Abstract
Comparative genomics is a powerful means to gain insight into the evolutionary processes that shape the genomes of related species. As the number of sequenced genomes increases, the development of software to perform accurate cross-species analyses becomes indispensable. However, many implementations that have the ability to compare multiple genomes exhibit unfavorable computational and memory requirements, limiting the number of genomes that can be analyzed in one run. Here, we present a software package to unveil genomic homology based on the identification of conservation of gene content and gene order (collinearity), i-ADHoRe 3.0, and its application to eukaryotic genomes. The use of efficient algorithms and support for parallel computing enable the analysis of large-scale data sets. Unlike other tools, i-ADHoRe can process the Ensembl data set, containing 49 species, in 1 h. Furthermore, the profile search is more sensitive to detect degenerate genomic homology than chaining pairwise collinearity information based on transitive homology. From ultra-conserved collinear regions between mammals and birds, by integrating coexpression information and protein–protein interactions, we identified more than 400 regions in the human genome showing significant functional coherence. The different algorithmical improvements ensure that i-ADHoRe 3.0 will remain a powerful tool to study genome evolution.
INTRODUCTION
During their evolution, genomes have been altered at various levels. At the smallest scale, point mutations and small insertions and deletions (1) affect only a few nucleotides. Larger modifications include duplication, deletion, translocation or inversion of a single gene or genomic segment (2). At the largest scale, the entire genome can be doubled via genome duplication or merging (3–5). Identification of these structural rearrangements provides insight into how genomes have evolved and diverged over time. It is therefore of crucial importance to correctly determine chromosomal regions that are homologous (i.e. derived from a common ancestor), either within a genome, or between genomes of related species. Genomic homology can be inferred from collinearity, namely the conservation of both gene content and gene order. Synteny, though initially defined as ‘the property of being located on the same chromosome’ (6), is often used to indicate the conservation of gene content but not necessarily gene order (7). Like collinearity, synteny also points to homology between different genomic regions based on a number of shared genes (8,9).
Detection of collinear regions between the genomes of related species allows for the identification of chromosomal fusions and fissions, along with inverted or translocated regions. Additionally, gene loss and gain can be efficiently estimated, and cross-species genome analysis provides a framework for transferring gene annotation and biological information to newly sequenced genomes. Finally, orthologous intergenic sequences derived from collinear regions can be screened for conserved non-coding regions as a way to detect regulatory motifs and to identify various types of RNA genes (10). As both gene loss and different types of rearrangements accumulate over time, the resulting genome erosion gradually reduces the degree of collinearity between species. Therefore, gene order preserved over a large phylogenetic distance can imply a biological constraint (11).
Collinear regions within a genome can also hint at the occurrence of one or more rounds of whole-genome duplications (WGDs) (9,12). Based on within-genome collinearity, the loss of gene duplicates created during a WGD can be estimated (13–15), whereas the functions of genes retained in duplicate can be linked to lineage or species-specific adaptations, including specific pathways and biological processes. WGDs appear to have played a crucial role in the evolution of all major eukaryotic lineages and, particularly in plants, they are often associated with key events during evolution including fast adaptive radiation (4,16) and survival of mass extinction events (17). Additionally, gene family expansions critical for the pome fruit development in apple (Malus domestica) (18) have been linked to a recent WGD, whereas expansions in genes producing aromatic compounds have been observed in grapevine (Vitis vinifera) (19). Although remnants of several recent WGDs are abundant in the plant kingdom, WGDs in land vertebrates and fishes are seemingly much older (20,21). In vertebrates, the complex body plan is often attributed to the duplication of developmental genes during two WGDs 450 million years ago (Mya) (21). The first traces of a WGD have been unveiled in Saccharomyces cerevisiae based on comparative approaches (22). Additional proof for the WGD in brewer's yeast has been provided later by comparison with the genome of an unduplicated outgroup species, Kluyveromyces waltii (23). The more complex carbohydrate metabolism of S. cerevisiae and other post-duplication yeast species is probably a direct consequence of this duplication (24). Therefore, the discovery of large-scale duplications, through the study of collinear regions, has provided a remarkably detailed view on the genomic evolution and adaptation of various species.
Here, we focus on the accurate detection of homologous chromosomal segments both within and between the genomes of related species. Specifically, sensitive and accurate algorithms are needed for the identification and evolutionary analysis of duplicated regions that have undergone massive gene loss. Several tools, by means of various approaches, have recently been proposed (Supplementary Table S1). Whereas most tools only perform pairwise comparisons, the iterative Automatic Detection of Homologous Regions (i-ADHoRe) (25) was one of the first that simultaneously analyzed genomes of multiple species and allowed for the detection of highly diverged collinear regions. On the one hand, i-ADHoRe has been used in several genome projects to uncover the remnants of large-scale duplications [e.g. apple (M. domestica) (18), soybean (Glycine max) (26), Arabidopsis lyrata (27) and black cottonwood (Populus trichocarpa) (28)], and, on the other hand, to detect inter-species collinearity in yeasts (29) and Archaea (30). In contrast to tools that infer genomic homology through a multiple sequence alignment of complete genomic DNA sequences (31–34), i-ADHoRe detects genomic homology through the identification of gene collinearity and/or synteny. The core feature of i-ADHoRe 3.0, which is based on a new alignment algorithm (35) and improved statistical evaluation, is the ability to handle large numbers of genomes. Due to the further optimization of many algorithmic steps, the current version of i-ADHoRe 3.0 is roughly 30 times faster than the previous version. In addition, i-ADHoRe 3.0 can now take advantage of a parallel computing platform, reducing the runtime even further. For large data sets, the combination of improvements in the sequential algorithm and the parallelization results in overall speedup of a factor of 1000. Here, we demonstrate that i-ADHoRe is capable of processing much larger datasets than the current state-of-the-art tools. In particular, the complete Ensembl release 57 (36) data set that contains 49 eukaryotic genomes can be analyzed in 1 h (using 64 CPU cores), while producing highly accurate results.
MATERIALS AND METHODS
Data sets
The Arabidopsis thaliana and V. vinifera genomes together with gene family information were retrieved from PLAZA, an on-line plant comparative genomics resource (13) that provides gene families constructed with Tribe-MCL clustering (37) starting from an all-against-all BLAST (38) protein similarity search. The E-values and bit scores were saved, because these values are necessary for Cyntenator (39) and MCScan (40). The lengths for Carica papaya gene lists were also obtained via PLAZA. Animal genomes and families were downloaded from Ensembl (release 57) with the Ensembl Perl API (41). An all-against-all BLAST protein similarity search was done to obtain bit scores and E-values. An overview of all included species in both PLAZA and Ensembl is included in Supplementary Table S2.
Detection of collinearity
The initial steps of the algorithm (Supplementary Figure S1) are identical to i-ADHoRe 2.0; tandem duplicated genes are mapped to a single representative and for each pair of gene lists a gene homology matrix (GHM) is generated (Figure 1A). In this sparse matrix, pairs of homologous genes are represented as dots and as such collinear regions will appear as dense diagonals. Compared to the previous i-ADHoRe version, several major components of the algorithm were re-implemented for a better performance. First, the statistical validation of the clusters in the GHM was improved. To avoid inclusion of diagonals in the GHM generated merely by chance, the significance of each cluster is now estimated with a statistical model that takes into account the overall background density of the matrix. When multiple seeds (i.e. clusters with at least three homologous gene pairs that meet the initial criteria) were found, a correction for multiple hypothesis testing was done either with the Bonferroni or False Discovery Rate (FDR) (42,43) method.
Figure 1.
(A) GHM for the initial two segments (Seg I and Seg II). In a GHM, collinear regions will appear as dense diagonals. (B) Alignment of shared homologs between collinear regions; gaps are introduced to place as many homologous pairs in the same column as possible (35). The alignment (or ‘profile’) now contains the information of both segments. (C) Start of the iterative process, GHMs are now created with the profile and additional collinear regions can be found, e.g. Seg III. (D) Generation of a new profile. As long as additional segments can be found steps (C) and (D) are repeated. In this example (E) and (F) show how a single profile detects two additional segments that are mutually non-homologous (Seg IV and V), leading to a split in the detection process.
Significant collinear regions found during this initial detection were converted into a profile, both collinear regions were aligned, i.e. homologous genes are placed in the same column adding gaps where necessary (Figure 1B). Like in previous versions of i-ADHoRe this alignment can be done by progressively applying the Needleman–Wunsch (pNW) algorithm or a greedy graph (GG) based alignment strategy. In version 3.0, a novel alignment algorithm (GG2), described in Fostier et al. (35), was implemented. Using this aligned profile, a new search is performed (Figure 1C), here a GHM is created with the profile and all gene lists in the data set. Significant regions are added to a new profile and the profile search is repeated (Figure 1D). With a single profile, multiple segments can be found that are homologous to the profile but not necessarily to each other (Figure 1E and F). In this case, several profiles are generated and the detection algorithm continues detection with the longest profile first. Once no additional segments can be found the search continues with the next profile.
Additionally, the initial pairwise and profile searches can now be executed on a parallel computing platform (a multiprocessor/multicore systems or a computational cluster of networked computers). If N the number of gene lists provided as an input to i-ADHoRe, the N(N + 1)/2 pairwise comparisons could be processed independently and, hence, distributed over different processes. The size of each gene list was taken into account to ensure a good load balance between the processes. At the end of this step, the detected collinear regions are communicated [using the Message Passing Inteface (MPI)] among the processes. Similarly, a single profile search can be parallelized by distributing the N gene lists among the different processes, again taking the size of the chromosomes into account. At the end of every profile search, the detected collinear regions were again communicated between the processes. However, due to the much smaller task granularity of one single profile search, a good load balancing was more difficult to achieve. Methods accompanying the novel synteny mode (detection of genomic homology purely based on shared gene content) can be found in the Supplementary Methods S1 and Table S3.
Empirical estimation of false positive rates
False positive (FP) rates were calculated with permutation tests in which 100 randomized data sets were compared with a real reference data set. Tandem duplicated genes (homologs within a window of 70 genes) were removed prior to shuffling the reference data set to generate a randomized version. This pre-processing step guaranteed a comparable density in the randomized run because breaking up tandem-clusters artificially increased the GHM background density. All genes had their original orientation replaced with a randomly assigned one. The lengths of the original gene lists were maintained during the randomization, but genes could be moved from one gene list to another. To estimate the performance with different settings, a permutation test was carried out for each of the desired settings, generating parameter landscapes for Arabidopsis, human and yeast, with various combinations of q_value and gap_size parameters. Settings that yielded the maximum amount of anchor points, while maintaining a FP rate near the selected cut-off value were considered as optimal (Supplementary Methods S2).
Comparison with MCScan and Cyntenator
BLASTP pairs for MCScan were filtered and only the best five hits in each species were retained (40). Because in MCScan first proteins are clustered to group homologous genes in gene families, this step was excluded when monitoring runtimes for the different tools. Cyntenator was also run with filtered BLASTP output, retaining only the top five hits for each species if their bit score was within 95% of the highest bit score [as described in Rödelsperger et al. (44)]. The gap and mismatch penalties were set to −0.3, the threshold to 2 and the filter to 1000. i-ADHoRe was run with a gap_size of 30 and cluster_gap of 35, while keeping the prob_cutoff on 0.01 and the q_value on 0.75. GG2 was used as the alignment algorithm and correction for multiple hypothesis testing was done with FDR. The minimal number of anchor points in a cluster was set to five.
Detection of highly conserved regions enriched for coexpressing/interacting gene pairs
Phylogenetic profiles, describing the number of homologous regions per species present in a multiplicon, a set of mutually collinear regions, were generated for all multiplicons in the output from the high-quality Ensembl subset. Multiplicons with one human and one bird (either chicken or zebra finch) segment and with conserved segments of at least five other mammals were selected. From these regions, the human segment was identified and the genes collinear with genes from other segments were stored. Expression data were derived from COXPRESdb version c3.1 (45) and highly expressed gene pairs were selected based on a mutual rank below or equal to 50. Experimentally characterized interacting protein pairs (41 088 binary interactions for 9142 human genes) were downloaded from IntAct (46). Using Ensembl's BioMart tool, a conversion table was generated to map all gene identifiers in these data sets to the Ensembl genes. For each selected multiplicon, the length of the human segment and number of human collinear genes were determined. Then, the number of coexpressed or interacting pairs was counted. When at least one human gene pair was found, the statistical significance was tested with a permutation test. Over 10 000 iterations, a random segment from the human genome (with the tandem duplicated genes removed) was sampled with the same length as the selected multiplicon. From the random region, an equal number of genes was randomly selected as collinear and, the number of coexpressed or protein–protein interaction pairs in this gene set was established. The number of iterations in which a number of pairs was equal or larger than that found in the real data set were counted and used to calculate a P-value for each multiplicon. All regions with a P < 0.05 were considered significant.
Evaluation of low quality genomes
To artificially reduce the quality of the Arabidopsis genome, the gene list length distribution of the papaya genome was used as a template to split the Arabidopsis gene lists in fragments resembling a draft assembly. i-ADHoRe was executed on both the Arabidopsis genome and the artificial low-quality version. The collinear fractions were measured by enabling the write_stats option in i-ADHoRe. Supplementary Methods S3 describes an additional study that further addresses this issue, using various vertebrate genomes.
RESULTS AND DISCUSSION
The i-ADHoRe 3.0 algorithm
The detection strategy of i-ADHoRe 3.0 is shown in Supplementary Figure S1 (25,47). First, tandem duplicated genes are mapped onto one single representative gene, because tandem clusters can hinder the detection of diagonals (see further). Next, for each pair of chromosomes or scaffolds, a so-called gene homology matrix (GHM) is generated. A GHM is a sparse matrix in which homologous gene pairs are marked by dots and collinear regions appear as ‘diagonals’. For each detected diagonal, the statistical significance is evaluated (Figure 1A). Significant collinear regions are aligned into a profile (Figure 1B) that contains the combined gene content of the two collinear regions and can hence be used as a more sensitive probe to scan for additional collinear regions (Figure 1C and D). This step is iterated as long as new collinear regions are found and mutually homologous regions are grouped into a multiplicon. Even though the profile search requires an increased computational cost, it has proven its merits as a means to detect more degenerate genomic homology (12,25,48).
In order to deal with increasingly large data sets, various parts of the original i-ADHoRe code (47) have been replaced by equivalent algorithms with a reduced computational complexity. A first major improvement was the development of an efficient statistical model to estimate the significance of diagonals in the GHM, because the computational cost to calculate the exact P-value (49) increases exponentially with the number of gene pairs that shape the diagonal. The Arabidopsis thaliana data set was analyzed with different P-value thresholds and an empirical FP rate for each threshold was determined using permutation tests (Supplementary Figure S2). The combination of better heuristics and the implementation of a correction for multiple hypothesis testing (Bonferroni or FDR) resulted in a more realistic estimation of P-values and consequently improved the control of the FP rate compared to the previous statistical model. Benchmarks including other model organisms and the effects of using different parameter settings are reported in Supplementary Tables S4–S6.
In the iterative search procedure, additional collinear regions are identified and the corresponding profiles are updated in every step. Therefore, an accurate alignment algorithm is imperative for the sensitive discovery of more degenerate collinear regions (Figure 1C and D). Originally, i-ADHoRe relied on the progressive application of the pairwise Needleman–Wunsch (pNW) algorithm to align multiple homologous segments into profiles (47). Whereas with the Needleman–Wunsch algorithm an optimal pairwise alignment of two segments can be obtained, its quality quickly degrades due to the propagation of erroneous decisions in early alignment steps when additional segments are added (50). To resolve this issue, a greedy, graph-based (GG) aligner had been introduced into i-ADHoRe 2.0 that converted the alignment problem into a cycle-canceling problem in a graph (25). Whereas this implementation provided a viable solution for the ‘once a gap, always a gap’ problem, it was unable to outperform the pNW aligner in terms of number of correctly aligned homologous genes. In i-ADHoRe 3.0, a novel greedy, graph-based aligner (GG2) was featured that, by means of maximum flow calculations in the graph, resolved efficiently unalignable sections in the graph (conflicts). Even though this graph-based method is computationally more intensive than the application of the pNW aligner, fast heuristics allow this algorithm to be efficiently used (35).
Finally, two practical issues arise when multiple genomes are compared: the processing time and the memory requirements. Whereas the runtime increases super-linearly with the size of the data set, i.e. faster than the number of genomes that are analyzed, the memory requirements are mainly determined by the number of homologous gene pairs. To limit the runtime and, hence, facilitate the analysis of large-scale data sets, the two most time-consuming parts of the algorithm were parallelized (Figure 1, green boxes): the initial all-to-all pairwise comparison (every gene list versus every gene list) and the iterative profile searches (one profile versus every gene list). The parallelization of the all-to-all pairwise step revealed that by using a dataset of 31 high-quality genomes (Supplementary Table S2) and 64 CPU cores, a 46-fold increase in speed (Supplementary Figure S3) was observed. Searching additional collinear regions in a gene list using a profile is more difficult to parallelize, because of more intense communication requirements between the subtasks and hence a larger communication overhead. Overall, the runtime for the complete algorithm was reduced 32-fold on 64 cores, corresponding to a parallel efficiency (relative reduction in runtime compared to one with one single core, over the number of cores used) of ∼50%.
Evaluation of gene-based collinearity detection tools
When genomes with remnants of WGDs are dealt with or when highly diverged genomes are compared, gene loss and different types of rearrangements can interfere with the accurate detection of duplicated or homologous collinear regions (7,9). To the best of our knowledge, only Cyntenator (39), MCScan (40) and i-ADHoRe go beyond simple pairwise comparison and combine, via different approaches, information to find additional homologous regions. Cyntenator performs progressive pairwise combinations based on a user-defined species tree that strictly imposes the order in which genomes are compared. Only valid alignments including homologous regions from all species are retained to find collinearity with the next genome in line. Unlike the profile search of i-ADHoRe, in MCScan each chromosome is used as a reference and all pairwise collinear segments are mapped, followed by a multiple alignment procedure of homologous genes, inspired by the threaded blockset aligner (34). MCScan allows pairing regions that had initially not been detected based on their collinearity with the reference, a method referred to as ‘transitive homology’ (47). Unlike some tools (Supplementary Table S1), Cyntenator, MCScan and i-ADHoRe use ordered gene lists rather than the actual genome sequence. This level of abstraction allows for an efficient detection of collinearity. An additional advantage is that more diverged intergenic sequences do not interfere with the discovery of ancient collinearity or synteny.
To benchmark the application of a profile search versus transitive homology mapping of pairwise collinear segments, i-ADHoRe and MCScan were executed on the Arabidopsis thaliana data set to identify degenerated duplicated segments. Cyntenator was excluded from this experiment, because it does not allow detection of internal duplications. Figure 2 shows the number of genes present in regions with a certain level, indicating the total number of homologous segments. Although i-ADHoRe and MCScan use very different approaches, the number of genes in collinear regions was comparable (23 912 and 24 559, respectively), but the profile search enabled i-ADHoRe to group more genes in regions with level four (4499 versus 2669 genes), five (1223 versus 891) and six (1318 versus 340). This result implies that the more advanced profile search allows for a more sensitive detection of collinear regions compared to the progressive chaining in MCScan.
Figure 2.
Distribution of the fraction of genes (n) found in sets of homologous genomic segments (multiplicons) with different levels (m) by MCScan and i-ADHoRe, respectively. Level 1 indicates the fraction of genes that was not found in any collinear region. The cumulative curve (i.e. the sum of all genes with the indicated level or lower) remains lower for i-ADHoRe, indicating that a larger fraction of the genome could be grouped into higher level multiplicons.
To evaluate the discovery of inter-species collinearity, the three tools were applied to analyze a small subset of the genomes available in Ensembl, namely human (Homo sapiens) (51), chimpanzee (Pan troglodytes) (52), mouse (Mus musculus) (53), chicken (Gallus gallus) (54) and pufferfish (Tetraodon nigroviridis) (55). For each gene, all overlapping homologous segments were retrieved and the highest number of species found in one single alignment (or multiplicon) was scored. In contrast to Cyntenator, MCScan and i-ADHoRe collapse tandem genes into one single representative and, therefore, reported fewer genes. The predefined species order applied by Cyntenator to compare genomes forms a major drawback for large-scale analyses including multiple species. For instance, a region that is collinear between human and mouse, but for which the homologous counterpart in the chimpanzee lineage was lost, will not be reported because only collinear regions from the first pairwise comparison (i.e. human and chimpanzee) are retained to identify additional collinearity in mouse. Therefore, a fair comparison was possible only for regions in which collinearity was conserved in all five species. Whereas using MCScan and Cyntenator, 416 and 498 genes were assigned to such regions, respectively, the profile search applied by i-ADHoRe allocated 3296 genes in multiplicons containing regions conserved in all five species (Supplementary Figure S4).
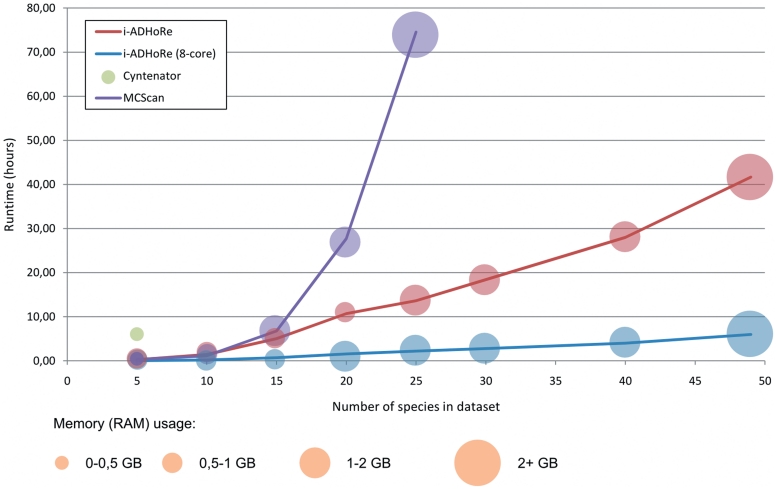
Fast algorithms that exhibit a favorable computational complexity are imperative to keep pace with the ever-increasing number of available genomes. Therefore, the runtime of all three programs was first monitored on the data set of the five species. i-ADHoRe, the only tool that takes advantage of a parallel environment, was executed using a single and eight threads, respectively, on a multicore machine. Because, MCScan first clusters proteins into gene families, a step not part of the actual collinearity detection algorithm, the program runtime was measured without this pre-processing step (Figure 3). Whereas Cyntenator required 6.25 h to analyze the five genomes, MCScan and i-ADHoRe were considerably faster, analyzing the data set in 19 and 14 min, respectively. When i-ADHoRe was run with eight cores, the runtime was reduced to only 3 min.
Figure 3.
Runtime and memory usage comparison of Cyntenator (39), MCScan (40) and i-ADHoRe (this study). Each tool was run on subsets of the Ensembl data set each including a different number of species.
In a second experiment, the maximum number of genomes that could be analyzed was determined by processing data sets of gradually increased size (Figure 3). Only i-ADHoRe succeeded in analyzing the complete Ensembl data set covering 49 species (832 666 genes). Although Cyntenator could analyze up to 17 high-coverage genomes (39), the detection approach based on the strict usage of a guidance species tree posed a problem for data sets that include genomes sequenced at low coverage. As a result, inclusion of low-coverage or fractionated genomes into a large data set quickly eroded the amount of collinearity found, abruptly terminating the algorithm and leading to missing data when 10 or more genomes were included in the benchmark data set. For MCScan, the largest possible data set that could be analyzed in 48 h included 20 species (Figure 3); although within 168 h also 30 species could be covered, this duration however is impractically long for the efficient processing of extremely large data sets. In contrast, i-ADHoRe finished the full Ensembl data set covering 49 genomes within 42 h using a single CPU core. This runtime could be reduced to 6 h using the eight cores (88% efficiency). Finally, when using eight such machines (i.e. 64 cores in total) that are connected through a fast communication network (Infiniband), the runtime decreased to 40 min (50% efficiency; Supplementary Figure S3). An additional advantage of i-ADHoRe is that gene families rather than individual homologous gene pairs can be used to construct the GHM, whereas in both Cyntenator and MCScan, per query gene, a limit of five homologous genes in each other species (based on BLAST hits) is suggested. Furthermore, the usage of gene families is a more memory-efficient alternative than storage of all homologous gene pairs covering multiple genomes. Although for small data sets, i-ADHoRe utilizes more memory than MCScan and Cyntentor, the required memory scales linearly with the total number of genes and remains below that of MCScan once the data sets include 20 or more genomes (Figure 3).
Biological significance of ultra-conserved multispecies collinearity
Starting from 25 293 genomic scaffolds present in the Ensembl data set, 319 245 multiplicons were identified, some of which contained homologous regions from more than 20 species. The ‘largest’ multiplicon contained 33 segments from 22 species and included several homeobox Dlx proteins. Several HOX gene clusters including homeobox transcription factors were also found in a few high-level multiplicons (HOX D, level 28; HOX C and HOX D, level 22; HOX A and HOX D, level 25; HOX B and HOX D, level 20). This region is known to be highly conserved across species because these genes, involved in development of the body plan, require correct order to function (56). The HOX cluster was duplicated and retained during two rounds of WGD in the ancestor of the vertebrates over 450 Mya (21), and since then the HOX A, HOX B, HOX C and HOX D clusters diverged significantly (57).
Many genes coding for interacting proteins are robust against rearrangements (11) and clusters of coexpressed genes conserved between human and mouse have been reported (58). Given the large set of species, regions where gene order is strongly conserved over a large phylogenetic distance were delineated (see ‘Materials and Methods’ section). Next, we assessed whether genes in these strongly conserved regions showed significant functional clustering. Briefly, experimental protein–protein interaction data and coexpression information were used to determine whether a highly conserved region was significantly enriched for interacting proteins or genes showing coordinated expression profiles. Coexpression is frequently used as a strong indicator for functionally related genes (‘guilt by association’). Also, interacting protein pairs are known to have a high chance to be involved in the same biological process (59). From the output of the high-quality subset, multiplicons with a strong conservation between either chicken or the songbird zebra finch (Taeniopygia guttata) (60), human, and at least five other mammals were extracted. Out of these 2863 multiplicons, 466 regions containing 2424 human genes, were significantly enriched (P < 0.05) for coexpressed pairs and/or gene pairs coding for interacting proteins (Figure 4). Mapping of these regions to a chromosome conservation plot depicting collinearity with all the 23 species included revealed that these regions are often among the most conserved in the genome (Supplementary Figure S5). A full list of conserved regions showing functional clustering, including the human genes within these regions and P-values for functional enrichment, can be found in Supplementary Table S7.
Figure 4.
Gene order alignment of collinear regions conserved over a large phylogenetic distance (human–chicken). Species to which the segments belong are given on every line by the boxes on the right: Homo sapiens (ho_sa), Pan troglodytes (pa_tr), Pongo pygmaeus (po_py), Macaca mulatta (ma_mu), Mus musculus (mu_mu), Rattus norvegicus (ra_no), Cavia porcellus (ca_po), Bos taurus (bo_ta), Equus caballus (eq_ca), Canis familiaris (ca_fa), Monodelphis domestica (mo_do) and Gallus gallus (ga_ga). Arrows indicate coding genes and their orientation. Homologous genes are depicted in the same color. Coexpressed gene pairs are linked by black curved lines, of which the thickness of the line corresponds to the coexpression level (based on the mutual rank of the human genes in CoXPRESDB). Blue curved lines link pairs of genes coding for interacting proteins (in human). This region was found to be significantly enriched for coexpressed genes and, therefore, a biological constraint might cause gene order in this region to be retained.
Significant enrichment of coexpressed and interacting protein pairs points toward an evolutionary constraint to conserve gene order in these regions. These results provide further evidence that gene order in vertebrates is non-random and might play a considerable role in regulation of gene expression. However, the precise mechanism of the observed coexpression remains an open question, because transcription factors, chromatin modifications (61), and long-range enhancers are likely candidates to play a role in this process.
Performance on low-coverage and fractionated genomes
Whereas new techniques greatly speed up the sequencing of novel genomes at a low cost, short-read lengths make it difficult to assemble reads into full chromosomes without genetic maps or a finishing phase (30,62,63). The same is true for genomes sequenced with traditional Sanger methods and low coverage (2×) (64). Consequently, these genomes are generally released as a collection of annotated scaffolds instead of being assembled into complete chromosomes or pseudomolecules. Draft-quality genomes are usually sequenced to get an overview of the overall gene content and, because most gaps occur in repeat regions, the majority of the genes are present despite the low coverage. However, for studies focusing on genome organization and evolution, inclusion of these low-quality genomes can become problematic (64). Consequently, we expect a genome sequence provided as a large set of unassembled scaffolds to interfere with the accurate detection of collinearity (Supplementary Table S8).
For instance, to estimate the impact of low-quality genomes on the detection of WGDs, the Arabidopsis genome was used as a reference and randomly cut into several artificial scaffolds with a length distribution comparable to the papaya (Carica papaya) genome (65) that is available as 4635 scaffolds (containing on an average six genes). The papaya genome was selected as a template because it is a draft version, sequenced up to 3× coverage, and without assembly in pseudomolecules. The Arabidopsis genome and the low-quality version were analyzed without any additional genomes. As expected, the number of genes that could be analyzed decreased because scaffolds with less than five genes were discarded (overall 17.47% gene loss). Whereas in the full genome 20 898 genes were found in duplicated regions (87.47%), this number decreased to only 10 091 (42.23% of all genes or 51.17% for the genes located only on scaffolds of sufficient length) in the low-quality version. Additionally, a drop in maximum level was observed: in the original genome up to seven homoeologous segments could be grouped whereas in the low-quality version the maximum level was five. However, including a genome reflecting the ancestral genome organization, like that of grapevine, can improve the number of genes found in collinear regions considerably. With grapevine included, 18% more Arabidopsis genes could be found in regions with level two to five (counting only Arabidopsis segments). Despite this increase in the number of genes found in duplicated regions, no more than five Arabidopsis segments grouped together after adding grapevine to the dataset. Because the maximum level is often used as a proxy for the number of large-scale duplication events (12), this result implied highly fragmented genomes organized in thousands of scaffolds are prone to misinterpretation when certain aspects of genome evolution are studied.
CONCLUSION
We show that the novel version of i-ADHoRe represents a major improvement over the current state-of-the-art algorithms and can be successfully applied to some of the largest data sets currently available.
As new sequencing initiatives such as the 1000 human genome project (66), the 1001 Arabidopsis genomes (67) and the 10 000 vertebrate genomes (68) will continue to generate many more genome sequences, the improved scalability of i-ADHoRe is imperative to keep runtimes acceptable. The support for parallel computing platforms ensures that i-ADHoRe 3.0 will efficiently detect genomic homology and will be instrumental to unveil genome evolution in the different kingdoms of life.
AVAILABILITY
The i-ADHoRe 3.0 software package is available from http://bioinformatics.psb.ugent.be/software. Source code, documentation and example data sets are provided in the package.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–8, Supplementary Figures 1–5, Supplementary Methods 1–3 and Supplementary References [69–86].
FUNDING
Ghent University (Multidisciplinary Research Partnership ‘Bioinformatics: from nucleotides to networks’); Interuniversity Attraction Poles Programme [IUAP P6/25] [initiated by the Belgian State, Science Policy Office (BioMaGNet)]; Agency for Innovation by Science and Technology in Flanders (predoctoral fellowship to S.P.); Postdoctoral Fellow of the Research Foundation-Flanders (to K.V.). Funding for Open Access Charge: Ghent University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Martine De Cock for help preparing the manuscript. The computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by Ghent University.
REFERENCES
- 1.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Hurles M. Gene duplication: the genomic trade in spare parts. PLoS Biol. 2004;2:E206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 4.Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. The flowering world: a tale of duplications. Trends Plant Sci. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 6.Passarge E, Horsthemke B, Farber RA. Incorrect use of the term synteny. Nat. Genet. 1999;23:387. doi: 10.1038/70486. [DOI] [PubMed] [Google Scholar]
- 7.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 9.Van de Peer Y. Computational approaches to unveiling ancient genome duplications. Nat. Rev. Genet. 2004;5:752–763. doi: 10.1038/nrg1449. [DOI] [PubMed] [Google Scholar]
- 10.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino T, McLysaght A. Interacting gene clusters and the evolution of the vertebrate immune system. Mol. Biol. Evol. 2008;25:1855–1862. doi: 10.1093/molbev/msn137. [DOI] [PubMed] [Google Scholar]
- 12.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne KP, Wolfe KH. Consistent patterns of rate asymmetry and gene loss indicate widespread neofunctionalization of yeast genes after whole-genome duplication. Genetics. 2007;175:1341–1350. doi: 10.1534/genetics.106.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 17.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl Acad. Sci. USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 19.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 20.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl Acad. Sci. USA. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 23.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 24.Scannell DR, Butler G, Wolfe KH. Yeast genome evolution–the origin of the species. Yeast. 2007;24:929–942. doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- 25.Simillion C, Janssens K, Sterck L, Van de Peer Y. i-ADHoRe 2.0: an improved tool to detect degenerated genomic homology using genomic profiles. Bioinformatics. 2008;24:127–128. doi: 10.1093/bioinformatics/btm449. [DOI] [PubMed] [Google Scholar]
- 26.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 27.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 29.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 30.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004;14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dewey CN. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol. Biol. 2007;395:221–236. doi: 10.1007/978-1-59745-514-5_14. [DOI] [PubMed] [Google Scholar]
- 33.Dewey CN, Pachter L. Evolution at the nucleotide level: the problem of multiple whole-genome alignment. Hum. Mol. Genet. 2006;15(Spec No. 1):R51–R56. doi: 10.1093/hmg/ddl056. [DOI] [PubMed] [Google Scholar]
- 34.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fostier J, Proost S, Dhoedt B, Saeys Y, Demeester P, Van de Peer Y, Vandepoele K. A greedy, graph-based algorithm for the alignment of multiple homologous gene lists. Bioinformatics. 2011;27:749–756. doi: 10.1093/bioinformatics/btr008. [DOI] [PubMed] [Google Scholar]
- 36.Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates G, Cox T, Cunningham F, et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodelsperger C, Dieterich C. CYNTENATOR: progressive gene order alignment of 17 vertebrate genomes. PLoS One. 2010;5:e8861. doi: 10.1371/journal.pone.0008861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiplyaligned angiosperm gene maps. Genome Res. 2008;18:1944–54. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Series B. 1995;57:289–300. [Google Scholar]
- 43.Dudoit S, van der Laan MJ. Multiple Testing Procedures with Applications to Genomics. New York: Springer; 2008. [Google Scholar]
- 44.Rodelsperger C, Dieterich C. Syntenator: multiple gene order alignments with a gene-specific scoring function. Algorithms Mol. Biol. 2008;3:14. doi: 10.1186/1748-7188-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–D82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2009;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simillion C, Vandepoele K, Saeys Y, Van de Peer Y. Building genomic profiles for uncovering segmental homology in the twilight zone. Genome Res. 2004;14:1095–1106. doi: 10.1101/gr.2179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandepoele K, Simillion C, Van de Peer Y. Detecting the undetectable: uncovering duplicated segments in Arabidopsis by comparison with rice. Trends Genet. 2002;18:606–608. doi: 10.1016/s0168-9525(02)02796-8. [DOI] [PubMed] [Google Scholar]
- 49.Durand D, Sankoff D. Tests for gene clustering. J. Comput. Biol. 2003;10:453–482. doi: 10.1089/10665270360688129. [DOI] [PubMed] [Google Scholar]
- 50.Feng DF, Doolittle RF. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 51.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 52.The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 53.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 54.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 55.Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 56.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 57.Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 58.Singer GA, Lloyd AT, Huminiecki LB, Wolfe KH. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol. Biol. Evol. 2005;22:767–775. doi: 10.1093/molbev/msi062. [DOI] [PubMed] [Google Scholar]
- 59.De Bodt S, Proost S, Vandepoele K, Rouze P, Van de Peer Y. Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genomics. 2009;10:288. doi: 10.1186/1471-2164-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu C. Chromatin remodeling and the control of gene expression. J. Biol. Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 62.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 63.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milinkovitch MC, Helaers R, Depiereux E, Tzika AC, Gabaldon T. 2x genomes–depth does matter. Genome Biol. 2010;11:R16. doi: 10.1186/gb-2010-11-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haussler D, O'Brien SJ, Ryder OA, Barker FK, Clamp M, Crawford AJ, Hanner R, Hanotte O, Johnson WE, McGuire JA, et al. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J. Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Proost S, Pattyn P, Gerats T, Van de Peer Y. Journey through the past: 150 million years of plant genome evolution. Plant J. 2011;66:58–65. doi: 10.1111/j.1365-313X.2011.04521.x. [DOI] [PubMed] [Google Scholar]
- 70.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandepoele K, Saeys Y, Simillion C, Raes J, Van De Peer Y. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 2002;12:1792–1801. doi: 10.1101/gr.400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hampson S, McLysaght A, Gaut B, Baldi P. LineUp: statistical detection of chromosomal homology with application to plant comparative genomics. Genome Res. 2003;13:999–1010. doi: 10.1101/gr.814403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hampson SE, Gaut BS, Baldi P. Statistical detection of chromosomal homology using shared-gene density alone. Bioinformatics. 2005;21:1339–1348. doi: 10.1093/bioinformatics/bti168. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, Shi X, Li Z, Zhu Q, Kong L, Tang W, Ge S, Luo J. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics. 2006;7:447. doi: 10.1186/1471-2105-7-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calabrese PP, Chakravarty S, Vision TJ. Fast identification and statistical evaluation of segmental homologies in comparative maps. Bioinformatics. 2003;19(Suppl. 1):i74–i80. doi: 10.1093/bioinformatics/btg1008. [DOI] [PubMed] [Google Scholar]
- 78.Pavesi G, Mauri G, Iannelli F, Gissi C, Pesole G. GeneSyn: a tool for detecting conserved gene order across genomes. Bioinformatics. 2004;20:1472–1474. doi: 10.1093/bioinformatics/bth102. [DOI] [PubMed] [Google Scholar]
- 79.Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- 80.Hachiya T, Osana Y, Popendorf K, Sakakibara Y. Accurate identification of orthologous segments among multiple genomes. Bioinformatics. 2009;25:853–860. doi: 10.1093/bioinformatics/btp070. [DOI] [PubMed] [Google Scholar]
- 81.Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: a system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16:1159–1168. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soderlund C, Bomhoff M, Nelson WM. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011;39:e68. doi: 10.1093/nar/gkr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cannon SB, Kozik A, Chan B, Michelmore R, Young ND. DiagHunter and GenoPix2D: programs for genomic comparisons, large-scale homology discovery and visualization. Genome Biol. 2003;4:R68. doi: 10.1186/gb-2003-4-10-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sinha AU, Meller J. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinformatics. 2007;8:82. doi: 10.1186/1471-2105-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang H, Lyons E, Pedersen B, Schnable JC, Paterson AH, Freeling M. Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics. 2011;12:102. doi: 10.1186/1471-2105-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pham SK, Pevzner PA. DRIMM-Synteny: decomposing genomes into evolutionary conserved segments. Bioinformatics. 2010;26:2509–2516. doi: 10.1093/bioinformatics/btq465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.