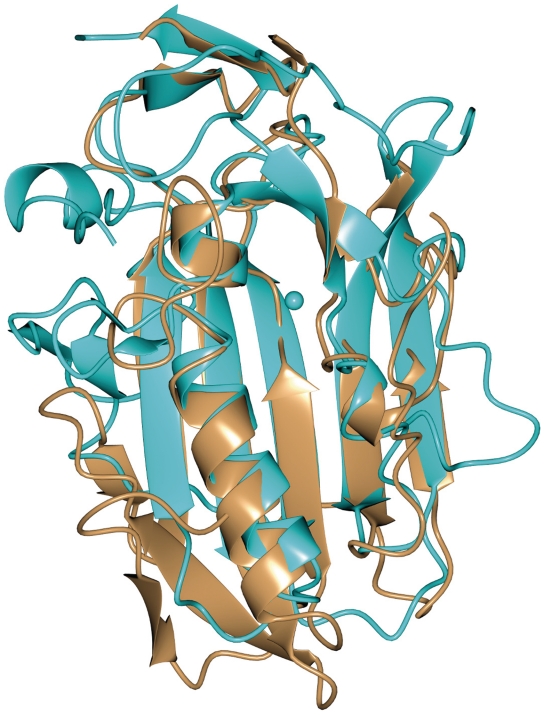
The closest structural homologues related to Spd1 are EndA from S. pneumoniae, with a Z-score of 7.8 and rmsd of 1.8 Å, based on SSM server search (48) (PDB code 3OWV), followed by NucA from Anabaena sp. (Z-score 6.4, rmsd of 2.1 Å, PDB code 1ZM8), Sm endonuclease from S. marcenscens (Z-score of 5.8 and rmsd of 1.9 Å, PDB code 1QL0) and EndoG endonuclease (Z-score of 5.6, rmsd of 2.3 Å, PDB code 3ISM). Despite a low sequence similarity between these proteins, there is strong conservation of the DRGH (Figure 7) sequence motif; in the Spd1 sequence there is an Asparagine (Asn119) instead of Aspartic acid (Figure 7). The structural similarity between these nucleases is mostly pronounced in the β-strands forming the central sheet and in the ββα-Me motif comprised of two short β-strands and a long α-helix running along the central β-sheet. Further away from the catalytic centre Spd1 and EndA share the highest resemblance in their secondary architectures (Figure 6). They both contain a small C-terminal β-sheet composed of two anti-parallel strands, which are situated on the top of the central core of the structure. The disordered loop of Spd1 (from Asn94 to Ser114) corresponds to the missing region in EndA (Tyr124 to Ser136). The proximity of this loop to the putative DNA binding site and mutagenesis studies in EndA suggest the residues located in this region may synergistically bind the DNA. Another feature characteristic to Spd1 and EndA is the ‘finger-loop’ interruption of the long α-helix that is not observed in any other described nuclease structure. The active site architecture of all ββα-Me nucleases is very similar. Bound metal ions identified in described structures are Mg2+ in EndoA, Serratia, I-PpoI, Vvn and nucleases, Ca2+ in T4 endo VII and Mn2+ in NucA and I-HmuI nucleases. In all of these structures, the metal ion is hydrated by a cluster of neighbouring water molecules. In the Spd1 structure, there are only two water molecules in the proximity of Mg2+. The side chain oxygen of Asn145 is a direct ligand for the metal ion and is highly conserved among ββα-Me nucleases. A strong resemblance is also seen for the catalytic Histidine (His121 in Spd1, His160 in EndA, His124 in NucA, His89 in Serratia, His98 in I-PpoI, His80 in Vvn nuclease, His41 in T4 Endo VII and H143 in mitochondrial EndoG). Glu164 that is facing the magnesium ion, corresponds to Glu127 and Glu163 in Serratia endonuclease and NucA, respectively, and to Glu205 in EndA. These Glutamate residues are involved in positioning two water molecules to interact with the metal ion and form a metal cluster. This may explain why mutating Glu164 in Spd1 has a detrimental effect on nuclease activity, even though in the crystal structure it is >6 Å away from the magnesium ion. Arg61 in the I-PpoI structure interacts with the nucleotide phosphate backbone. The same has been observed for the corresponding Arg99 in Vvn nuclease. Equivalent residues in NucA and Serratia are Arg93 and Arg57, respectively. In the Spd1 structure, the corresponding Arg90 is flipped 180° and points outside. Gln150 in NucA positions the neighbouring water molecule to interact with Mn2+. It corresponds to Gln114 in Serratia nuclease, Gln186 in EndA, Glu114 in I-PpoI and 79 in Vvn nuclease. Spd1 is the only structure with a Methionine in this position (Met140).
Figure 7.
Protein sequence alignment showing Spd1 in comparison to its closest known structural homologues: S. pyogenes nuclease (Spd1), pdb code 2xgr; S. pneumoniae nuclease (EndA), pdb code 3owv; Anabaena sp. nuclease (NucA), pdb code 1zm8; Drosophila melanogaster nuclease (EndoG), pdb code 3ism; Serratia marcescens nuclease (smn), pdb code 1smn. Secondary structure elements are taken from the Spd1 structure and indicated by the following, β-beta strand; α-alpha helix; TT-beta-turn; η-310 helix; grey star-residues modelled with an alternate conformation. Conserved residues are shown highlighted in red; areas of high homology are boxed in blue. Note Spd1 shows a variant asparagine residue at position 118, resulting in a NRGH catalytic motif, which was previously considered to be a conserved DRGH motif.
Figure 6.
WT Spd1 overlaid with EndA from S. pneumoniae (PDB code 3OWV) showing the structural similarity between the nucleases. Notable structures are the β-strands forming the central sheet, the ββα-Me motif comprised of two short β-strands and a long α-helix running along the central β-sheet.
Active site architecture
DNA Binding
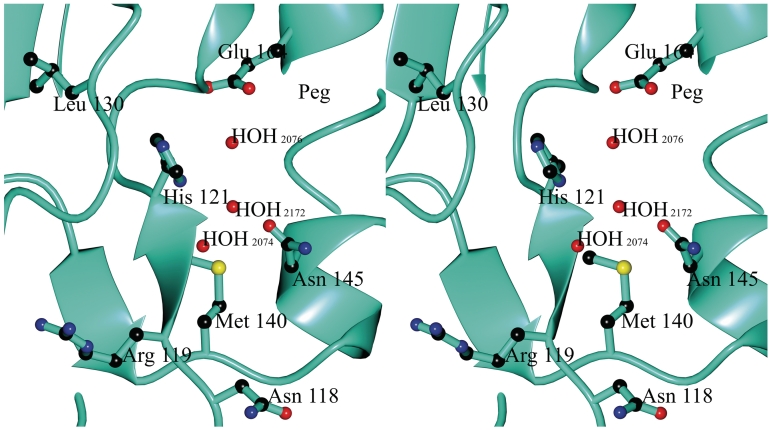
The crystal structure of Spd1 reveals a ββα-metal finger motif, which is comprised of β-sheets 4 and 5 (green, Figure 1a), which lie adjacent to the smaller α1 helix. These motifs harbour residues involved in metal ion coordination and catalysis. Mutagenic studies and structure comparisons with other published nucleases allowed us to identify catalytically important residues. It has been shown before that an active site His (His121 equivalent) is crucial for S. pyogenes nuclease activity (49). Mutations His121Ala and His121Asp abrogate Spd1 activity (Figure 2). In the Spd1 structure, His121 is located at the C-terminus of strand β5 (Figure 8). The main-chain oxygen of Leu130 makes a hydrogen bond with the other imidazole nitrogen of His121 (2.9 Å distance), suggesting this residue is deprotonated in the structure. On the opposite side, in the helical motif, there is Asn145 that is likely to coordinate the active site metal ion directly through its side-chain oxygen atom. Mutations of this residue to Ala and Gln curtailed Spd1 activity (Figure 2). Glu164 is crucial for activity, even though it is 6.7 Å away from active centre and does not make direct contacts in the active site environment.
Figure 8.
Stereo representation of the WT Spd1 active site showing the positions of the key active site constituents (the Arg90 from the symmetry molecule has been removed for clarity): Waters 2074, 2076, 2076 and polyethylene glycol and residues Asn118, Arg119, His121, Val123, Leu130, Met140 and Asn145.
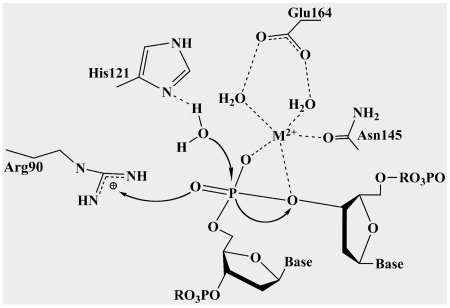
Proposed reaction mechanism
Similarities between the active sites of the discussed nucleases and high conservation of the ββα-Me motif suggest that Spd1 may use the same mechanism for DNA hydrolysis. We propose that His121 would act as a general base, removing a proton from a neighbouring water molecule that would then attack the scissile phosphodiester bond (Figure 9). The divalent magnesium ion would act as a Lewis acid positioning the water molecule to donate a proton to a departing 3′-oxygen. Arg90 would change its position and orient in the direction of cleaved 5′-phosphate to make a hydrogen bond, as has been shown to occur in the Vvn–DNA complex and in the I-PpoI structures. This would stabilize the product in order to prevent the reverse reaction (20). The role of Asn145 and Glu164 in coordinating the essential metal ion (Glu164 indirectly through a water molecule) agrees with the mutagenesis experiments that prove the crucial role of these residues, as Asn145Ala, Asn145Gln and Glu164Ala mutants appear to diminish Spd1 activity.
Figure 9.
Schematic diagram of the proposed mechanism of Spd1. His121 acts as a general base, removing a proton from a neighbouring water molecule; this attacks the scissile nucleotide phosphate. The divalent metal ion acts as a Lewis acid stabilizing the negative charge on the nucleotide phosphate transition state and the leaving group. The metal ion is coordinated by the side-chain oxygen of a conserved Asparagine (Asn145) and a Glutamic acid (Glu164). Arginine 90 is involved in positioning the substrate around the cleavage point.
The biological role of Spd1
The precise biological role of Spd1 in S. pyogenes SF370 and the SF370.1 prophage/phage is not fully understood. At the point of prophage induction, there exists a three way interplay between the phage, the bacterium and the human host. The phage-mediated acquisition of both toxin and DNase genes as a factor enhancing virulence appears to be a common theme, and it is tempting to speculate that their co-expression is evolutionarily advantageous by synergistically acting against the immune system to promote survival of the phage and bacterium. It has been suggested that phage-encoded streptococcal DNases may promote the survival of the phage by liquefying pus and cellular material to reduce viscosity and aid bacterial and phage particle dissemination (8). This idea may be extended and it is proposed that the occurrence of the toxin and DNase genes may give the phage and bacterium (prophage) a selective advantage by avoiding clearance by the NETs. There is some evidence that the expression of phage-encoded DNase extends beyond the Streptococcus genus. A BLAST search using Spd1 (http://blast.ncbi.nlm.nih.gov/Blast.cgi) reveals that Pro-phage homologues can be found in the lactobacillus genus exemplified by a homologue from Lactococcus lactis sharing a 46.25% identity to Spd1 (8,9). This indicates that phage-encoded DNases enhance the survival of the phage in an environment that is not normally associated with the human immune system and so points to broader evolutionary mechanism beyond the host pathogen interaction.
In this study, we have described a method for the cloning, expression and purification of a DNase from S. pyogenes in WT and mutant forms. We have gone on to present its X-ray structure, shown the location of key active site residues and that His121, Asn45 and Glu164, are essential in maintaining activity. Our work has shown two crystal forms, a WT catalytically blocked dimer and a monomeric Asn145Ala mutant. The results from the SEC-MALLS experiments show that Spd1 behaves as a monomer in solution in the presence and absence of the divalent metal ion Mg2+. The digestion of plasmid DNA shows three distinct phases, (i) relaxation (ii), partial linearization and (iii) plasmid degradation. This triphasic digestion pattern points to an underlying complex substrate interaction at a molecular level. Due to the role of streptococcal DNases in disease progression, these proteins represent a target for inhibition. The provision of these structures and data will be of interest to those wishing to perform future mechanistic studies and epitope mapping to aid vaccination design.
ACCESSION NUMBER
PDB ID Code: 2XGR and 2XH3 (EMBL Protein Data Bank in Europe, http://www.ebi.ac.uk/pdbe/).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S3.
FUNDING
Royal Society (grant number: UF100116) and the Biotechnology and Biological Sciences Research Council (grant number: DTA A0031002). Funding for open access charge: The Royal Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the Diamond Light Source, Didcot and the ESRF, Grenoble for the provision of excellent beamlines and Dr Andrew P. Leech of the Technology facility, University of York for his kind assistance with the SEC-MALLs and continuous DNase assays.
REFERENCES
- 1.Rangarajan ES, Shankar V. Sugar non-specific endonucleases. FEMS Microbiol. Rev. 2001;25:583–613. doi: 10.1111/j.1574-6976.2001.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 2.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl Acad. Sci. USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 4.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 5.Scott J, Thompson-Mayberry P, Lahmamsi S, King CJ, McShan WM. Phage-associated mutator phenotype in group A streptococcus. J. Bacteriol. 2008;190:6290–6301. doi: 10.1128/JB.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vojtek I, Pirzada ZA, Henriques-Normark B, Mastny M, Janapatla RP, Charpentier E. Lysogenic transfer of group A Streptococcus superantigen gene among Streptococci. J. Infect. Dis. 2008;197:225–234. doi: 10.1086/524687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl Acad. Sci. USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broudy TB, Pancholi V, Fischetti VA. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect. Immun. 2002;70:2805–2811. doi: 10.1128/IAI.70.6.2805-2811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canchaya C, Desiere F, McShan WM, Ferretti JJ, Parkhill J, Brussow H. Genome analysis of an inducible prophage and prophage remnants integrated in the Streptococcus pyogenes strain SF370. Virology. 2002;302:245–258. doi: 10.1006/viro.2002.1570. [DOI] [PubMed] [Google Scholar]
- 10.Broudy TB, Pancholi V, Fischetti VA. Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells. Infect. Immun. 2001;69:1440–1443. doi: 10.1128/IAI.69.3.1440-1443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green NM, Beres SB, Graviss EA, Allison JE, McGeer AJ, Vuopio-Varkila J, LeFebvre RB, Musser JM. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J. Clin. Microbiol. 2005;43:4083–4091. doi: 10.1128/JCM.43.8.4083-4091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 2005;187:3311–3318. doi: 10.1128/JB.187.10.3311-3318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhlmann UC, Moore GR, James R, Kleanthous C, Hemmings AM. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- 14.Flick K, Jurica M, Monnat R, Stoddard B. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 15.Shlyapnikov SV, Lunin VV, Perbandt M, Polyakov KM, Lunin VY, Levdikov VM, Betzel C, Mikhailov AM. Atomic structure of the Serratia marcescens endonuclease at 1.1 A resolution and the enzyme reaction mechanism. Acta Crystallogr. D Biol. Crystallogr. 2000;56:567–572. doi: 10.1107/s090744490000322x. [DOI] [PubMed] [Google Scholar]
- 16.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 17.Raaijmakers H, Törö I, Birkenbihl R, Kemper B, Suck D. Conformational flexibility in T4 endonuclease VII revealed by crystallography: implications for substrate binding and cleavage. J. Mol. Biol. 2001;308:311–323. doi: 10.1006/jmbi.2001.4592. [DOI] [PubMed] [Google Scholar]
- 18.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh M, Meiss G, Pingoud A, London R, Pedersen L. Structural insights into the mechanism of nuclease A, a betabeta alpha metal nuclease from Anabaena. J. Biol. Chem. 2005;280:27990–27997. doi: 10.1074/jbc.M501798200. [DOI] [PubMed] [Google Scholar]
- 20.Li CL, Hor LI, Chang ZF, Tsai LC, Yang WZ, Yuan HS. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altermark B, Smalas A, Willassen N, Helland R. The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1387–1391. doi: 10.1107/S0907444906034196. [DOI] [PubMed] [Google Scholar]
- 22.Scholz SR, Korn C, Bujnicki JM, Gimadutdinow O, Pingoud A, Meiss G. Experimental evidence for a beta beta alpha-Me-finger nuclease motif to represent the active site of the caspase-activated DNase. Biochemistry. 2003;42:9288–9294. doi: 10.1021/bi0348765. [DOI] [PubMed] [Google Scholar]
- 23.Moon AF, Midon M, Meiss G, Pingoud A, London RE, Pedersen LC. Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae. Nucleic Acids Res. 2011;39:2943–2953. doi: 10.1093/nar/gkq1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fogg MJ, Wilkinson AJ. Higher-throughput approaches to crystallization and crystal structure determination. Biochem. Soc. Trans. 2008;36:771–775. doi: 10.1042/BST0360771. [DOI] [PubMed] [Google Scholar]
- 25.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 27.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org. [Google Scholar]
- 36.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 37.Aziz RK, Ismail SA, Park HW, Kotb M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol. Microbiol. 2004;54:184–197. doi: 10.1111/j.1365-2958.2004.04255.x. [DOI] [PubMed] [Google Scholar]
- 38.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- 40.Tolun G, Myers RS. A real-time DNase assay (ReDA) based on PicoGreen fluorescence. Nucleic Acids Res. 2003;31:e111. doi: 10.1093/nar/gng111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Midon M, Schafer P, Pingoud A, Ghosh M, Moon AF, Cuneo MJ, London RE, Meiss G. Mutational and biochemical analysis of the DNA-entry nuclease EndA from Streptococcus pneumoniae. Nucleic Acids Res. 2011;39:623–634. doi: 10.1093/nar/gkq802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 43.Gololobov GV, Chernova EA, Schourov DV, Smirnov IV, Kudelina IA, Gabibov AG. Cleavage of supercoiled plasmid DNA by autoantibody Fab fragment: application of the flow linear dichroism technique. Proc. Natl Acad. Sci. USA. 1995;92:254–257. doi: 10.1073/pnas.92.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller MD, Krause KL. Identification of the Serratia endonuclease dimer: structural basis and implications for catalysis. Protein. Sci. 1996;5:24–33. doi: 10.1002/pro.5560050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franke I, Meiss G, Pingoud A. On the advantage of being a dimer, a case study using the dimeric Serratia nuclease and the monomeric nuclease from Anabaena sp. strain PCC 7120. J. Biol. Chem. 1999;274:825–832. doi: 10.1074/jbc.274.2.825. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh M, Meiss G, Pingoud AM, London RE, Pedersen LC. The nuclease a-inhibitor complex is characterized by a novel metal ion bridge. J. Biol. Chem. 2007;282:5682–5690. doi: 10.1074/jbc.M605986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suck D, Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986;321:620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 48.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 49.Iwasaki M, Igarashi H, Yutsudo T. Mitogenic factor secreted by Streptococcus pyogenes is a heat-stable nuclease requiring His122 for activity. Microbiology. 1997;143(Pt 7):2449–2455. doi: 10.1099/00221287-143-7-2449. [DOI] [PubMed] [Google Scholar]