Abstract
Due to ongoing advances in sequencing technologies, billions of nucleotide sequences are now produced on a daily basis. A major challenge is to visualize these data for further downstream analysis. To this end, we present GenomeView, a stand-alone genome browser specifically designed to visualize and manipulate a multitude of genomics data. GenomeView enables users to dynamically browse high volumes of aligned short-read data, with dynamic navigation and semantic zooming, from the whole genome level to the single nucleotide. At the same time, the tool enables visualization of whole genome alignments of dozens of genomes relative to a reference sequence. GenomeView is unique in its capability to interactively handle huge data sets consisting of tens of aligned genomes, thousands of annotation features and millions of mapped short reads both as viewer and editor. GenomeView is freely available as an open source software package.
INTRODUCTION
Because of decreasing costs and increasing performance, so-called high-throughput sequencing or next-generation sequencing (NGS) machines produce millions of sequences at dozens of genome institutes around the world (1–4). The applications of NGS data are manifold. For instance, NGS is used for the efficient sampling of genomic diversity in viral and bacterial populations in large metagenomics projects (5). Another popular application of NGS is the re-sequencing of genomes, such as the 1000 human genomes project (http://www.1000genomes.org/) or the 1001 Arabidopsis genome project (http://www.1001genomes.org/). Genome (re)sequencing is important for polymorphism detection (6), structural variation analysis (7) and cancer allele detection (8). Two other recent applications of NGS, RNA-seq and ChIP-seq, are promising alternatives for microarrays (9). In RNA-seq, EST or cDNA samples are sequenced and mapped to a reference genome, providing a unique insight into the transcriptome (10,11). ChIP-seq provides a viable alternative to ChIP-on-chip microarrays to map transcription factor binding sites in vivo (12).
Last, but not least, apart from resequencing the genomes of species for which a reference genome sequence is already available, hundreds of complete genomes from a wide variety of organisms are currently being sequenced through NGS as well (13–16). However, problems regarding assembly still need to be overcome due to the limited length of the reads generally obtained from NGS (17,18).
When multiple complete genomes are available, depending on their phylogenetic distance, these genomes can be globally aligned (19) to study genome structure and genome evolution by looking for colinearity, insertions and deletions, and genome rearrangements. Examples of such whole genome multiple alignments for 45 vertebrate genomes, 5 worm genomes and 12 insect genomes are available for instance from the UCSC web site (http://genome.ucsc.edu/).
The production of these large amounts of sequence data has created a great need for visualization. Visual inspection of biological data is of great importance since it can help researchers to communicate results, to generate new hypotheses and to provide insights in biological processes (20). Many analyses are done computationally, but often there are steps that require human judgement. In this case, visualization can be extremely valuable as a sanity check on newly generated results, or can provide a valuable complement to the automated methods to plan new experiments (21). There are some resources available to browse mapped short reads, multiple alignments or genome annotation data (Table 1), but interactive browsers that comprehensively support the different data types are rare and suffer from several drawbacks such as speed, resolution, user-friendliness, proprietary file formats, cost and limited integration and extension options. Especially at the scale that these data are currently being generated, the choice of appropriate software tools to adequately handle these data, is very limited (21).
Table 1.
Biographical information and website addresses for a broad range of visualization tools for genome centric data
aEnvironment, can be either web based (web) or stand-alone (SA). Web-based applications require a server to function, stand-alone programs can work without.
To address these challenges, we present GenomeView, a genome browser that can handle a broad range of the new sequence data types resulting from NGS, in a user-friendly and intuitive manner. GenomeView is designed to browse sequences, annotations, multiple sequence alignments and NGS data all at once and on a genome-wide scale. It is a high-speed, stand-alone, interactive browser that gives the user access to a high-level overview of the data, but is equally capable to zoom-in down to a single nucleotide using semantic zooming. In contrast to regular zoom, semantic zoom does not only change the size of a graphical representation, but modifies the selection and structure of data to be displayed, which provides more useful information to the user. We also provide examples of how GenomeView can be integrated in existing projects and compare it with other state-of-the-art tools for vizualization.
MATERIALS AND METHODS
GenomeView is designed according to the Model-View-Controller architecture, which isolates the data from the representation and the control elements. This allows independent testing and development of the different components of the application.
Data management
Data management is done by a library, called JAnnot, which is developed in conjunction with GenomeView. JAnnot can also be used as an independent sequence analysis framework. The file types that JAnnot supports are listed in Table 2. The most common formats for genomics data are included. Most file-types are supported as read-only, except for the major annotation formats EMBL and GFF. While JAnnot supports multiple short-read alignment file formats, we strongly recommend users to convert their mappings to the BAM format described by Li et al. (41) using the SAMtools package. In a very short time, this format has gained broad support and seems to have become the de-facto standard for short-read alignments. GenomeView will automatically create index files and request the user to preprocess particular file format to more efficient alternatives.
Table 2.
Supported file formats in JAnnot, an up to date list is available at http://genomeview.org/content/data-formats
| Description | File types |
|---|---|
| Sequence and annotation | EMBL, Genbank |
| Sequence | fasta |
| Annotation | GFF, BED, Blast, GeneMark, PTT, TransTermHP TBL (Tair) |
| Multiple alignment | ClustalW, MAF, multi-fasta |
| Short-read alignment | BAM, MAQ/MapView |
| Continuous values/coverage | wiggle, TDF |
Data can either be loaded from a local file, or straight from a URL that points to a file on a web server. For remote files, we have implemented Secure Sockets Layer (SSL) encryption and authentication (http-basic) protocols that are supported by most modern web servers, ensuring that data are transferred encrypted from the server to GenomeView and only to people who are authenticated with credentials provided by the owner of the data.
When saving data loaded from a URL, GenomeView uses an http-post to send the data with changes back. This can be used in conjunction with a web service that handles this post to set up a gene curation platform. Because GenomeView can readily load data from a web server, it is straightforward to integrate GenomeView in existing websites as a visualization front-end. The full specification on how to implement the integration and further instructions to interact with existing data is described in detail in the manual on the website (http://genomeview.org/content/integration).
Availability and distribution
GenomeView is made available as Open Source Software. The code is licensed under the GNU GPL version 3. It is both available as binary and source code distributions. To run GenomeView, Java 6u10+ is required, which can be obtained free of charge for all major platforms (Windows, Linux, Mac OS and others) and is installed already on many systems. We recommend users to have at least 1 Gb of available memory and a dual-core processor for optimal performance.
GenomeView is distributed as a Java Web Start application, as a Java Applet and as a Java component. Java Web Start provides a platform-independent and secure deployment technology that enables us to deploy GenomeView to end-users by making it available on a standard web server. With any web browser, users can launch the application and be confident they always have the most recent version. This deployment is available for other labs to use GenomeView as standalone application or to integrate GenomeView in their web site without the need to set up their own local installation. Besides the actual program, we provide a user manual, a mailing list to discuss issues, a bug tracker, instructional videos and sample data for most track types for a number of different organisms.
Website URL: http://genomeview.org
RESULTS
The GenomeView Interface
Figure 1 illustrates the organization of the different components in GenomeView. There are two main panels within the Graphical User Interface (GUI), one containing all visualization tracks, and the other presenting the user with information about the data and selected features. The visualization panel is organized in separate tracks which are exemplified in Figures 2 through 7. We discuss the different track types in more detail in the next paragraphs.
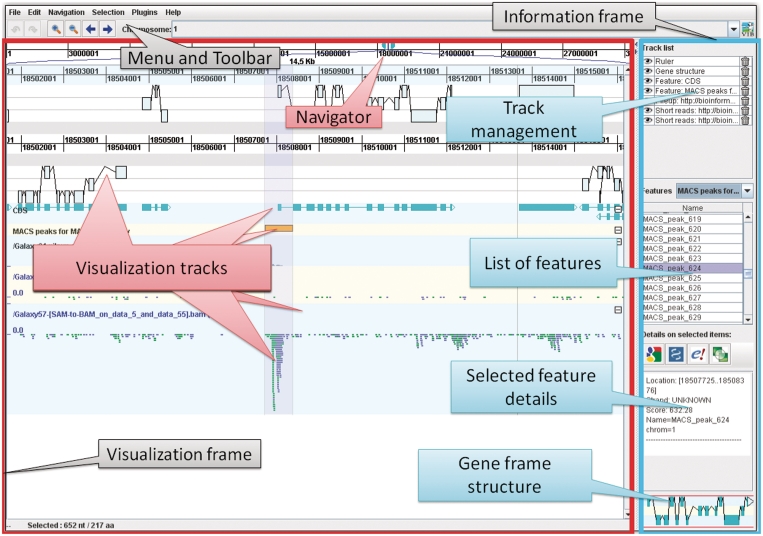
Figure 1.
Overview of the GUI layout of GenomeView. The top of the screen displays the menu and toolbar which provide access to all functions of GenomeView. The user interface is divided into two main frames with on the left the visualization frame and on the right the information frame. The visualization frame is further divided into a navigation bar at the top (indicated with ‘Navigator’), which allows users to quickly browse through the genome, and a large area that contains the various visualization tracks. The information frame contains four panels that provide management options and information about the data that is currently loaded. Navigation is entirely continuous, in contrast to step-wise navigation found in most other browsers. Furthermore, the zooming is semantic which means that the representation of the data changes according to zoom level and the amount of data on-screen.
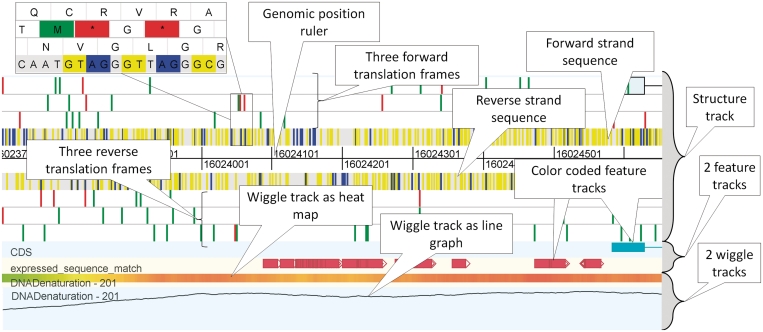
Figure 2.
Overview of some standard genome browser tracks. The structure track, which forms the upper part of the figure contains three forward and three reverse translation frames, as well as the forward and reverse strand sequence and the genomic ruler. In the structure track, start and stop codons and splice sites are indicated with a color (see inset for detailed zoom). Beneath the structure track there are two feature tracks that display annotation features using colored blocks. At the bottom of the figure there are two wiggle tracks, one depicted as heat map, the other as a line graph. The data in this figure is part of human chromosome 21, release hg18.
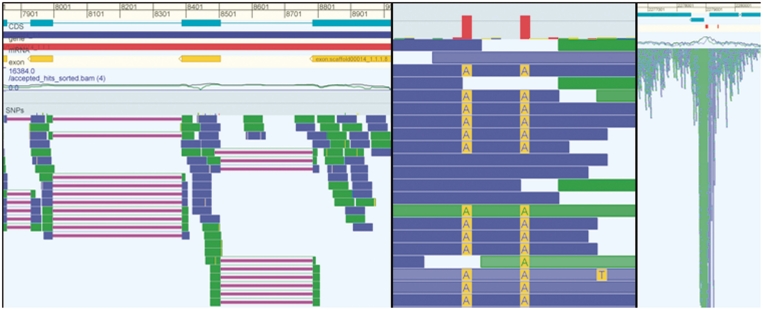
Figure 7.
The three panels in this figure show examples for different data types. The left panel shows a eukaryote genome with RNA-seq data. The reads in this RNA-seq experiment have been aligned with gaps (purple lines) and it is clear that many reads identify/confirm the splice junctions of the annotated CDS (cyan). The middle panel shows data from a resequencing project and shows a detailed view of two SNPs. The right panel shows data from a ChIP-seq experiment and it is obvious that the peak in the mapped sequencing data corresponds to the annotated binding sites (red blocks right above the coverage graph).
Classic genome browser tracks
Figure 2 illustrates the different track types that GenomeView provides in terms of typical genome browser tracks. The top track is called the stucture track. This track shows both strands of the sequence as well as potential splice sites. For both strands, also the three potential reading frames are displayed, plus potential start and stop codons, indicated in green and red, respectively. While we discuss the colors as they appear in the default color scheme, almost all colors are configurable.
Below the structure track there are two feature tracks which contain annotation features. These tracks will show the typical annotation associated with a sequence, such as CDS, genes, exons and many others. There can be multiple annotation tracks, each containing one type of feature. Annotation features are displayed as colored blocks, and when the structure consists of multiple locations, the blocks are connected with lines. GenomeView can also provide so-called wiggle tracks for showing continuous valued properties (see Figure 2).
Multiple alignment tracks
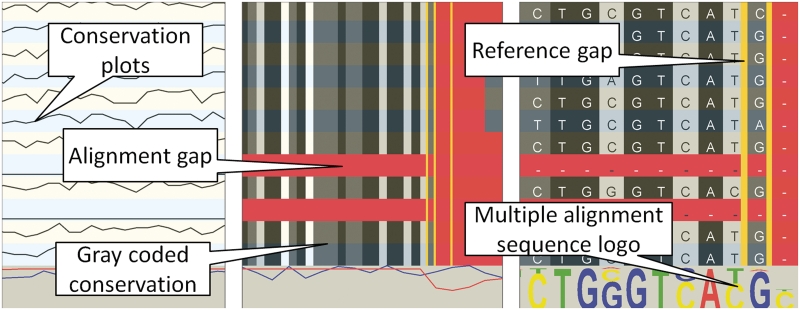
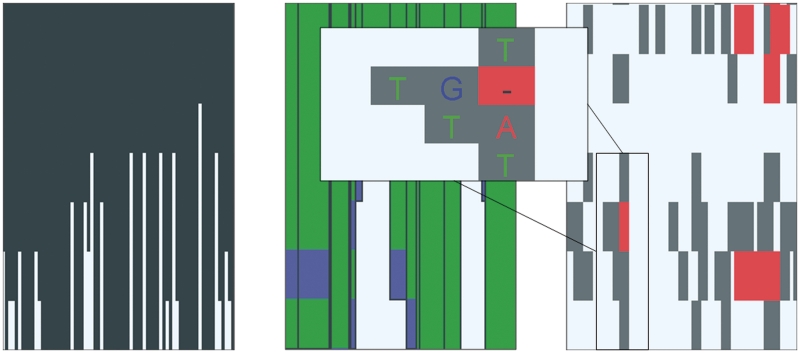
Multiple alignments in GenomeView are typically whole-genome sequence alignments. The type of track that is displayed depends on the data format. For instance, when importing a multiple sequence alignment from ClustalW or from a multi-fasta file, the alignment track is displayed as one line per aligned sequence plus one additional line at the bottom that shows the global conservation and coverage. Figure 3 shows such a multiple alignment track at three different zoom levels. When zoomed out, the tracks only show conservation plots. However, when zooming in, the conservation becomes color coded and finally individual nucleotides will be shown. At that point, the multiple alignment is also summarized as a sequence logo which allows users a quick overview of conserved sequences. The primary application domain for this type of track is the alignment of closely related genomes that have a nearly one-to-one nucleotide relationship.
Figure 3.
Overview of the basic multiple alignment track. Each line represents another organism. The left panel shows a large area and the multiple alignment tracks show conservation plots between the aligned sequence and the reference. When zooming in (the two rightmost panels), the conservation over the organisms is gray-coded in each track (black=100%, dark gray >75%, light gray >50% and white <50% conservation). Gaps in the aligned sequence are indicated in red, gaps in the reference are indicated with vertical yellow lines. The rightmost zoom is down to the nucleotide level and shows a sequence logo for each position in the alignment.
When loading data from a file in the multiple alignment format (MAF, http://genome.ucsc.edu/FAQ/FAQformat.html#format5), the multiple alignment is loaded as a MAF track as shown in Figure 4 for different zoom levels. This track is better suited to browse multiple alignments for large genomes, like for example vertebrate, insect or plant genomes. Zoomed out, this track shows the overall conservation. When zooming in, rearrangements in the aligned genomes are shown color coded and finally, again individual nucleotides can be seen. The mismatches are highlighted for easy discovery.
Figure 4.
Overview of the advanced multiple alignment track for large genomes. At a high-level perspective (left panel) the histogram-like plot shows the number of segments that align to the corresponding region in the reference genome. Essentially, this is a conservation plot. When zooming in, more details emerge (center panel). The height of the block is still relative to the number of species that contain that particular alignment block. The colors of the lines in each alignment block indicate the strand used in the alignment. Blue segments are aligned to the reverse strand, while green segments are mapped to the forward strand. When hovering over the block, a pop-up will appear that shows the organism name for each line in the alignment block. Zooming in further will provide more detailed information on the multiple alignments (right panel). At this level, mismatches are indicated with gray blocks and gaps with red blocks. When zooming in even further (inset), the gray blocks will contain the letter of the mismatched nucleotide in the aligned organism.
Short-read alignments
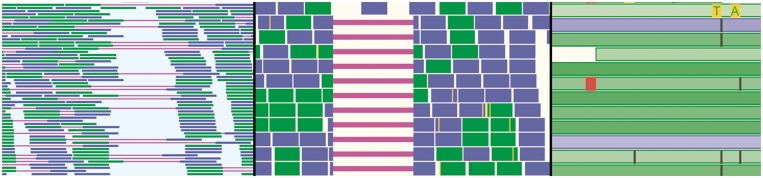
GenomeView supports multiple short-read mapping formats coming from the different NGS technologies. Figure 5 shows a short-read alignment track at various zoom levels with different data sets to show different features of GenomeView. The detailed view can be collapsed, or will collapse automatically when zooming out to a larger region (by default 25 000 nt). Before the NGS sequence reads can be shown in GenomeView they have to be aligned to a reference sequence with any of the short-read aligners that are available.
Figure 5.
Overview of the short-read visualization track. This track shows the alignment of individual reads. Green and blue indicate reads mapping to the forward and reverse strand respectively. Purple lines are used to connect paired-end reads (left panel), or two parts of a read that has been aligned over a splice-junction (center panel). Yellow indicates mismatches between the read and the reference genome. Black and red are used to indicate indels (right panel). The brightness of the color of a read indicates the mapping quality assigned by the mapping software used in producing this short-read alignment. The lighter a read, the lower the mapping quality. In case of paired-end sequencing there is a second color scheme (not shown) to indicate directionality of the read.
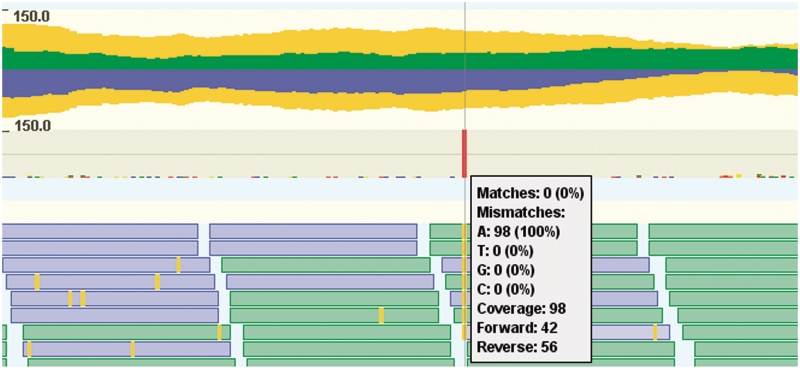
While being able to browse individual reads can be extremely valuable in many studies, for others it is sufficient to see a summary of the sequencing data. An important summary for NGS data is the read coverage, i.e. how many reads align to a particular position. This can be accomplished in GenomeView using the pile up track, depicted in Figure 6. This track shows both the coverage, as well as the consensus nucleotide composition of the reads.
Figure 6.
Overview of the pileup visualization track. This track consists of two parts: the top part is a read coverage plot, while the bottom part shows the consensus sequence summary of individual nucleotides. The read coverage plot consists of three plots, one for the reads mapping to the forward (green, above central axis), one for the reverse strand (blue, belows central axis) and one for the total coverage (yellow, mirrored above and below the central axis). The second part of the pile up track provides a more detailed view that displays a summary of the individual nucleotides as colored bar charts. Centrally in the figure, there is one red bar, indicating a single nucleotide polymorphism (SNP). This is confirmed by hovering the mouse over the pileup track, which gives a textual summary of the coverage and nucleotide frequencies. There are some reads (blue and green boxes) from the short-read track at the bottom of the figure overlapping with the pop-up with detailed information.
GenomeView is agnostic about the experiment type that is represented in your NGS data set. Figure 7 shows examples for RNA-seq, resequencing data and ChIP-seq. This figure also illustrates the benefit of visualizing data: it makes RNA-seq or ChIP-seq experiments much easier to understand and interpret.
Integration
Even though GenomeView is a stand-alone application, it is fairly straightforward to integrate it as a viewer or editor in another environment. The GenomeView website provides detailed information that guides website developers through the different steps to present their data in GenomeView. In essence one needs to construct a hyperlink (URL) to GenomeView that contains a pointer to the data and configuration that needs to be loaded. The data can be in any of the supported file formats (see overview in Table 2). Many of these formats have been used for many years, with the notable exception of the BAM format (41), which was only recently conceived to handle the massive number of sequence reads generated by NGS methods. Even though this format is relatively new, it is already wide-spread and has emerged as the leading format for NGS mappings.
Once this URL is constructed, GenomeView will start and fetch the required data. Even though it works with flat files, it does support indexing for mosts formats and can retrieve just the small part of a file that is needed for the current view. This is accomplished through support for HTTP range queries. Essentially, this allows GenomeView to load data from the web server for a particular genomic region first and fetch more as needed. This approach thus provides virtually instant access to a particular region while also providing the user the ability to explore the initial region as if the entire data set had been loaded all at once. To illustrate the value and opportunities of integration of a visualization tool into a data platform, we discuss two case studies in which GenomeView is used as viewer or editor for a third-party platform.
GenomeView and integration with the Tuberculosis Database
The Tuberculosis Database (TBDB, http://www.tbdb.org/) is an online database providing integrated access to genome sequence, expression data and literature information for Mycobacterium tuberculosis and related actinomycetes (44). Among the data being hosted at TBDB is next generation short-read sequencing data for a M. tuberculosis phylogeographic diversity sequencing project. This project builds on existing models of TB global population structure (45) by re-sequencing 31 TB strains that have been carefully selected as representatives of the global diversity of M. tuberculosis. Sequence polymorphisms between these strains were then detected by alignment to the H37Rv reference genome sequence.
GenomeView has been integrated in TBDB as the primary visualization tool for short-read alignment data. As described above, GenomeView provides a dynamic and interactive genome browser-style visualization of the reference genome, features of the genome (e.g. genes) and aligned reads. With GenomeView, TBDB users may zoom from a full genome view down to a single nucleotide. By providing access to the underlying read alignments, GenomeView allows TBDB users to verify reported polymorphisms, look for possible missed polymorphisms and visualize regions with low coverage where possible polymorphisms cannot be identified. The integration within TBDB highlights the ability of GenomeView to rapidly visualize large-scale short-read data sets over a network connection.
GenomeView is also used as a primary tool for analyzing NGS data as part of an National Institute of Allergy and Infectious Diseases funded contract for Systems Biology for tuberculosis. This project is applying a range of profiling techniques to reconstruct the regulatory and metabolic network of M. tuberculosis. A substantial challenge of this project is the management and visualization of large-scale data sets, including short-read data sets. Within this project, GenomeView is being used to both visualize and analyze RNA-seq and ChIP-seq data.
In addition to internal uses for data analysis and visualization, GenomeView is also being used by the TB Systems Biology project to host RNA-seq and ChIP-seq data publically through a web-based interface (http://www.broadinstitute.org/annotation/tbsysbio/resources.html).
GenomeView as an annotation curator tool
Besides a tool for visualizing NGS and comparative data sets, GenomeView is also designed to assist manual gene annotation and curation. It has all capabilities a genome curation expert would expect from an annotation editor, such as the possibility to modify gene coordinates, indicate missing start or stop codons, correct splice sites, add functional annotation to genes, identify and annotate new genes and merge or split genes. For instance, GenomeView is integrated as a viewer and an annotation editor in the BOGAS genome curation platform (http://bioinformatics.psb.ugent.be/webtools/bogas/). Registered genome curators use the BOGAS website to go to their assigned loci and then start GenomeView to correct gene models. Typically this involves correcting splice-sites, merging single exon genes and splitting fused genes. During these tasks the curators have immediate access to all the data used in the predictions like RNA-seq, multiple alignments, blast hits and coding potential graphs. Finally, they save their data in GenomeView, which updates the data on the BOGAS server by using a webservice.
GenomeView can also be used as an independent annotation editor without the support of the BOGAS platform. Users can load their own annotation data from any of the supported file formats (see Table 2). They can change, add or remove annotations and save them back to the original file, or as a new locally stored GFF or EMBL file. This can be useful to quickly bookmark interesting locations in the genome for later retrieval.
Plug-ins
To make GenomeView as small, efficient and maintainable as possible, the core code only provides the basic browsing and editing functionality. All other functions can be added as plug-ins. The Java Plug-in Framework (JPF) is used to manage plug-ins (http://jpf.sourceforge.net). JPF provides a runtime engine that dynamically discovers and loads plug-ins. It maintains a registry of available plug-ins and the functions they provide.
We actively support two plug-ins (http://genomeview.org/plugins) at the moment and several others are in development and can be retrieved from the code repository. The first officially supported plug-in contains a collection of properties that can be calculated from the DNA sequence. These so-called sequence-dependent properties include GC-content, physical properties of DNA and many others.
Plug-ins are available from the website, which also has step-by-step instructions on how to download and install plug-ins. The website also has basic documentation to get you started with developing your own plug-in.
Comparison with other genome browsers
Table 1 lists 24 genome browsers, genome editors and a plethora of other visualization tools for genomics data with citation and website information. In the next paragraphs we highlight features that are beneficial to a large group of researchers working with genome centric data. We did not perform a one-on-one comparison of a list of features as the NGS visualization field is evolving so fast, this comparison would be outdated within months, if not weeks.
There are generally two types of genome browsers. First of all there are the web-based systems like Ensembl and UCSC that have the advantage that they can do a lot of work on the server side, without the user noticing. A drawback of this approach is that you need to be online to access the data and that the experience is less interactive. You have to wait for the page to reload if you want to move around. A second problem has to do with visualizing personal data. Either you have to set up your own server, which is hardly trivial, or you need to send your data to a remote server not under your control, which may not be possible for medical data. The second type of systems, including GenomeView, covers most other genome browsers. They are standalone applications that do all the heavy lifting on the user computer, but have the advantage they still work offline and can be used with local data.
Most of the tools in Table 1 focus on either NGS data, annotation or multiple alignments. The notable exceptions are IGV and GenomeView, which both allow you to visualize all three in one tool. Enabling scientists to integratively explore their different data sources is key in gaining new knowledge. In many respects IGV and GenomeView have some similarities, but we like to highlight a number of differences from our perspective.
The first big difference is that GenomeView is also an annotation editor. This has major repercussions on the internal handling of data and the efficiency of rendering algorithms. Annotations can be modified by the user and therefore the visualizations cannot be pre-rendered or cached (as is done in most tools). Allowing users to change the data requires architectural decisions from the very beginning of a project. Putting in editing capabilities as an afterthought would be nearly impossible and would require an almost rewrite of the software.
GenomeView has a richer representation of NGS data than most, if not all, current NGS tools. It shows visual clues about insertions, deletions, read-pairs, alignments over splice junctions, the DNA strand to which a read maps, directional information whether the read came from a sense or anti-sense transcript as well as detailed information about the individual reads. Besides visualization of raw read data, GenomeView also has a rich visualization of the summary NGS data, as visualized with a coverage plot (Figure 6).
Integrating GenomeView as viewer in third-party websites is straightforward, either as an applet or a webstart application. GenomeView uses almost exclusively community standardized file formats, that make it easy to exchange data with other software. It is possible to control GenomeView from the website in which it is embedded using JavaScript. Data security is provided throught http authentication and SSL encryption.
GenomeView and Savant are the only two tools that provide users the ability to extend the platform with custom analytical modules using a plug-in architecture.
GenomeView is suitable to handle a wide variety of genomics data and is designed for commodity hardware. It will work fine on a modest desktop computer. Our test system has a dual-core 1 GHz processor and 1 Gb of available memory. Reference sequences with associated annotation, multiple alignments and short-read mappings including mammalian sized genomes are no problem to browse with GenomeView on a regular desktop computer. GenomeView is able to load larger data sets than most other stand-alone tools described in Table 1. This is done by using community standards for indexing of data files (41). Semantic zooming allows GenomeView to handle extremely large data sets elegantly, while still presenting the user with an informative view.
DISCUSSION
GenomeView has been designed with a number of criteria in mind. First, our aim was to cover a broad range of data types that can be displayed, the rationale being to be able to show any type of data that can be mapped to a reference sequence. Types of data that are currently supported include sequence, annotation, short-read alignments, multiple alignments, genome colinearity and expression data. Our second aim was to make the tool as user-friendly as possible. This means that the tool itself has a very basic user interface with only the essentials. It also means that it is straightforward to integrate or connect GenomeView with your existing data sources. Additional functions are made available through plug-ins which the user can install as needed.
GenomeView is an interactive tool that allows you to take a quick glance at a genome. As such it can easily handle complete chromosomes and remains fast with dozens of aligned genomes, thousands of annotation features and millions of mapped short reads.
Preloaded demo instances for Caenorhabditis elegans, Drosophila melanogaster, Bacillus anthracis and the iDEA challenge (http://www.illumina.com/landing/idea/) data sets are presented to the user the first time they start GenomeView. These demos contain a reference sequence, a gene annotation and at least one other data type. The extra data is typically one of: a multiple alignment, re-sequencing data or RNA-seq. GenomeView instances for 22 plant genomes are already made available through GenomeView in collaboration with the PLAZA platform, a resource for plant comparative genomics (46). We are making available new genomes on a regular basis and users can request new genomes to be included. Currently we have 40 genomes pre-loaded (7 demo, 2 bacterial, 22 plant and 9 animal genomes) and we continue to expand this number.
Because human interpretation is extremely valuable throughout a project, visual methods are the key complement to automatic analyses. They enable researchers to inspect the data, create hypotheses, perform much needed visual evaluation on any preliminary results and keep an eye on further downstream results.
CONCLUSION
In conclusion, GenomeView provides an attractive way to present the results and data to the scientific community for any genomics or sequence analysis project. GenomeView has the ability to export high-resolution images of the visualized data, as illustrated by the figures throughout the manuscript.
A recent review by Nielsen et al. (21) distinguishes three core user tasks in visualizing genomes: (i) analyzing NGS data, (ii) browsing annotations and experimental data and (iii) comparing sequences from different organisms or individuals. GenomeView is well-suited for each of these core tasks. Furthermore, the authors discuss a number of challenges with current genome visualization methods. GenomeView tackles several of the challenges raised in this review.
The first point brought forward by Nielsen et al. (21) was that a visualization platform is a good start, but it would even be better to allow scientists to perform interactive analyses on their data. The GenomeView plug-in architecture allows scientists to develop and perform on-the-fly-analyses within GenomeView. A second point of concern is the increasingly large amount of sensitive information. In particular personal genomic information requires protection. As such there is an emerging need for data security. To the best of our knowledge GenomeView is the only tool that supports SSL encryption and authentication when loading data from a webserver. Authentication and encryption are both needed to protect sensitive information. A final point that was raised by the authors concerns indels, both in short-read alignments and in genome alignments. In both cases GenomeView is capable of visualizing such indels.
GenomeView provides a huge interactive visualization range in terms of data types compared to any other tool, while still going from a multi-mega base chromosome overview down to the single nucleotide within that chromosome. While GenomeView is very well suited to handle the data that are available now, the future will hold a whole new set of challenges. Visualizing even more types of data and ever larger sets will require us to keep improving the existing capabilities. GenomeView has been under constant development for the past 3 years and will remain so to stay current with new developments as they happen. Keeping up with the pace that sequencing methods evolve will prove to be an interesting challenge, especially in light of the several thousands to tens of thousands of genomes currently underway for humans, vertebrates and plants. This will challenge us to think about new techniques and paradigms to visualize data sets of this magnitude, but also to think about technical improvements to algorithms and data structures.
FUNDING
Research Foundation Flanders (FWO) (to T.A. and Y.S.); Belgian American Education Foundation (to T.A.); Ghent University (Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks”); Interuniversity Attraction Poles Programme (IUAP P6/25), initiated by the Belgian State, Science Policy Office (BioMaGNet). Funding for open access charge: Ghent University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marguerat S, Wilhelm BT, Bhler J. Next-generation sequencing: applications beyond genomes. Biochem. Soc. Trans. 2008;36(Pt 5):1091–1096. doi: 10.1042/BST0361091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster SC. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 4.Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. MetaHIT Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 7.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, et al. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 9.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bhler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 10.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 11.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 12.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez D, Franois P, Farinelli L, Osters M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imelfort M, Edwards D. De novo sequencing of plant genomes using second-generation technologies. Brief Bioinform. 2009;10:609–618. doi: 10.1093/bib/bbp039. [DOI] [PubMed] [Google Scholar]
- 16.Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg LA, Bouffard P, Burt DW, Crasta O, Crooijmans RPMA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinform. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Donoghue SI, Gavin A-C, Gehlenborg N, Goodsell DS, Hrich J-K, Nielsen CB, North C, Olson AJ, Procter JB, Shattuck DW, et al. Visualizing biological data-now and in the future. Nat. Methods. 2010;7(Suppl. 3):S2–S4. doi: 10.1038/nmeth.f.301. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen CB, Cantor M, Dubchak I, Gordon D, Wang T. Visualizing genomes: techniques and challenges. Nat. Methods. 2010;7(Suppl. 3):S5–S15. doi: 10.1038/nmeth.1422. [DOI] [PubMed] [Google Scholar]
- 22.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis SE, Searle SMJ, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0082. research0082–0082.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engels R, Yu T, Burge C, Mesirov JP, DeCaprio D, Galagan JE. Combo: a whole genome comparative browser. Bioinformatics. 2006;22:1782–1783. doi: 10.1093/bioinformatics/btl193. [DOI] [PubMed] [Google Scholar]
- 25.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 26.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 27.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 28.Huang W, Marth G. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–1543. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staden R. The Staden sequence analysis package. Mol. Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 31.Huntley D, Tang YA, Nesterova TB, Butcher S, Brockdorff N. Genome Environment Browser (GEB): a dynamic browser for visualising high-throughput experimental data in the context of genome features. BMC Bioinformatics. 2008;9:501. doi: 10.1186/1471-2105-9-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Istrail S, Sutton G, Florea L, Halpern A, Mobarry C, Lippert R, Walenz B, Shatkay H, Dew I, Miller J. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl Acad. Sci. USA. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson JT, Thorvaldsdttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–2132. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao H, Guo H, Wang J, Zhou R, Lu X, Shi S. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics. 2009;25:1554–1555. doi: 10.1093/bioinformatics/btp255. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiume M, Williams V, Brook A, Brudno M. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–1944. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet–next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 44.Galagan JE, Sisk P, Stolte C, Weiner B, Koehrsen M, Wymore F, Reddy TBK, Zucker JD, Engels R, Gellesch M, et al. TB database 2010: overview and update. Tuberculosis (Edinb) 2010;90:225–235. doi: 10.1016/j.tube.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]