The molecular chaperone Hsp90 regulates morphogenesis of the leading human fungal pathogen Candida albicans. Hsp90 inhibition induces filaments with a delay in mitotic exit mediated by the checkpoint protein Bub2. Hsp90 depletion destabilizes the cyclin-dependent kinase Cdc28, providing a link between Hsp90, cell cycle regulation, and morphogenesis.
Abstract
The trimorphic fungus Candida albicans is the leading cause of systemic candidiasis, a disease with poor prognosis affecting immunocompromised individuals. The capacity of C. albicans to transition between morphological states is a key determinant of its ability to cause life-threatening infection. Recently the molecular chaperone heat shock protein 90 (Hsp90) was implicated as a major regulator of temperature-dependent C. albicans morphogenesis; compromising Hsp90 function induces filamentation and relieves repression of Ras1–protein kinase A (PKA) signaling, although the mechanism involved remains unknown. Here we demonstrate that filaments generated by compromise of Hsp90 function are neither pseudohyphae nor hyphae but closely resemble filaments formed in response to cell cycle arrest. Closer examination revealed that these filaments exhibit a delay in mitotic exit mediated by the checkpoint protein Bub2. Furthermore, Hsp90 inhibition also led to a distinct morphology with defects in cytokinesis. We found that the cyclin-dependent kinase Cdc28 was destabilized in response to depletion of Hsp90 and that Cdc28 physically interacts with Hsp90, implicating this major cell cycle regulator as a novel Hsp90 client protein in C. albicans. Taken together, our results suggest that Hsp90 is instrumental in the regulation of cell division during yeast-form growth in C. albicans and exerts its major effects during late cell cycle events.
INTRODUCTION
The yeast Candida albicans is an important opportunistic human pathogen. It is the leading cause of systemic candidiasis, the fourth-most-common hospital-acquired bloodstream infection in the United States (Pfaller and Diekema, 2007). Normally living commensally in the gastrointestinal flora, this fungus is capable of causing oral, esophageal, and vaginal mucosal infections, as well as life-threatening systemic disease in immunocompromised individuals. C. albicans is also emerging as an important model organism for the study of fungal morphogenesis. This trimorphic fungus can grow as unicellular budding yeast and in two filamentous, multicellular forms: hyphae and pseudohyphae. These three morphotypes are characterized by differences in shape, cell cycle progression, and control of polarized growth (Sudbery et al., 2004; Sudbery, 2011; Berman, 2006; Shapiro et al., 2011). C. albicans is highly morphologically plastic and switches readily between forms in response to external cues. The capacity to undergo morphogenesis is important for C. albicans virulence, such that mutants that cannot undergo morphological transitions often have attenuated virulence (Lo et al., 1997; Murad et al., 2001; Gow et al., 2002; Banerjee et al., 2008; Carlisle et al., 2009; Noble et al., 2010; Shapiro et al., 2011; Sudbery, 2011). Filaments are further implicated in pathogenicity, as many filament-specific genes encode virulence factors such as proteases and adhesins (Kumamoto and Vinces, 2005; Shapiro et al., 2011; Sudbery, 2011).
Pseudohyphae and hyphae are distinct morphologies. Pseudohyphae resemble chains of elongated yeast cells; they are wider than hyphae and have clear constrictions at cellular junctions. Hyphae are thinner, lack a constriction between the mother and daughter cell, and have unconstricted septa. One of the major features that distinguish true hyphae from yeast or pseudohyphae is their mode of cell cycle progression (Sudbery et al., 2004; Berman, 2006). In both yeast and pseudohyphae, budding is coordinated with the onset of DNA replication (Hazan et al., 2002; Warenda and Konopka, 2002). A ring of septin proteins forms at the bud site prior to bud emergence. This ring later acts as both a scaffold and diffusion barrier for the assembly and localization of machinery required for cytokinesis (Longtine and Bi, 2003; Dobbelaere and Barral, 2004; Finger, 2005). Nuclear division occurs across the bud neck, followed by the formation and contraction of an actomyosin ring, deposition of a chitinous septum, and, in the case of yeast-form growth, separation of the mother and daughter cells (Lippincott and Li, 1998). Pseudohyphae are characterized by an extended period of polarized growth, more time spent in G2 phase, and the persistent attachment of mother and daughter cells following nuclear division (Berman, 2006). In hyphal growth, in contrast, evagination of a long, thin germ tube occurs prior to the initiation of DNA replication, and nuclear division takes place across a septin ring that forms some distance down the germ tube (Sudbery, 2001; Hazan et al., 2002; Finley and Berman, 2005). As in pseudohyphae, following septum deposition at the site of nuclear division, mother and daughter cells remain attached. Despite the many distinctions between pseudohyphae and hyphae, a common transcriptional regulatory mechanism specifies their fate in a dosage-dependent manner (Carlisle et al., 2009).
Cell shape in C. albicans is tightly linked to regulation of cell cycle progression, and disruption of components of the cell cycle regulatory machinery has dramatic morphological consequences (Berman, 2006; Shapiro et al., 2011). Cell cycle progression in C. albicans is regulated through the association of the cyclin-dependent kinase (CDK) Cdc28 with different cyclins. Regulation of these cyclins is complex and differs among morphotypes. The G1 cyclin Cln3, for example, is required to maintain yeast-form growth, whereas another G1 cyclin, Ccn1, is required for hyphal extension (Loeb et al., 1999; Bachewich and Whiteway, 2005; Chapa y Lazo et al., 2005). The two C. albicans mitotic cyclins, Clb2 and Clb4, both negatively regulate polarized growth, and depletion of either results in filamentous growth (Bensen et al., 2005). Depletion of the CDK Cdc28 results in variable filamentous morphologies, and other major regulators of cell cycle progression, including the polo-like kinase Cdc5 and the CDK inhibitor Sol1, also have a strong influence on growth morphology (Bachewich et al., 2003; Atir-Lande et al., 2005; Umeyama et al., 2006). Finally, activation of cell cycle checkpoints through DNA damage, DNA replication stress, or spindle damage stall cell cycle progression and cause strongly polarized growth (Shi et al., 2007).
Recently the molecular chaperone heat shock protein 90 (Hsp90) has been implicated as an important regulator of the temperature-dependent switch between yeast and filamentous growth in C. albicans (Shapiro et al., 2009). Impairing Hsp90 function in the absence of other hypha-inducing cues causes a transition from yeast to filamentous growth that is contingent on Ras1–protein kinase A (PKA) signaling, a pathway involved in hyphal induction in response to most cues (Liu, 2001; Shapiro et al., 2011). Hsp90 is a key temperature sensor governing morphogenesis, given that reduction of Hsp90 function bypasses the requirement for elevated temperature for cues such as serum to induce filamentous growth and that Hsp90 function is partially compromised in conditions that induce hyphal morphogenesis (Shapiro et al., 2009). Hsp90 is an essential molecular chaperone that stabilizes a diverse but select set of inherently unstable client proteins, including many regulators of cell signaling (Wandinger et al., 2008; Taipale et al., 2010). Among Hsp90's many roles in the cell, it is indispensable for appropriate regulation of cell cycle progression in many organisms. This feature has sparked interested in its potential utility as a cancer drug target. Inhibition of Hsp90 causes arrest in different stages of the cell cycle in a variety of cancer cell lines, and this is often accompanied by the induction of apoptosis (Srethapakdi et al., 2000; Hostein et al., 2001; de Carcer, 2004; Georgakis et al., 2006; Garcia-Morales et al., 2007; Watanabe et al., 2009). Hsp90's role in cell cycle progression appears to be conserved in the fungal kingdom. In the budding yeast Saccharomyces cerevisiae, Hsp90 is required for cell cycle progression during environmental stress, and in the fission yeast Schizosaccharomyces pombe Hsp90 interacts with cell cycle machinery (Aligue et al., 1994; Munoz and Jimenez, 1999; McClellan et al., 2007). Several key regulators of cell cycle progression have been implicated as Hsp90 interactors, including the mammalian tyrosine kinase Wee1 and its budding yeast homologue Swe1, the polo-like kinase Plk1 and its yeast homologue Cdc5, and the cyclin-dependent kinase Cdc2 and its budding yeast homologue Cdc28 (Aligue et al., 1994; Munoz and Jimenez, 1999; de Carcer et al., 2001; Goes and Martin, 2001; Mort-Bontemps-Soret et al., 2002; Mollapour et al., 2010). Recently it was demonstrated that Hsp90 itself is subject to posttranslational modification in a cell cycle–dependent manner. In S. cerevisiae, during S phase of the cell cycle the tyrosine kinase Swe1 phosphorylates a residue in the N-terminus of Hsp90 that facilitates chaperone interaction with numerous client proteins (Mollapour et al., 2010).
A role for Hsp90 in C. albicans cell cycle progression has not been established, and properties of filaments induced upon Hsp90 inhibition remain enigmatic. Here we demonstrate that filaments generated by Hsp90 compromise are distinct from either pseudohyphae or hyphae but rather closely resemble filaments induced by cell cycle arrest. Consistent with this finding, we establish that Hsp90 is instrumental for progression through the cell cycle during yeast-form growth in C. albicans and demonstrate that cell cycle arrest may play a role in induction of filamentation in response to compromised Hsp90 function. We also identify the CDK Cdc28 as a client protein of Hsp90. The only other two Hsp90 client proteins identified to date include the protein phosphatase calcineurin and the MAP kinase Mkc1, which are key regulators of the evolution and maintenance of antifungal drug resistance (Singh et al., 2009; LaFayette et al., 2010). Thus we provide a molecular link between Hsp90, cell cycle regulation, and morphogenesis and identify the third Hsp90 client protein in C. albicans and the first with a function in cell cycle progression.
RESULTS
Filaments resulting from compromised Hsp90 function are neither pseudohyphae nor hyphae
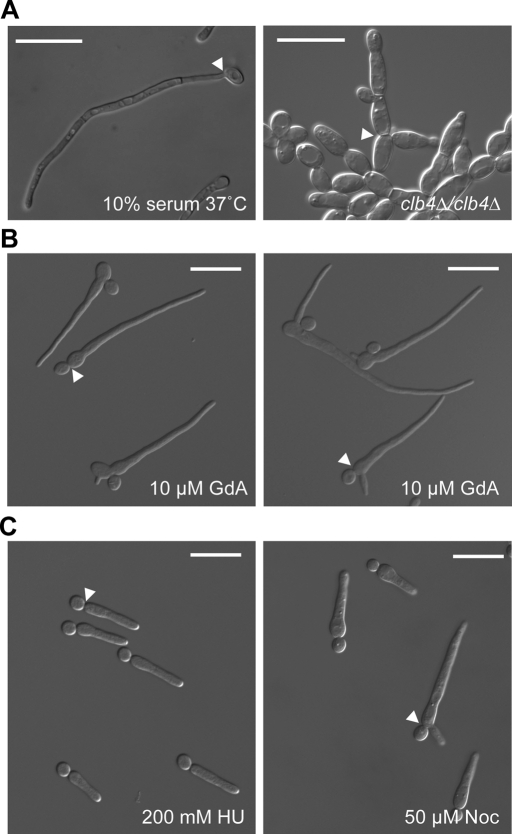
We first attempted to define the growth morphology of filaments resulting from compromised Hsp90 function. We analyzed filaments induced by growth in the presence of the Hsp90 inhibitor geldanamycin (Whitesell et al., 1994), which inhibits Hsp90 function by binding with high affinity to Hsp90's unusual ATP-binding pocket, according to the canonical definitions of pseudohyphae and hyphae (Sudbery et al., 2004; Figure 1). Like pseudohyphae, filaments induced by geldanamycin had a clear constriction between the filament and the attached yeast-form cell. Unlike pseudohyphae, however, the highly elongated filaments had no obvious subsequent constrictions at cell junctions. In fact, the walls of the filament were approximately parallel and more closely resembled hyphae than pseudohyphae. When the filaments were compared with other documented filamentous morphologies of C. albicans, it became clear that they bore a striking resemblance to filaments induced by cell cycle arrest, specifically at S, G2, and M phases of the cell cycle (Bachewich et al., 2003, 2005; Bensen et al., 2005; Shi et al., 2007).
FIGURE 1:
Filaments induced by Hsp90 inhibition most closely resemble filaments induced by cell cycle arrest. (A) C. albicans hyphae (10% serum, 37°C) and pseudohyphae (clb4Δ/clb4Δ). Bud necks are indicated by white arrowheads. (B) C. albicans treated with the Hsp90 inhibitor GdA. Note that bud necks (white arrowheads) have a constriction similar to pseudohyphae, whereas the walls of the filament are approximately parallel, similar to hyphae. (C) C. albicans elongating in response to arrest in S phase (HU treatment) or M phase (Noc treatment) bear a striking resemblance to GdA-induced filaments. Bud necks are indicated by white arrowheads. Scale bars, 20 μm.
Response of C. albicans to Hsp90 inhibition is variable and results in two distinct morphologies
To more closely examine the morphology of individual filaments and to assess how they form in response to Hsp90 inhibition, we selected three time points representing early, middle, and later stages of induction: 4, 6, and 8 h of treatment with geldanamycin. Beyond 8 h, filaments were generally too long and intertwined to allow for analysis of individuals. After 4 h of treatment, filaments were short and resembled an elongated, tapered yeast cell attached to another yeast cell (Figure 2A). Because polarized growth is generally restricted to the daughter cell in C. albicans, the elongating cell was presumed to be the daughter cell. After 6 and 8 h of treatment, filaments were longer, and some had begun to form additional branches (Figure 2A). The number of branches in an individual filament showed some correlation with the degree of Hsp90 compromise, with higher doses of geldanamycin favoring more highly branched filaments (Supplemental Figure S1).
FIGURE 2:
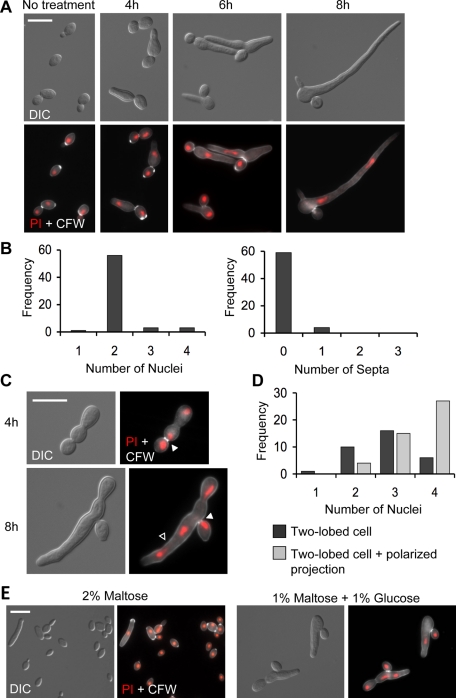
Hsp90 inhibition produces two distinct morphologies. (A) Filaments stained with PI (DNA, red) and CFW (chitin, white) after 4, 6, and 8 h of treatment with GdA. (B) Frequency histograms of the number of nuclei and number of septa in representative cells following 8 h of GdA treatment. (C) Cells of a two-lobed morphology stained as described in A. Solid white arrowheads indicate bud necks, and open arrowhead indicates a polarized projection growing off of a two-lobed cell. (D) Frequency histograms of the number of nuclei in representative two-lobed cells with and without polarized projections. (E) Genetic depletion of Hsp90 phenocopies pharmacological inhibition of Hsp90. MAL2p-HSP90/hsp90Δ grown in media containing 2% maltose (promoter induced) or 1% maltose and 1% glucose (promoter repressed) for 6 h. In response to glucose, both binucleate filaments and two-lobed cells are present. Scale bars, 10 μm.
To examine the distribution of septa and nuclei, a protocol was developed to simultaneously stain DNA and the cell wall using dyes with distinct spectral properties. Specifically, nuclei were stained using the red fluorescent nucleic acid stain propidium iodide, and chitin in the cell wall was stained with calcofluor white. Septa are composed mainly of chitin, and calcofluor white gives particularly bright bands of staining at septal junctions (Supplemental Figure S2). After 8 h of treatment, almost all filaments contained two nuclei and no septa (Figure 2B). C. albicans hyphae of similar average length induced by a standard hypha-inducing cue (N-acetyl glucosamine) were analyzed by the same criteria and found to contain more varied numbers of nuclei and septa, most ranging from one to three (Supplemental Figure S2). Of interest, geldanamycin-induced filaments almost always contained exactly two nuclei even in filaments with multiple branches (Supplemental Figure S1). This is intriguing, as lateral branching in C. albicans hyphae occurs when subapical cells become competent to reenter the cell cycle and is thus correlated with nuclear division (Barelle et al., 2003). It appears that filament branching and nuclear division are uncoupled in the case of geldanamycin-induced filamentation.
A second, distinct morphology was found in geldanamycin-treated cultures (Figure 2C). This form consists of a single yeast cell, believed to be the mother cell, attached to a daughter cell consisting of two lobes with an obvious constriction between them. This constriction was consistently wider than the mother/daughter cell junction. At later time points, these two-lobed cells could also form highly polarized projections (Figure 2C, open arrowhead). Calcofluor white staining revealed that the daughter cell constriction contained no septum, although the mother/daughter cell junction tended to have brighter staining. Propidium iodide staining suggested that this form is multinucleate (Figure 2D). Cells contained between two and four discernible nuclei.
To confirm that both of these morphologies were a specific result of compromised Hsp90 function rather than an off-target effect of geldanamycin, we used a strain in which levels of Hsp90 can be genetically depleted. In this strain, one allele of HSP90 is deleted and the remaining allele placed under the control of the MAL2 promoter (Shapiro et al., 2009; Figure 2E). This promoter is induced when cells are grown in the presence of maltose and repressed in the presence of glucose. After 6 h of growth, cells grown in maltose were primarily of the budding yeast form, whereas cells grown in glucose displayed both of the prominent morphologies found in response to geldanamycin treatment—elongated, binucleate cells and multinucleate, two-lobed cells (Figure 2E).
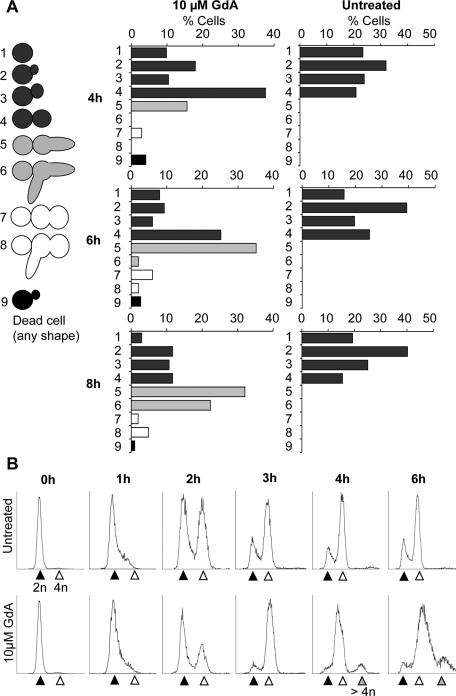
To quantify the relative frequencies of the different phenotypes over time, a randomly selected population of cells was analyzed and scored for morphology after 4, 6, and 8 h of geldanamycin treatment (Figure 3A). This analysis revealed that filaments were the most prominent response, with the two-lobed morphology forming a much smaller proportion of the total cell population. In addition, a population of yeast cells was present at all time points. The results of this experiment also gave clues about the temporal dynamics of morphological transitions. Notably, at 4 and 6 h there was an enrichment of large-budded yeast cells compared with the untreated sample, and this occurred prior to the emergence of a large population of filaments at later time points (Figure 3A). This observation supports the notion that filaments may represent a yeast-form cell that has become hyperelongated. In addition, filaments with multiple branches and two-lobed cells with polarized projections only emerged at later time points.
FIGURE 3:
Hsp90 inhibition induces cells with distinct morphological states and higher ploidy. (A) Categories of observed morphologies and frequency histograms showing percentage of cells in each morphological category after 4, 6, and 8 h of growth with GdA treatment, as indicated. Cells were selected at random. At, 4 h, n = 173; 6 h, n = 151; and 8 h, n = 103. (B) FACS analysis of nuclear DNA content identifies major populations of 4n and higher-ploidy DNA content following GdA treatment. The 2n peak is indicated by the black arrowhead, 4n by white, and higher ploidy by gray. Samples were fixed after 2, 4, and 6 h of treatment as indicated and stained with propidium iodide. The y-axes indicate the number of recorded events, and the x-axes are arbitrary units of fluorescence intensity.
To monitor cell cycle progression and confirm the presence of discrete binucleate and multinucleate populations, fluorescence-activated cell sorting (FACS) analysis was performed to analyze nuclear DNA content in individual cells. Small, unbudded G1 cells were isolated, and half of the population was left untreated and half was treated with geldanamycin. After 2 h of treatment, a higher proportion of the geldanamycin-treated population had 2n nuclear content compared with the untreated sample (Figure 3B, black arrowheads). By 3 and 4 h, the geldanamycin-treated population is highly enriched for 4n cells and, in addition, a third peak indicative of cells at higher ploidy appeared after 4 h in the geldanamycin-treated samples (Figure 3B, gray arrowheads). At 6 h there was not an appreciable recovery of the 2n peak compared with the untreated control, the 4n peak (Figure 3B, white arrowheads) represented the largest proportion of the population, and the higher-ploidy peak persisted and became larger. We hypothesized that the substantial 4n peak accounted for both large-budded yeast cells and binucleate filaments and that the higher-ploidy peak accounted for the multinucleate, two-lobed cells. To confirm this, populations of cells larger than typical yeast cells with either 4n or 8n DNA content were sorted by FACS and examined under the microscope (Supplemental Figure S3). Consistent with our hypothesis, the 8n group was highly enriched for two-lobed cells, and this cell type was found almost exclusively in this group. It was surprising, however, that a large population of binucleate filaments was also present in the 8n group. This either may be due to increased background staining due to larger cell size or may indicate that nuclei in these cells have undergone a second round of DNA replication and are of higher ploidy.
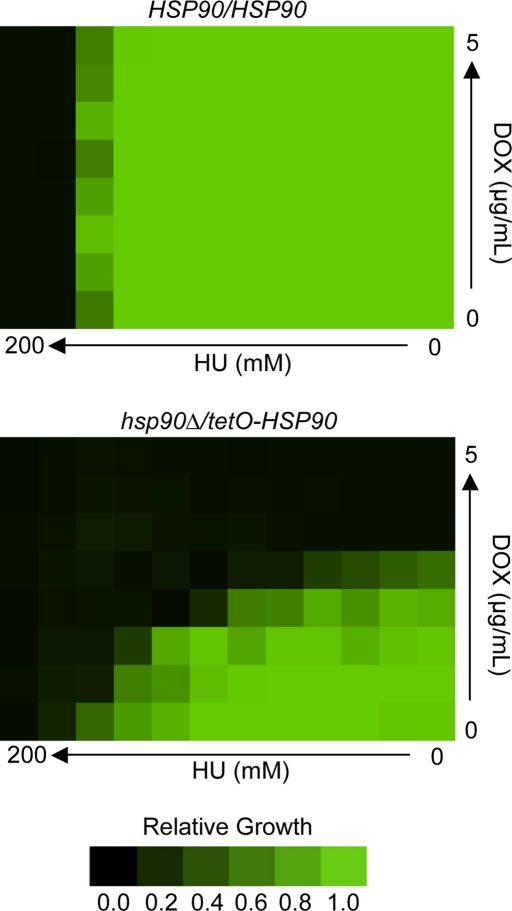
Notably, after 4 h of geldanamycin treatment, the large 4n peak in the FACS analysis appears shifted toward lower DNA content values (Figure 3B), suggesting that cells may not be completing S phase efficiently. The delay in the appearance of the 4n peak after 2 h in the geldanamycin-treated population further suggests that cells may be delayed in their entry or progression through S phase. We reasoned that if Hsp90 compromise resulted in delayed progression through S phase, cells depleted of Hsp90 would be hypersensitive to hydroxyurea, a drug that inhibits ribonucleotide reductase and arrests DNA replication. We used a strain with one allele of HSP90 deleted and the other under the control of a tetracycline-repressible promoter (Shapiro et al., 2009). When this strain is exposed to the tetracycline analogue doxycyline, HSP90 expression is diminished. A checkerboard assay was performed in which a fixed concentration of cells was exposed to concentration gradients of both doxycycline and hydroxyurea across a 96-well plate (Figure 4). The combined activity of the two drugs was synergistic, suggesting that cells with decreased levels of Hsp90 were hypersensitive to hydroxyurea.
FIGURE 4:
Depletion of Hsp90 confers hypersensitivity to the DNA replication inhibitor hydroxyurea. Checkerboard assays were performed with gradients of HU and the tetracycline analogue doxycycline (DOX) to mediate transcriptional repression of HSP90 in the hsp90Δ/tetO-HSP90 strain. HU and DOX were diluted twofold across wells of a 96-well plate, which were then inoculated with equal concentrations of cells. Green represents full growth, and black represents no growth relative to the drug-free control well. The wild-type strain (HSP90/HSP90) shows no hypersensitivity to HU at any concentration of DOX, whereas a strain with its only allele of HSP90 under the control of a tetracycline-repressible promoter (hsp90Δ/tetO-HSP90) shows dose-dependent hypersensitivity with increasing concentrations of DOX.
Cells with the two-lobed morphology show defects in cell division
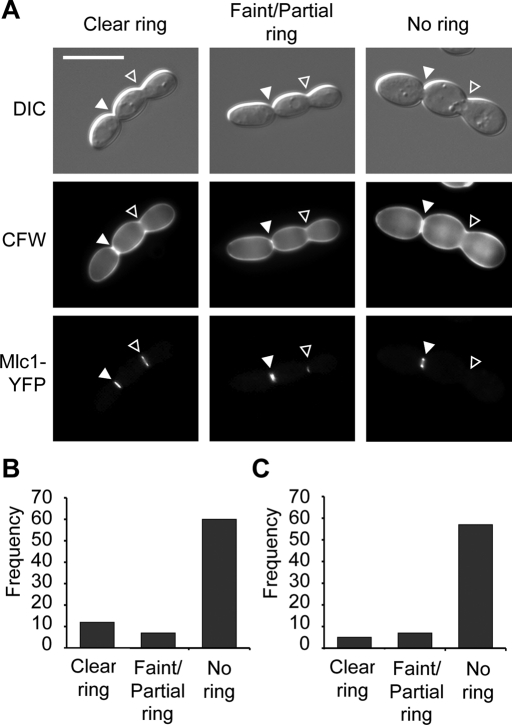
The two-lobed form is characterized by an aseptate constriction in the daughter cell. This constriction was consistently devoid of septa even when nuclear material was visible on either side (Figure 2C), suggesting that these cells may have defects in cytokinesis. In budding yeast, cytokinesis involves the formation and contraction of an actomyosin ring, followed by septum deposition (Lippincott and Li, 1998). In C. albicans mutants with defects in cytokinesis, sometimes an actomyosin ring forms and persists through subsequent cell cycles, whereas in other cases it forms and subsequently breaks down (Li et al., 2008; Gonzalez-Novo et al., 2009). In both cases the ring is unable to contract, resulting in wider cell junctions similar to the constriction in the two-lobed daughter cell (Figures 2C and 5). To assess actomyosin ring formation in cells with the two-lobed morphologies, a strain harboring a yellow fluorescent protein (YFP)–tagged version of the actomyosin ring marker Mlc1 (Crampin et al., 2005) was treated with geldanamycin. It is striking that nearly all cells examined (96%, n = 79) had Mlc1-YFP fluorescence at the bud neck between mother and daughter cell (Figure 5A, white arrowheads). The presence of Mlc1-YFP fluorescence at the daughter cell constriction, however, was variable. The majority of cells had no ring, whereas some cells contained bright, faint, or partial rings (Figure 5B). This could indicate that rings initially form and then break down without contraction, that rings are delayed in their formation, or that cells of this form have variable ability to assemble contractile actomyosin rings.
FIGURE 5:
The two-lobed form has defects in cytokinesis. (A) The mother/daughter cell junction (solid white arrowheads) nearly always contained a clear actomyosin ring (Mlc1-YFP) and septin ring (Cdc3-YFP; not shown), whereas the daughter cell constriction (open arrowheads) could have a clear, faint, partial, or absent ring. Cells were stained with CFW to confirm that the daughter cell constriction did not contain a septum regardless of the presence or absence of an actomyosin or septin ring. (B) Frequency histograms of actomyosin ring appearance at the daughter cell constriction based on the categories shown in A. n = 79. (C) Frequency histograms of septin ring appearance at the daughter cell constriction based on the categories shown in A. n = 69. Scale bar, 10 μm.
Septins are another important component of the cytokinesis machinery in budding yeast. One of their functions is to serve as a scaffold for the assembly of actomyosin rings (Longtine and Bi, 2003). A strain with a YFP-tagged allele of the septin Cdc3 was used to test whether septin ring formation was defective in the two-lobed morphology (Finley and Berman, 2005). Similar to Mlc1-YFP fluorescence, Cdc3-YFP was almost ubiquitously present at the junction between mother and daughter cells (97%, n = 69) but its presence was variable at the daughter cell constriction, with the majority of cells having no visible septin ring (Figure 5C). The presence of a septin ring at the mother/daughter cell junction and its absence from the daughter cell constriction is intriguing. Mother and daughter cells remained attached after sonication, and it was not uncommon to observe anucleate mother cells or nuclei spanning the mother/daughter cell junction, indicating nuclear migration across the site (data not shown). This strongly suggests that these two cells are continuous and that both constrictions have defects in cytokinesis despite their differing appearances. It may also suggest, however, that different processes are affected at each of the two sites. At the mother/daughter cell junction septum deposition appears to be the primary defect, whereas septin ring establishment appears to be aberrant at the daughter cell constriction.
Binucleate filaments exhibit a delay in mitotic exit
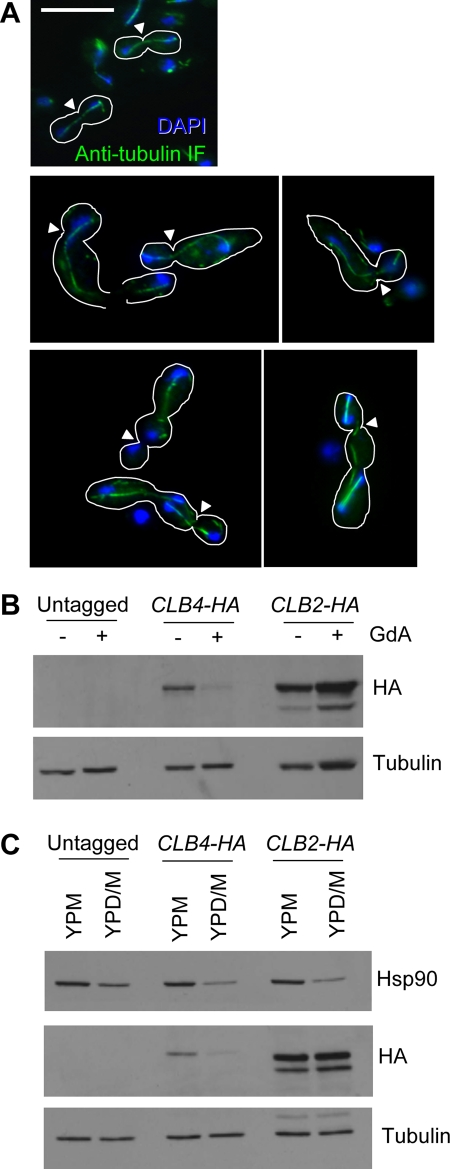
Because filaments generated by inhibition of Hsp90 most often contained exactly two nuclei and no septa (Figure 2) and grow in a morphology markedly similar that induced by cell cycle arrest (Figure 1), we wanted to determine whether there was further evidence for a delay in a specific part of the cell cycle in these filaments. We assessed spindle morphology using an indirect immunofluorescence technique to visualize tubulin. Cells were examined at early stages of induction, since an extensive microtubule network in highly elongated cells obscured any cell cycle–related structures (Supplemental Figure S5). It is striking that a majority of cells (76%, n = 85) had elongated microtubule structures between the two nuclei that resembled mitotic spindles (Figure 6A), suggesting that these cells may experience a delay in late mitosis after sister chromatid separation but prior to spindle breakdown. Two-lobed cells were also examined. It is remarkable that many of these cells (62%, n = 24) contained at least one, sometimes two, elongated spindles between pairs of nuclei, indicating that this cell type may also be subject to cell cycle delay (Figure 6A).
FIGURE 6:
Cells treated with GdA show evidence of mitotic arrest. (A) Tubulin (green) was visualized using an indirect immunofluorescence protocol, and nuclei were stained with DAPI (blue). Cell shapes are outlined in white, and bud necks are indicated by white arrowheads. Top, mitotic spindles in untreated cells. In cells elongating in response to GdA treatment (middle), nuclei are connected by elongated microtubule structures resembling mitotic spindles. Similar structures were observed in the two-lobed form (bottom). Scale bar, 10 μm. (B) The two C. albicans mitotic cyclins, Clb2 and Clb4, behave differently in response to Hsp90 inhibition. Western blot using stains with HA-tagged alleles of the mitotic cyclins Clb2 and Clb4. In response to Hsp90 inhibition with GdA for 5 h, levels of Clb4 are reduced and levels of Clb2 are not. Tubulin served as a loading control. (C) Western blot as in B using the MAL2p-HSP90/hsp90Δ strain with HA-tagged Clb2 or Clb4 grown for 5.5 h in either 2% maltose (YPM; promoter induced) or 1% maltose and 1% glucose (YPD/M; promoter repressed). Levels of Hsp90 were assessed using an anti-Hsp90 antibody, and tubulin served as a loading control. Genetic depletion of Hsp90 phenocopied pharmacological inhibition.
Elongation of C. albicans cells in response to cell cycle arrest usually represents a terminal phenotype. Although the cells continue to elongate for some time, they never divide and eventually die (Berman, 2006). To establish whether geldanamycin-induced filaments were subject to the same fate, cultures were grown for 24 h and then assayed for cell viability and signs of cell division. Viability was assayed by incubation with propidium iodide, a dye that is easily excluded by healthy, metabolically active cells and is readily taken up by dead or dying cells. Cell division was assayed using calcofluor white staining to look for evidence of septum deposition. Although many filaments took up the propidium iodide stain, some were easily able to exclude it after 24 h (Supplemental Figure S4B). Furthermore, septa were easily detected in many filaments after 24 h of growth, suggesting that by this time point, cell division had resumed in many filaments (Supplemental Figure S4A). Taken together, these results suggest that cells are able to overcome the temporary mitotic exit block induced by Hsp90 inhibition in the conditions used in our experiments, and many remain viable after 24 h of growth.
Polarized growth initiated by cell cycle arrest typically results in cytoskeletal organization reminiscent of growing hyphae (Bachewich et al., 2003; Bensen et al., 2005; Andaluz et al., 2006; Umeyama et al., 2006). Consistent with these reports, geldanamycin-induced filaments also exhibited the extensive helical microtubule networks and highly polarized actin characteristic of hyphal growth (Supplemental Figure S5B). Of interest, in filaments with multiple branches not all tips had equal intensity of staining, with some tips having no evidence of polarized actin. This may indicate that the multiple branches are not all actively growing at the same time.
To further explore Hsp90's impact on cell cycle progression, we examined the dynamics of the two C. albicans mitotic cyclins, Clb2 and Clb4. These proteins are expressed from S phase through G2 and M phases and are degraded at the end of mitosis (Bensen et al., 2005; Cote et al., 2009). Strains harboring hemagglutinin (HA)-tagged Clb2 or Clb4 (Bensen et al., 2005) were used to monitor changes in protein levels in response to either pharmacological compromise of Hsp90 function or genetic reduction of Hsp90 levels. Of interest, the two cyclins behaved quite differently (Figure 6). Levels of Clb4 were markedly lower in conditions in which Hsp90 was compromised compared with controls, whereas levels of Clb2 were unchanged. Clb4 plays a major role in S phase progression, whereas Clb2 functions in G2/M phase and is elevated in cells arrested in late mitosis (Bensen et al., 2005; Chou et al., 2011; Ofir and Kornitzer, 2010). Furthermore, degradation of both of these cyclins is necessary for the cell to exit from mitosis (Bensen et al., 2005). Thus the persistence of Clb2 in the absence of Clb4 may support a delay in mitotic completion. Alternatively, Hsp90 could be required for Clb4 stability. This would be consistent with the possibility of slowed S phase progression observed in the FACS analysis (Figure 3B). However, this is less likely to contribute to Hsp90's impact on morphogenesis. as CLB4 deletion does not phenocopy Hsp90 inhibition (Bensen et al., 2005).
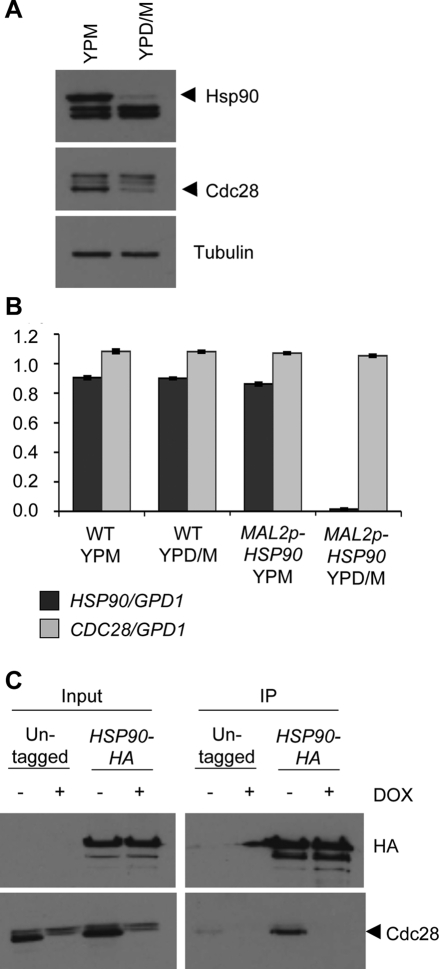
Cyclins regulate cell cycle progression through their association with CDKs. In S. cerevisiae, the CDK Cdc28 has been shown to interact with both Hsp90 and one of its cochaperones, Cdc37 (Mort-Bontemps-Soret et al., 2002; Mollapour et al., 2010). In C. albicans, Cdc28 is normally present at constant levels throughout the cell cycle, and its depletion was demonstrated to cause filamentous growth (Umeyama et al., 2006). Unlike the response to geldanamycin, these filaments have variable morphologies and do not consistently arrest with a specific number of nuclei. Nevertheless, Cdc28 is an attractive candidate for an Hsp90 client protein that may play a role in Hsp90's influence on cell cycle progression. To test whether stability of Cdc28 was affected by Hsp90 depletion, an antibody raised against a conserved CDK motif, PSTAIRE, was used to detect Cdc28 by Western blot. In response to depletion of Hsp90, Cdc28 levels were markedly reduced (Figure 7A). Quantitative RT-PCR demonstrated that CDC28 transcript levels remained stable when Hsp90 was depleted, suggesting that Cdc28 depletion was most likely due to protein instability (Figure 7B). To determine whether Cdc28 physically interacts with Hsp90 in C. albicans, a coimmunoprecipitation assay was performed. HA tags were added to the C-terminus of Hsp90 in a strain in which one allele of CDC28 was deleted and the other was placed under the control of a tetracycline-repressible promoter. Immunoprecipitation of HA-tagged Hsp90 with HA agarose beads copurified a band detected by the anti-PSTAIRE antibody, based on strong enrichment relative to the untagged control strain (Figure 7C). This band disappeared when cells were treated with doxycycline to deplete Cdc28, confirming that this band corresponded to Cdc28.
FIGURE 7:
Cdc28 is a client protein of Hsp90 in C. albicans. (A) The MAL2p-HSP90/hsp90Δ strain was grown in rich medium with 2% maltose (YPM; promoter induced) or 1% maltose and 1% glucose (YPD/M; promoter repressed) for 7.5 h. Protein extracts were subject to Western blot with an Hsp90 antibody to confirm Hsp90 depletion, a PSTAIRE antibody against a conserved cyclin-dependent kinase motif to detect Cdc28, and a tubulin antibody as a loading control. In conditions that repress Hsp90 expression, Cdc28 levels are reduced. (B) Quantitative RT-PCR of CDC28 transcript levels in wild-type (WT) and MAL2p-HSP90/hsp90Δ. CDC28 transcript levels are normalized to GPD1 transcript, and error bars are ± SD of three technical replicates. (C) Coimmunoprecipitation (IP) of Hsp90-HA and Cdc28 in the cdc28Δ/tetO-CDC28 background. Hsp90-HA was immunoprecipitated with immobilized anti-HA antibody, and Cdc28 was detected using the anti-PSTAIRE antibody. Western blot of IP inputs (left) shows depletion of Cdc28 when either untagged and HA-tagged strains were treated with doxycycline (DOX). When Hsp90-HA was immunoprecipitated, a band detected by the anti-PSTAIRE antibody was highly enriched in the Hsp90-HA strain compared with the untagged control strain. This band is not present in the DOX-treated sample, confirming that this band corresponds to Cdc28.
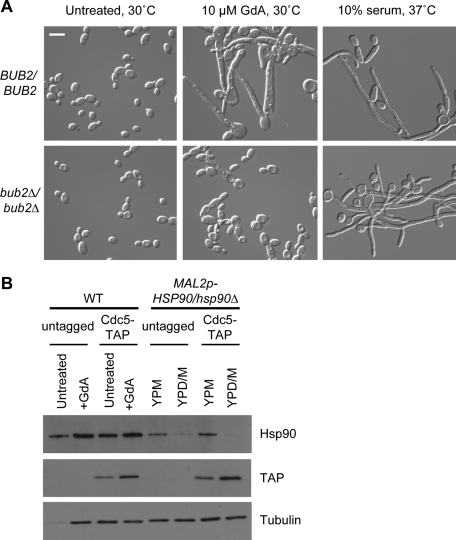
Events involved in mitotic exit are poorly understood in C. albicans, but in S. cerevisiae, this process has been well studied and involves two major signaling cascades: the Cdc14 early anaphase release (FEAR) in early anaphase and the mitotic exit network (MEN) in late anaphase (de Gramont and Cohen-Fix, 2005). Central to the MEN is the Bub2/Bfa1 checkpoint complex. When this complex is activated, signaling through the MEN is inhibited and cells cannot progress to G1 of the next cell cycle (Wang et al., 2000). To see whether activation of this checkpoint plays a role in filamentation in response to Hsp90 compromise, a strain with both copies of BUB2 deleted (Bachewich et al., 2005) was treated with geldanamycin. The bub2Δ/Δ mutant did not form the characteristic long, tapered filaments and two-lobed cells typically seen in response to geldanamycin (Figure 8A). Instead, the cultures primarily contained yeast with some pseudohyphae and chains of unseparated cells. bub2Δ/Δ cells responded normally to growth in 10% serum at 37°C by forming hyphae, ruling out the possibility of a general filamentation defect. The dampened phenotypic response of this strain to geldanamycin suggests that the Bub2/Bfa1 checkpoint may be involved in filamentation in response to Hsp90 compromise.
FIGURE 8:
The mitotic exit checkpoint protein Bub2 is involved in the morphological response to Hsp90 inhibition, whereas the polo-like kinase Cdc5 is not. (A) Morphology of wild-type and bub2Δ/bub2Δ cells treated with GdA at 30°C or with 10% serum at 37°C. (B) Western blot of strains with TAP-tagged Cdc5 in a wild-type or MAL2p-HSP90/hsp90Δ background. Strains were grown in conditions with Hsp90 fully functional (untreated or YPM) or with Hsp90 function or expression compromised (GdA or YPD/M) for 6 h. When Hsp90 was inhibited or depleted, levels of Cdc5 detected with a TAP antibody were elevated. Hsp90 levels were assessed to ensure depletion in the MAL2p-HSP90/hsp90Δ background, and tubulin was included as a loading control. Scale bar, 10 μm.
In S. cerevisiae, the polo-like kinase Cdc5 is expressed during G2/M phase and regulates numerous steps of mitosis (van Vugt and Medema, 2005). In late mitosis it phosphorylates and inactivates the Bub2/Bfa1 checkpoint to facilitate activation of the MEN (Hu et al., 2001). Depletion of Cdc5 in C. albicans results in cell elongation due to arrest in earlier events of mitosis and is likely involved in mitotic exit as well (Bachewich et al., 2003; Chou et al., 2011). Furthermore, a link between Cdc5 and Bub2 has been established. Deletion of BUB2 diminishes the cell cycle arrest and cell elongation phenotype typically observed upon Cdc5 depletion, suggesting that regulation of the Bub2/Bfa1 checkpoint by Cdc5 is conserved in C. albicans (Bachewich et al., 2005). To determine whether destabilization of Cdc5 could be causing activation of the Bub2/Bfa1 checkpoint, strains with Cdc5 tandem affinity purification (TAP) tagged were subject to pharmacological inhibition or genetic depletion of Hsp90. Of interest, rather than being depleted, levels of Cdc5 were noticeably elevated compared with controls (Figure 8B). Therefore destabilization and degradation of Cdc5 cannot explain the observed phenotype. Cdc5 accumulation was reported in C. albicans during mitotic arrest due to depletion or deletion of the major MEN targets Cdc20 and Cdh1 (Chou et al., 2011). Thus elevated Cdc5 levels in response to Hsp90 depletion are likely due to the large number of cells arrested in mitosis.
DISCUSSION
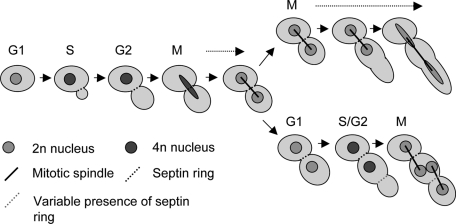
Our results establish an entirely new role for Hsp90 in regulating morphogenesis of the pathogenic fungus C. albicans. Hsp90 inhibition produces two prominent morphologies with properties that suggest previously unidentified roles for the chaperone in cell division and cell cycle progression. One morphology is binucleate filaments, which exhibit a delay in late mitosis (Figures 2 and 6). Given the phenotypic similarity between these filaments and those induced by cell cycle arrest, cell cycle delay could be the causative agent of filament induction, with the daughter cell representing a hyperelongated yeast-form cell (Figure 9, top path). The second morphology is the multinucleate, two-lobed form, which primarily has defects in cytokinesis (Figures 2 and 5). The multiple nuclei suggest that this cell type may undergo one successful round of nuclear division before becoming delayed in mitosis during its second round (Figures 6 and 9, bottom path), which could explain the formation of polarized projections off this form at later stages. In addition, FACS analysis (Figure 3B) and hydroxyurea sensitivity (Figure 4) suggest that S phase progression may be slowed down when Hsp90 function is compromised.
FIGURE 9:
Stochasticity determines the morphological consequence of Hsp90 inhibition. The outcome of the first nuclear division determines whether a filament or two-lobed cell will occur. Failure in spindle breakdown results in cell elongation and the formation of a binucleate filament. Successful spindle breakdown followed by a failure in cell separation leads to the two-lobed morphology, which may subsequently experience a delay in mitotic exit akin to the binucleate filaments.
That Hsp90 may influence more than one process in cell cycle progression is not surprising, since it interacts with ∼10% of the S. cerevisiae proteome (Zhao et al., 2005). One possibility is that when Hsp90 is inhibited in asynchronous cultures, cells at different stages of the cell cycle have different outcomes. This is unlikely, however, because when synchronous G1 cells were collected by elutriation for FACS analysis, both cell types were present following geldanamycin treatment (data not shown). The other, more likely possibility is that stochasticity determines which morphology will occur, favoring a higher proportion of filaments (Figure 3A). We propose that the moment a cell's morphological fate is decided occurs at the time of spindle breakdown in late anaphase/early telophase. If this process is not successful, the result is a binucleate filament. If, however, the cell is able to exit mitosis, the daughter cell may fail to separate from the mother cell and proceed to the next cell cycle, where cytokinetic defects lead to the production of the two-lobed form (Figure 9). Given that a population of yeast cells typically persists after Hsp90 inhibition, the final possibility is that both mitotic exit and cell separation proceed normally and a new G1 yeast cell is formed (not depicted).
Hsp90's influence on cell cycle progression must be considered in the context of its role as a molecular chaperone. We demonstrated that levels of Cdc28, the CDK whose mammalian and S. cerevisiae homologues are known Hsp90 interactors (Mort-Bontemps-Soret et al., 2002; Watanabe et al., 2009; Mollapour et al., 2010), are reduced in response to depletion of Hsp90 and that Hsp90 and Cdc28 physically interact in C. albicans (Figure 7). This implicates Cdc28 as a client protein of Hsp90 in C. albicans, one of very few such proteins identified. Reduction of Cdc28 levels in C. albicans results in filamentous growth, but in contrast to Hsp90 inhibition, in which a very specific binucleate phenotype is obtained, reduced Cdc28 leads to a mixture of pseudohyphae, hyphal-like filaments, and hyperelongated aseptate cells with one or two nuclei (Umeyama et al., 2006). This is in contrast to the phenotype observed in response to Hsp90 inhibition, in which only two distinct morphologies occur. Of note, Cdc28 depletion in response to compromised Hsp90 function was not complete (Figure 7), indicating there is likely residual Cdc28 function. The reduction in Cdc28 levels may not be sufficient to cause a phenotype on its own but may sensitize the cell to further cell cycle perturbations. Thus, although Hsp90 affects stability of a major cell cycle regulator that operates during all stages of the cell cycle, the specific defects observed in S phase and mitotic exit upon Hsp90 compromise suggest the involvement of other Hsp90 client proteins critical for these phases of the cell cycle. When these client proteins that remain to be identified are unable to function due to Hsp90 compromise, the cell may be unable to cope with the additional stress of reduced Cdc28 levels, culminating in prominent cell cycle arrest phenotypes.
Events involved in mitotic exit have been poorly studied in C. albicans but are well established in S. cerevisiae. In S. cerevisiae, Cdc15 and Dbf2, two kinases in the MEN cascade, were identified in screens for Hsp90 interactors, and bub2 null mutants are hypersensitive to Hsp90 inhibitors, as are strains lacking the major MEN targets Sic1 and Cdh1 (Zhao et al., 2005; McClellan et al., 2007). cdc15 null mutants are also hypersensitive to geldanamycin in C. albicans (S. Diezmann and L. E. Cowen, unpublished results). We have established a link between Hsp90 and the MEN in C. albicans by demonstrating that the major checkpoint protein Bub2 is involved in the response of C. albicans to Hsp90 inhibition (Figure 8). Supporting this connection are clear phenotypic similarities between morphologies found in our studies and C. albicans strains that, for example, cannot degrade Clb2, have the C. albicans homologue of SIC1 deleted, or have Dbf2 levels depleted (Atir-Lande et al., 2005; Bensen et al., 2005; Gonzalez-Novo et al., 2009; Chou et al., 2011). There does appear, however, to be considerable divergence between S. cerevisiae and C. albicans MEN architecture, so it would be impractical to make any predictions as to how Hsp90 interacts with this pathway based on the S. cerevisiae model (Bensen et al., 2005; Clemente-Blanco et al., 2006; Gonzalez-Novo et al., 2009; Chou et al., 2011).
Although the C. albicans Bub2/Bfa1 checkpoint may play a role in mitotic delay in response to Hsp90 inhibition, Cdc5, a kinase that inactivates Bub2/Bfa1, does not. Levels of Cdc5 increase in response to Hsp90 inhibition, suggesting that it is not a major Hsp90 client protein in C. albicans (Figure 8). This is intriguing, as Plk1, the mammalian homologue of Cdc5, is an Hsp90 client protein, and Cdc5 interacts with an Hsp90 cochaperone, Cdc37, in S. cerevisiae (de Carcer et al., 2001; de Carcer, 2004; Mort-Bontemps-Soret et al., 2002). Of note, Cdc37 has been demonstrated to function both in consort with and independent of Hsp90, so Cdc5's function may not necessarily rely on Hsp90 in fungi (MacLean and Picard, 2003). In S. cerevisiae, an additional, unidentified kinase also regulates Bub2/Bfa1, and this kinase may represent the more likely candidate for a fungal Hsp90 client protein (Hu and Elledge, 2002). Another possibility is that Hsp90 compromise may result in a genotoxic stress that causes activation of the Bub2/Bfa1 checkpoint. In S. cerevisiae this checkpoint is involved in response to both DNA and spindle damage, but its roles in these processes in C. albicans have not been studied (Wang et al., 2000; Hu et al., 2001).
Hsp90 regulates temperature-dependent morphogenesis by exerting a repressive effect on Ras1-PKA signaling, which is alleviated upon temperature elevation (Shapiro et al., 2009). In S. cerevisiae strains with modest functional defects in their mitotic exit machinery, an increase in Ras-PKA signaling greatly enhances the severity of these phenotypes (Irniger et al., 2000). A similar relationship exists in S. pombe (Yamashita et al., 1996). This phenomenon may be conserved in C. albicans and play an important role in the effect of Hsp90 inhibition on mitotic exit. By both facilitating MEN signaling and inhibiting Ras1-PKA signaling, Hsp90 would be poised to regulate mitotic exit through two pathways. The critical assumption for this model is that, in the context of hyphal growth, additional factors exist to promote activity of the APC/C even in the presence of activated Ras1-PKA signaling, and these factors simply are not activated when Hsp90 is pharmacologically or genetically compromised.
Understanding C. albicans biology is crucial to our comprehension of how this organism causes disease and for the development of new therapeutic strategies. Hsp90 is an important molecular chaperone that governs key virulence traits in C. albicans (Cowen and Lindquist, 2005; Shapiro et al., 2009; Singh et al., 2009). It is instrumental in the evolution and maintenance of antifungal drug resistance, and compromise of C. albicans Hsp90 function enhances the efficacy of the two leading classes of antifungal drugs for the treatment of invasive fungal infections—the azoles and echinocandins—in a mouse model of disseminated disease (Cowen et al., 2009; Singh et al., 2009). Depletion of Hsp90 also leads to clearance of kidney fungal burden in a mouse model of candidiasis (Shapiro et al., 2009). The finding that Hsp90 inhibitors cause major delays in cell cycle progression and induce multiple cell morphologies may also factor into the efficacy of this combinatorial treatments. This work highlights the importance of Hsp90 in fungal growth and development through its role in chaperoning major regulators of cell signaling, illuminates novel circuitry through which Hsp90 regulates morphogenesis, and provides a new facet to the therapeutic potential of targeting Hsp90 in the treatment of life-threatening fungal disease.
MATERIALS AND METHODS
General culturing techniques, strains, drugs, and primer sequences
C. albicans strains used in this study are listed in Table 1. Strains were maintained on solid (agar-containing) yeast extract peptone media containing dextrose (YPD) or maltose (YPM). Strains that were auxotrophic for uridine were maintained on YPD + 80 μg/ml uridine. Unless otherwise noted, liquid media were inoculated with cultures from solid media plates and grown to saturation overnight at 30°C with shaking prior to use. This saturated liquid culture was then diluted for use in subsequent experimental procedures as indicated. Geldanamycin (GdA; A.G. Scientific, San Diego, CA, and Cedarlane, Hornby, Canada) was formulated as a 5 mM stock in dimethyl sulfoxide and used at a final concentration of between 10 and 30 μM. Cell dimensions were measured using AxioVision 4.7 (Zeiss, Thornwood, NY). Hydroxyurea (HU; BioShop, Burlington, Canada) was formulated as a 2 M stock in water and used at a final concentration of 200 mM. Nocodazole (Noc; Sigma-Aldrich, St. Louis, MO) was formulated as a 20 mM stock in DMSO and used at a final concentration of 50 μM. Doxycycline (BD Biosciences, San Diego, CA) was formulated in a 20 mg/ml stock in water and used at a final concentration of 20 μg/ml.
TABLE 1:
C. albicans strains used in this study.
| Strain name | Genotype | Source |
|---|---|---|
| CaLC192 | ura3Δ::imm434/ura3Δ::imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG | Blankenship and Heitman (2005) |
| CaLC235 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG | Bachewich et al., 2005 |
| CaLC239 | arg4Δ/arg4Δ his1Δ/his1Δ URA3/ura3Δ::imm434 IRO1/iro1Δ::imm434 | Noble and Johnson (2005) |
| CaLC432 | CaLC239, HIS1/his1::TAR-FRT, hsp90::CdHIS1/NAT-MAL2p-HSP90 | Shapiro et al. (2009) |
| CaLC955 | CaLC192, CDC3/CDC3-YFP-URA3 | Finley and Berman (2005) |
| CaLC960 | CaLC192, TUB2/TUB2-GFP-HIS1 | Finley and Berman (2005) |
| CaLC1221 | CaLC192, clb4::HIS1/CLB4-HA:URA3 | Bensen et al. (2005) |
| CaLC1222 | CaLC192, clb4::HIS1/CLB2-HA:URA3 | Bensen et al. (2005) |
| CaLC1411 | CaLC239, HIS1/his1::TAR-FRT, hsp90::CdHIS1/FRT-tetO-HSP90 | Shapiro et al. (2009) |
| CaLC1589 | CaLC235, bub2::URA3/bub2::HIS1 | Bachewich et al., 2005 |
| CaLC1641 | CaLC192, MLC1/MLC1-YFP:URA3 | Crampin et al. (2005) |
| CaLC2021 | CaLC432, CDC5/CDC5-TAP:ARG4 | This study |
| CaLC2028 | CaLC432, CLB2/CLB2-HA:ARG4 | This study |
| CaLC2029 | CaLC432, CLB4/CLB4-HA:ARG4 | This study |
| CaLC2085 | HIS1/his1::TAR-FRT, TetOp-CDC28/cdc28::FRT, HSP90/HSP90-HA | This study |
Strain construction
For CaLC2085, the CDC28 knockout construct was made by amplifying the nourseothricin (NAT)-flipper cassette from plasmid pLC49 using primers oLC1887/oLC1888. This construct was transformed into a prototrophic strain with the tetracycline-repressible transactivator integrated at the HIS1 locus (CaLC205, identical to CaLC206 [ Shapiro et al., 2009], but in the SC5314 background). NAT-resistant transformants were tested for proper integration of the construct by PCR using primer pairs oLC1895/oLC275 and oLC274/oLC1896. The NAT marker was excised. The tetO-CDC28 construct was made by PCR amplifying NAT-tetO from plasmid pLC330 using primer pair oLC1889/oLC1890. This construct was transformed into CaLC205 with one allele of CDC28 deleted. NAT-resistant transformants were tested for proper integration by PCR using primer pairs oLC1895/oLC275 and oLC274/oLC1477. Transformants were additionally tested by PCR with primer pair oLC1895/oLC1896 to verify presence of the deleted allele and oLC1895/oLC1477 to confirm no additional wild-type alleles of CDC28. The NAT marker was excised. To make the HA-tagging construct for Hsp90 (pLC617), sequence within HSP90 (including the HA tag located in primer oLC1637) was PCR amplified from genomic DNA using primer pairs oLC313 and oLC1637. The PCR product was cloned into pLC49 at KpnI and ApaI. Bacterial transformants were PCR tested using primers oLC275 and oLC313. Sequence downstream of HSP90 was PCR amplified with primers oLC318 and oLC319. The PCR product was cloned into the vector with HSP90 sequence at SacI and SacII. Bacterial transformants were PCR tested with primers oLC274 and oLC319. To HA tag HSP90, pLC617 was digested with KpnI and SacI. The digested DNA was transformed in to CaLC1905, and NAT-resistant transformants were tested for proper integration by PCR using primer pairs oLC275/323 and oLC274/335. The NAT marker was excised.
Other HA and TAP tags were added to the C-terminus of proteins of interest using a PCR-based tagging method (Lavoie et al., 2008). Oligonucleotides used for strain construction are listed in Table 2. For CaLC2028 and CaLC2029, tagging constructs were PCR amplified from plasmids pLC576 (pFA-HA-ARG4), using primer pairs oLC2011/2012 and oLC2013/2014 containing sequence homologous to the C-terminus and downstream region of CLB2 and CLB4 respectively (plasmids used in this study are listed in Table 3). This construct was transformed into CaLC432 using a standard polyethylene glycol–lithium acetate protocol. Prototrophic transformants were selected on synthetic defined medium lacking arginine. Transformants were tested for correct upstream and downstream integration of constructs by colony PCR using primer pairs oLC1164/2029 and oLC1594/1165 (CLB2) or oLC1166/2029 and oLC1594/1167 (CLB4). For CaLC2021, the tagging construct was PCR amplified from plasmid pLC573 (pFA-TAP-ARG4) using primers oLC2025/2026 containing sequence homologous to the C-terminus and downstream region of CDC5. The construct was transformed as described earlier, and prototrophic transformants were tested for correct upstream and downstream integration by colony PCR using primer pairs oLC2027/1593 and oLC2028/1594.
TABLE 2:
Oligonucleotides used in this study.
| Primer name | Sequence (5′–3′) |
|---|---|
| oLC274 | CTGTCAAGGAGGGTATTCTGG |
| oLC275 | AAAGTCAAAGTTCCAAGGGG |
| oLC313 | GGGGTACCAAGGCTCAAGCTTTGAGAGACACC |
| oLC318 | TCCCCGCGGACACCAGAAGGGCTACAGTT |
| oLC319 | CGAGCTCTCTCTATGTTATGTTACTGG |
| oLC323 | AAAAGCTGCTAGAGAAAAGG |
| oLC335 | GTTGGTTGTTGATTACTCC |
| oLC752 | AGTATGTGGAGCTTTACTGGGA |
| oLC753 | CAGAAACACCAGCAACATCTTC |
| oLC754 | CCATCTGATATCACTCAAGATG |
| oLC755 | AGTGATAAACACTCTACGGACG |
| oLC1164 | CGACCCATAGGCTAACATTAG |
| oLC1165 | CTACACCTCACCAGAGACCT |
| oLC1166 | GAGAACTGTCGCCATGCTGA |
| oLC1167 | GGCTTGGAAATCTGGGATAA |
| oLC1476 | GGTATCGAGCTCCCGAAATC |
| oLC1477 | GGAACTGCTTCACTCAAAGG |
| oLC1593 | TAAACTTTGGATGAAGGCG |
| oLC1594 | ATGTTGGCTACTGATTTAGCTG |
| oLC1637 | TTGCGGGCCCTCAGGCGGCCGGAGCGTAATCTGGAACGTCATATGGATAATCAACTTCTTCCATAGCAG |
| oLC1887 | TCAAAACAACATGTTTACTAACCAACTATAGAACACACACATCCCAAGCCAAGACCAACACTTATTGCAAGGAAACAGCTATGACCATG |
| oLC1888 | ATCTGACAGTTTGATTTTTTTTCCGTTTTTTCTTTTCGATGTCGATATTTTATTGAGGAGGCGACAAGATGTAAAACGACGGCCAG |
| oLC1889 | TCAAAACAACATGTTTACTAACCAACTATAGAACACACACATCCCAAGCCAAGACCAACACTTATTGCAACGAGGAAGTTCCTATACTTT |
| oLC1890 | CTTTATAAACAACCCCATAAGTACCTTCTCCGACTTTTTCTTGACGTTGATAATCAGATAACTCTACCATCGACTATTTATATTTGTATG |
| oLC1895 | GCTACTCCTTTTTCTTAAAC |
| oLC1896 | CCTGATTAGTGTGAGACATTAC |
| oLC2011 | CGACCCATAGGCTAACATTAGAAGATGATGACGAAGAAGAAGAAATAGTGGTAGCAGAAGCAGAAGAGCCCGGGTACCCATACGATGT |
| oLC2012 | GGTAATGCACATAACTCATGTTCATCTTCTTTCATTTCCTCATTTATGCATTGTAAAGATAAGAACTCGATGAATTCGAGCTCGTT |
| oLC2013 | CAAGGAAAGAAGGTATAGAAAAAGTTCACTTTTTGTTCAAGAATATTTTCGTCACATAATGTCCCAGAGTCCCGGGTACCCATACGATGT |
| oLC2014 | CTTCAACAAATCAGTACTTAACCAATGCATAGAATTAATGATTTTGGTTAGATTGAATAAAATGTTCGATGAATTCGAGCTCGTT |
| oLC2025 | GTTCCGGACTGTATGGAGAAGATAATGGTCATCAAAGAAGCTATCAAGAAAAAAGCATTTAAAGAAGCTGGTCGACGGATCCCCGGGTT |
| oLC2026 | GAAAGTAATAACGAGAAGTAACTATATAATACAAAAGAGAAAATATTGCAAATATCAAGTAGCGTTGTCTTCGATGAATTCGAGCTCGTT |
| oLC2027 | GAGACGCTACACCAGAACTC |
| oLC2028 | ATTAGCAACGTGAAAGTGGC |
| oLC2029 | GGCGAGGTATTGGATAGTTC |
TABLE 3:
Plasmids used in this study.
| Plasmid | Description (backbone) | Source |
|---|---|---|
| pLC49 | FLP-CaNAT, ampR | Shen et al. (2005) |
| pLC330 | tetO-CaHSP90, NAT, ampR (pLC49) | Shapiro et al. (2009) |
| pLC617 | CaHSP90-HA, NAT, ampR (pLC49) | This study |
General imaging techniques
Imaging was performed on a Zeiss Imager M1 upright microscope and AxioCam MRm with AxioVision 4.7 software. An X-Cite series 120 light source with ET green fluorescent protein (GFP), 4′,6-diamidino-2-phenylindole (DAPI) hybrid, and ET HQ tetramethylrhodamine isothiocyanate (TRITC)/DsRED filter sets from Chroma Technology (Bellows Falls, VT) were used for fluorescence microscopy. Calcofluor white and DAPI were viewed under the DAPI hybrid filter, GFP- and YFP-tagged proteins and fluorescein isothiocyanate (FITC) under the GFP filter, and propidium iodide and rhodamine–phalloidin under the ET HQ TRITC/DsRED filter. Cells with fluorescent tags were pelleted and resuspended in phosphate-buffered saline (PBS), pH 7.4, prior to viewing.
Propidium iodide and calcofluor white costaining of DNA and chitin
Overnight cultures were diluted to OD600 of 0.2 in relevant media with indicated treatments. At collection time, ∼5 × 107 cells were centrifuged for 5 min at 3000 rpm. The pellet was resuspended in 70% ethanol and fixed overnight at 4°C with gentle agitation. Ethanol-fixed cells were washed twice with PBS, pH 7.4, then treated with 5 mg/ml pepsin (Sigma-Aldrich) in 55 mM HCl for 20 min at room temperature. Cells were washed twice with PBS and then treated with 100 μg/ml RNase A (Fermentas, Burlington, Canada) at 37°C for at least 2 h. Cells were washed twice with PBS and resuspended in the same. Propidium iodide (PI; Sigma-Aldrich) was added to final concentration of 10 μg/ml and cells incubated at room temperature for 1 h with gentle shaking. Cells were spun down and the pellet resuspended in fresh PBS. Calcofluor white (CFW; Sigma-Aldrich) was added to a final concentration of 1 μg/ml, and cells were incubated at room temperature with gentle shaking for 10 min. After washing once in PBS, cells were sonicated at low setting for 5 s, spun down, resuspended in 200 μl of immunofluorescence mounting solution (IFM; 10% 10 mg/ml p-phenylenediamine in PBS, 90% glycerol, pH adjusted to ∼9.0 using carbonate/bicarbonate buffer), and stored at −20°C.
Rhodamine–phalloidin staining of F-actin
Overnight cultures were diluted to OD600 of 0.2 in relevant media with treatments indicated. At collection time, 1.25 ml of 16% formaldehyde (Ted Pella, Redding, CA) was added to a 3.75-ml culture to obtain a final concentration of 4%. Cultures were incubated at growth temperature for 10 min. Cells were pelleted by centrifugation and resuspended in PBS and 16% formaldehyde was added to achieve a final concentration of 4%. Cells were fixed for 1 h at room temperature with occasional mixing. Following fixation, cells were washed twice in PBS and resuspended in the same. Rhodamine–phalloidin (Biotium, Hayward, CA) was added to a final concentration of 0.66 μM. Cells were incubated in the presence of the dye overnight at 4°C protected from light. Following overnight incubation, cells were washed twice in PBS, sonicated at low setting for 5 s, spun down, and resuspended in IFM containing 50 ng/ml DAPI (Fluka, Sigma-Aldrich). Stained samples were stored at −20°C.
Indirect immunofluorescence of tubulin
Overnight cultures were diluted to OD600 of 0.2 in relevant media with indicated treatments. At collection time, 16% formaldehyde was added to 3.75 ml of culture to a final concentration of 4%. Cultures were incubated at growth temperature for 10 min. Cells were pelleted by centrifugation and resuspended in IF buffer (40 mM KPO4, pH 6.7, 500 μM MgCl2), and 16% formaldehyde was added to achieve a final concentration of 4%. Cells were fixed for 1 h at room temperature with occasional mixing. Following fixation, cells were washed twice in IF buffer and resuspended in the same containing 1.2 M sorbitol (sorbitol buffer) to a final concentration of 3 × 108 cells/ml. A total of 100 μl of this suspension was incubated for 30 min at room temperature with 0.6% β-mercaptoethanol and 15 U of lyticase enzyme (US Biological, Swampscott, MA) to partially digest cell walls. Following lyticase treatment, cells were washed gently with sorbitol buffer and resuspended in 200 μl of the same. Twenty microliters of this cell suspension was spotted onto 5-mm wells of a 10-well glass slide (Cel-Line, Thermo Scientific, Waltham, MA) coated with 0.1% poly-l-lysine (Sigma-Aldrich). Attached cells were permeabilized by plunging the slide into ice-cold (−20°C) methanol for 6 min, followed by ice-cold acetone for 30 s. Cells were blocked in block solution (PBS 1% milk, 0.5% bovine serum albumin, 10 μM octyl α/β-glucosides) for 30 min. Cells were then incubated in primary antibody (anti–α-tubulin; AbD Serotec, Raleigh, NC) diluted 1/1000 in block solution overnight at 4°C. Each well was washed five times with PBS, followed by incubation in FITC-conjugated anti-rat secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1/200 in block solution for 1 h at room temperature. Wells were washed five times with PBS. A small drop of IFM + DAPI was added to each well. The slide was covered with a coverslip and the edges sealed with clear nail polish. Finished slides were stored at −20°C.
Calcofluor white staining of chitin
Overnight cultures were diluted to OD600 of 0.2 in relevant media with treatments indicated. Calcofluor white was added directly to the culture medium to a final concentration of 1 μg/ml. Cultures were incubated with shaking at growth temperature for 10 min, pelleted by centrifugation, and suspended in PBS for viewing.
Propidium iodide viability assay
PI was added to cultures to a final concentration of 10 μg/ml and incubated for 10–20 min at room temperature. Cells were washed once with PBS and resuspended in the same prior to viewing.
Flow cytometry
For analysis of cell cycle progression, overnight cultures were inoculated in to 1 l of YPD and grown overnight at 25°C to mid-late log phase (OD600, ∼3–6). Unbudded G1 phase cells were isolated by elutriation using a J6-MI Beckman centrifuge (Beckman Coulter, Brea, CA) equipped with a JE-5.0 elutriation rotor and a large, 40-ml chamber at 2500 rpm with a flow rate of 16 ml/min. Collected cells were spun down and resuspended to a final concentration of 3 × 106 cells/ml. Cells were grown in YPD with indicated treatments. At least 106 cells were collected at each time point and fixed in 70% ethanol overnight at 4°C. Fixed cells were washed twice in PBS and treated with 5 mg/ml pepsin in 55 mM HCl for 20 min at room temperature. Cells were washed twice in PBS and then treated with 100 μg/ml RNase A (Fermentas) for 3 h at 37°C. Cells were incubated in 25 μg/ml PI overnight at 4°C. Prior to FACS analysis, cells were counted using a hemacytometer and each sample adjusted to contain 106 cells/ml in 25 μg/ml PI. Each sample was sonicated on low setting for 10 s. Cells were sorted on a FACSCalibur (BD Biosciences), and histograms represent at least 5000 ungated events. For sorting and collection of different populations, saturated overnight cultures were inoculated to OD600 0.1 in 30 ml of YPD with geldanamycin and grown for 6 h. Cells were fixed, treated, and sorted as indicated. Populations of interest were gated and collected for microscopic analysis.
Checkerboard assays
Checkerboard assays were set up in a total volume of 0.2 ml/well in a flat-bottomed, 96-well culture plate with twofold dilutions of hydroxyurea across the x-axis of the plate and twofold dilutions of doxycycline across the y-axis of the plate. HU gradient was from 200 mM to 0 in the following concentrations steps (in mM): 200, 100, 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.20. Doxycycline gradient was from 5 μg/ml down to 0 in the following concentration steps (in μg/ml): 5, 2.5, 1.3, 0.63, 0.31, 0.16, 0.078. Plates were inoculated with ∼103 cells and incubated in the dark at 30°C for 72 h. Growth was measured by absorbance at OD600, and values were normalized to the no-drug control well.
Western blot analysis
Overnight cultures were diluted to OD600 of 0.2 in relevant media with treatments indicated. Protein was extracted from ∼3 × 108 cells at early log phase. Strains in the CaLC432 background required longer incubation times for depletion of Hsp90 levels, and cells were typically collected at mid log phase. Cells were pelleted by centrifugation, washed with sterile water, and resuspended in lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100). An equal volume of 0.5-mm, acid-washed beads was added to each tube. Tubes were shaken on a BioSpec (Bartlesville, OK) minibeadbeater for six 1-min intervals. The beads and cell debris were pelleted by high-speed centrifugation and the supernatant removed for analysis. Protein concentration was determined using a Bradford reagent (Sigma-Aldrich) assay. Between 3 and 40 μg of protein was loaded in wells of a 10% SDS–PAGE gel. Separated proteins were transferred to a polyvinylidene fluoride membrane either overnight at 25 V or for 1 h at 100 V at 4°C. Membranes were blocked in 5% milk TBS or PBS containing 0.1% Tween-20 (TBST or PBST, respectively) at room temperature for 1 h and subsequently incubated in primary (1.5–2 h at room temperature or overnight at 4°C) and then secondary (1 h at room temperature) antibodies, with washes in TBST or PBST in between. Primary antibodies were diluted in PBST or TBST containing 2.5–5% milk at the following concentrations: anti-tubulin (AbD Serotec), 1:1000; anti-PSTAIRE (Santa Cruz Biotechnology, Santa Cruz, CA), 1:5000–1:10,000; anti-Hsp90 (Burt et al., 2003), 1:10,000, anti-HA (A. Cochrane, University of Toronto), 1:10–1:200; and anti-TAP (Thermo Scientific), 1:5000. Secondary antibodies (Bio-Rad, Hercules, CA) were diluted at 1:5000 in 5% milk TBST or PBST. Chemiluminescent detection reagent (0.1 M Tris, pH 8.5, 0.2 mM coumaric acid, 1.25 mM luminol, 0.01% hydrogen peroxide) or Western Lightning Plus ECL detection reagent (PerkinElmer, Waltham, MA) was used to detect protein bands. Following incubation of membranes with detection reagent, blots were exposed on Bioflex scientific imaging film (Clonex, Markham, Canada). Images shown are from films scanned using a Canoscan 4400F scanner (Canon, Lake Success, NY).
Coimmunoprecipitation assay
Overnight cultures were diluted to OD600 of 0.2 in relevant media with treatments indicated, grown for 24 h, and rediluted to OD600 of 0.2 with the same treatment. Protein was extracted from 50 ml of culture at early log phase. Cells were pelleted by centrifugation, washed with sterile water, and resuspended in lysis buffer J: 20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl, and 20% glycerol, with one protease inhibitor cocktail (complete, EDTA-free tablet; Roche Diagnostics, Indianapolis, IN) per 10 ml, 1 mM phenylmethylsulfonyl fluoride (EMD Chemicals, San Diego, CA), and 20 mM sodium molybdate (Sigma-Aldrich) added fresh before use. Acid-washed beads, 0.5 mm, were added to fill each tube. Tubes were shaken on a BioSpec minibeadbeater for two 4-min intervals with 1 min on ice in between. Holes were made in the bottom of tubes and the flowthrough collected by centrifugation. Protein concentration was determined using a Bradford Reagent (Sigma-Aldrich) assay. Six hundred μg of protein was subject to immunoprecipitation using the ProFound HA IP kit (Thermo Scientific) according to manufacturer's instructions. Input and immunoprecipitated samples were subject to Western blot as described.
Quantitative RT-PCR
Overnight cultures were diluted to OD600 of 0.2 in media indicated and grown overnight. The following day, cells were again diluted to OD600 of 0.2 in the same conditions and grown to mid log phase. Cultures were then pelleted at 3000 rpm for 5 min and frozen overnight at –80°C. RNA was isolated using the RxNeasy Kit and RNase-Free DNase (Qiagen, Valencia, CA), and cDNA synthesis was performed using the AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent Technologies, Santa Clara, CA). PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and the StepOnePlus Real-Time PCR System (Applied Biosystems) with the following cycling conditions: 95°C for 20 s; then 95°C for 3 min and 60°C for 30 s, for 40 cycles. Reactions were performed in triplicate, with oLC752 and oLC753 (GPD1); oLC754 and oLC755 (HSP90); and oLC1476 and oLC1477 (CDC28). Data were analyzed with StepOne software, version 2.2 (Applied Biosystems). Oligonucleotide sequences are listed in Table 2.
Supplementary Material
Acknowledgments
We thank C. Bachewich, J. Berman, J. Correa-Bordes, P. Sudbery, J. Heitman, and A. Johnson for strains; B. Larsen for an Hsp90 antibody and A. Cochrane for an HA antibody; M. Cook and M. Tyers for assistance with cell elutriation; and J. Heitman, R. Collins, S. Gray-Owen, J. Brill, and the Cowen lab members for helpful discussions. H.S. was supported by a Natural Sciences and Engineering Research Council of Canada CGS-M Award and a Canadian Institutes of Health Research Health Professional Student Research Award; R.S.S. is supported by a Natural Sciences and Engineering Research Council of Canada CGS-D Award; L.E.C. is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, a Canada Research Chair in Microbial Genomics and Infectious Disease, and Canadian Institutes of Health Research Operating Grant MOP-86452.
Abbreviations used:
- CDK
cyclin-dependent kinase
- CFW
calcofluor white
- DIC
differential interference contrast
- DOX
doxycycline
- FACS
fluorescence-activated cell sorting
- GdA
geldanamycin
- HA
hemagglutinin
- Hsp90
heat shock protein 90
- HU
hydroxyurea
- Noc
nocodazole
- PI
propidium iodide
- PKA
protein kinase A
- TAP
tandem affinity purification
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0729) on November 16, 2011.
REFERENCES
- Aligue R, Akhavan-Niak H, Russell P. A role for Hsp90 in cell cycle control: Wee1 tyrosine kinase activity requires interaction with Hsp90. EMBO J. 1994;13:6099–6106. doi: 10.1002/j.1460-2075.1994.tb06956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaluz E, Ciudad T, Gomez-Raja J, Calderone R, Larriba G. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol Microbiol. 2006;59:1452–1472. doi: 10.1111/j.1365-2958.2005.05038.x. [DOI] [PubMed] [Google Scholar]
- Atir-Lande A, Gildor T, Kornitzer D. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol Biol Cell. 2005;16:2772–2785. doi: 10.1091/mbc.E05-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, Nantel A, Whiteway M. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol Microbiol. 2005;57:942–959. doi: 10.1111/j.1365-2958.2005.04727.x. [DOI] [PubMed] [Google Scholar]
- Bachewich C, Thomas DY, Whiteway M. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol Biol Cell. 2003;14:2163–2180. doi: 10.1091/mbc.02-05-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4:95–102. doi: 10.1128/EC.4.1.95-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle CJ, Bohula EA, Kron SJ, Wessels D, Soll DR, Schäfer A, Brown AJ, Gow NA. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot Cell. 2003;2:398–410. doi: 10.1128/EC.2.3.398-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Clemente-Blanco A, Finley KR, Correa-Bordes J, Berman J. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol Biol Cell. 2005;16:3387–3400. doi: 10.1091/mbc.E04-12-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Heitman J. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect Immun. 2005;73:5767–5774. doi: 10.1128/IAI.73.9.5767-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt ET, Daly R, Hoganson D, Tsirulnikov Y, Essmann M, Larsen B. Isolation and partial characterization of Hsp90 from Candida albicans. Ann Clin Lab Sci. 2003;33:86–93. [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci USA. 2009;106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapa y Lazo B, Bates S, Sudbery P. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell. 2005;4:90–94. doi: 10.1128/EC.4.1.90-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H, Glory A, Bachewich C. Orthologues of the anaphase-promoting complex/cyclosome coactivators Cdc20p and Cdh1p are important for mitotic progression and morphogenesis in Candida albicans. Eukaryot Cell. 2011;10:696–709. doi: 10.1128/EC.00263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A, Gonzalez-Novo A, Machin F, Caballero-Lima D, Aragon L, Sanchez M, de Aldana CR, Jimenez J, Correa-Bordes J. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J Cell Sci. 2006;119:1130–1143. doi: 10.1242/jcs.02820. [DOI] [PubMed] [Google Scholar]
- Cote P, Hogues H, Whiteway M. Transcriptional analysis of the Candida albicans cell cycle. Mol Biol Cell. 2009;20:3363–3373. doi: 10.1091/mbc.E09-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- Cowen LE, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci USA. 2009;106:2818–2823. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, Sudbery P. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- de Carcer G. Heat shock protein 90 regulates the metaphase-anaphase transition in a polo-like kinase-dependent manner. Cancer Res. 2004;64:5106–5112. doi: 10.1158/0008-5472.CAN-03-2214. [DOI] [PubMed] [Google Scholar]
- de Carcer G, do Carmo Avides M, Lallena MJ, Glover DM, Gonzalez C. Requirement of Hsp90 for centrosomal function reflects its regulation of Polo kinase stability. EMBO J. 2001;20:2878–2884. doi: 10.1093/emboj/20.11.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gramont A, Cohen-Fix O. The many phases of anaphase. Trends Biochem Sci. 2005;30:559–568. doi: 10.1016/j.tibs.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Finger FP. Reining in cytokinesis with a septin corral. Bioessays. 2005;27:5–8. doi: 10.1002/bies.20167. [DOI] [PubMed] [Google Scholar]
- Finley KR, Berman J. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot Cell. 2005;4:1697–1711. doi: 10.1128/EC.4.10.1697-1711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Morales P, Carrasco-Garcia E, Ruiz-Rico P, Martinez-Mira R, Menendez-Gutierrez MP, Ferragut JA, Saceda M, Martinez-Lacaci I. Inhibition of Hsp90 function by ansamycins causes downregulation of cdc2 and cdc25c and G(2)/M arrest in glioblastoma cell lines. Oncogene. 2007;26:7185–7193. doi: 10.1038/sj.onc.1210534. [DOI] [PubMed] [Google Scholar]
- Georgakis GV, Li Y, Younes A. The heat shock protein 90 inhibitor 17-AAG induces cell cycle arrest and apoptosis in mantle cell lymphoma cell lines by depleting cyclin D1, Akt, Bid and activating caspase 9. Br J Haematol. 2006;135:68–71. doi: 10.1111/j.1365-2141.2006.06247.x. [DOI] [PubMed] [Google Scholar]
- Goes FS, Martin J. Hsp90 chaperone complexes are required for the activity and stability of yeast protein kinases Mik1, Wee1 and Swe1. Eur J Biochem. 2001;268:2281–2289. doi: 10.1046/j.1432-1327.2001.02105.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Novo A, Labrador L, Pablo-Hernando ME, Correa-Bordes J, Sanchez M, Jimenez J, Vazquez de Aldana CR. Dbf2 is essential for cytokinesis and correct mitotic spindle formation in Candida albicans. Mol Microbiol. 2009;72:1364–1378. doi: 10.1111/j.1365-2958.2009.06729.x. [DOI] [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- Hazan I, Sepulveda-Becerra M, Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol Biol Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostein I, Robertson D, DiStefano F, Workman P, Clarke PA. Inhibition of signal transduction by the Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin results in cytostasis and apoptosis. Cancer Res. 2001;61:4003–4009. [PubMed] [Google Scholar]
- Hu F, Elledge SJ. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle. 2002;1:351–355. [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Irniger S, Baumer M, Braus GH. Glucose and ras activity influence the ubiquitin ligases APC/C and SCF in Saccharomyces cerevisiae. Genetics. 2000;154:1509–1521. doi: 10.1093/genetics/154.4.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AA, Perfect JR, Cowen LE. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 2010;6:e1001069. doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CR, Wang YM, Wang Y. The IQGAP Iqg1 is a regulatory target of CDK for cytokinesis in Candida albicans. EMBO J. 2008;27:2998–3010. doi: 10.1038/emboj.2008.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol. 2001;4:728–735. doi: 10.1016/s1369-5274(01)00275-2. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol. 1999;19:4019–4027. doi: 10.1128/mcb.19.6.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- MacLean M, Picard D. Cdc37 goes beyond Hsp90 and kinases. Cell Stress Chaperones. 2003;8:114–119. doi: 10.1379/1466-1268(2003)008<0114:cgbhak>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Mollapour M, et al. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol Cell. 2010;37:333–343. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort-Bontemps-Soret M, Facca C, Faye G. Physical interaction of Cdc28 with Cdc37 in Saccharomyces cerevisiae. Mol Genet Genomics. 2002;267:447–458. doi: 10.1007/s00438-002-0676-3. [DOI] [PubMed] [Google Scholar]
- Munoz MJ, Jimenez J. Genetic interactions between Hsp90 and the Cdc2 mitotic machinery in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1999;261:242–250. doi: 10.1007/s004380050963. [DOI] [PubMed] [Google Scholar]
- Murad AM, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir A, Kornitzer D. Candida albicans cyclin Clb4 carries S-phase cyclin activity. Eukaryot Cell. 2010;9:1311–1319. doi: 10.1128/EC.00038-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev. 2011;75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Guo W, Kohler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun. 2005;73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell. 2007;18:815–826. doi: 10.1091/mbc.E06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srethapakdi M, Liu F, Tavorath R, Rosen N. Inhibition of Hsp90 function by ansamycins causes retinoblastoma gene product-dependent G1 arrest. Cancer Res. 2000;60:3940–3946. [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol. 2001;41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Umeyama T, Kaneko A, Niimi M, Uehara Y. Repression of CDC28 reduces the expression of the morphology-related transcription factors, Efg1p, Nrg1p, Rbf1p, Rim101p, Fkh2p and Tec1p and induces cell elongation in Candida albicans. Yeast. 2006;23:537–552. doi: 10.1002/yea.1373. [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13:2732–2746. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G, Behrns KE, Kim JS, Kim RD. Heat shock protein 90 inhibition abrogates hepatocellular cancer growth through cdc2-mediated G2/M cell cycle arrest and apoptosis. Cancer Chemother Pharmacol. 2009;64:433–443. doi: 10.1007/s00280-008-0888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature. 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.