Cyclin B accumulation in the early Xenopus embryo drives CDK1 oscillations. CDK1 directs a negative-feedback loop upon cyclin B synthesis through the repression of global protein translation, and this attenuation of cyclin B synthesis could improve the efficiency and robustness of this cell cycle oscillator.
Abstract
Cyclin B activates cyclin-dependent kinase 1 (CDK1) at mitosis, but conflicting views have emerged on the dynamics of its synthesis during embryonic cycles, ranging from continuous translation to rapid synthesis during mitosis. Here we show that a CDK1-mediated negative-feedback loop attenuates cyclin production before mitosis. Cyclin B plateaus before peak CDK1 activation, and proteasome inhibition caused minimal accumulation during mitosis. Inhibiting CDK1 permitted continual cyclin B synthesis, whereas adding nondegradable cyclin stalled it. Cycloheximide treatment before mitosis affected neither cyclin levels nor mitotic entry, corroborating this repression. Attenuated cyclin production collaborates with its destruction, since excess cyclin B1 mRNA accelerated cyclin synthesis and caused incomplete proteolysis and mitotic arrest. This repression involved neither adenylation nor the 3′ untranslated region, but it corresponded with a shift in cyclin B1 mRNA from polysome to nonpolysome fractions. A pulse-driven CDK1–anaphase-promoting complex (APC) model corroborated these results, revealing reduced cyclin levels during an oscillation and permitting more effective removal. This design also increased the robustness of the oscillator, with lessened sensitivity to changes in cyclin synthesis rate. Taken together, the results of this study underscore that attenuating cyclin synthesis late in interphase improves both the efficiency and robustness of the CDK1-APC oscillator.
INTRODUCTION
Protein synthesis is a process fundamental to cellular metabolism and proliferation, and both environmental and internal stimuli can lead to its up-regulation or suppression. Translation is an energy-intensive process, and it can be paused under a variety of circumstances. Cells exposed to a stress, such as glucose deficiency, for example, will transiently inhibit global translational initiation (Datta et al., 1999; Sonenberg and Hinnebusch, 2009; Spriggs et al., 2010). Global translational repression is also a normal occurrence during proliferation, concomitant with mitotic entry in an array of different systems, including yeast (Creanor and Mitchison, 1982), Xenopus laevis embryos and embryonic extracts (Kanki and Newport, 1991), and mammalian cells (Prescott and Bender, 1962; Salb and Marcus, 1965; Fan and Penman, 1970; Sivan et al., 2007). Numerous studies have shown that this mitotic suppression of protein synthesis corresponds to diminished rates of translational initiation (Bonneau and Sonenberg, 1987; Kanki and Newport, 1991; Pyronnet et al., 2001; Pyronnet and Sonenberg, 2001) and elongation (Sivan et al., 2007, 2011), but if and how this regulation influences the onset of M phase and cell cycle oscillations are not well understood.
Studies of cyclin B1—the activating binding partner of CDK1—reported differences in its M phase expression pattern during the cell cycle and in the mechanisms that underlie this control. Cyclin synthesis concomitant with M phase was described in Saccharomyces cerevisiae (Drapkin et al., 2009), Xenopus embryonic extract (Groisman et al., 2002), and mouse embryonic fibroblasts, with the last two attributed to CPE-binding protein–dependent polyadenylation of its mRNA (Groisman et al., 2002; Malureanu et al., 2010). Cytoplasmic adenylation can enable the translation of specific mRNAs upon global translational inhibition (Le Breton et al., 2005), but this is not the only means for up-regulating cyclin B1 production. For instance, in Drosophila early embryos, cyclin B1 translation is not increased by changes in poly(A)-tail length but is caused by a Pan Gu (PNG) kinase–mediated derepression of its synthesis (Vardy and Orr-Weaver, 2007). Variations in sensitivity to changes in cyclin B1 production have also been described. For example, the timing of Drosophila cell cycles is only minimally affected by significant increases in cyclin B1 dosage (Stiffler et al., 1999); alternatively, adding increasing concentrations of exogenous cyclin B RNA into Xenopus embryos (Hartley et al., 1996) or extracts (Pomerening et al., 2005) causes M phase arrest. Although there are indeed certain requirements for generating the appropriate stimulus for mitotic entry, it is not known why particular strategies are implemented to accomplish this and whether they are somehow advantageous to cell cycle oscillator design.
Although the regulation of cyclin B1 synthesis during the cell cycle might differ depending on organism and developmental context, whether it continues unimpeded during mitosis and how this might be advantageous to cell cycle oscillations remain open questions. One particularly compelling example of this is the reported increase in the cyclin B1 translation rate in M phase coincident with global translational repression during the X. laevis early embryonic cell cycle. On the seminal discovery of cyclin B1 oscillations corresponding to cell cycle progression, its synthesis was described to be continuous throughout the cell cycle (Evans et al., 1983). Later studies concluded that its production reached a maximum rate during mitosis as a result of polyadenylation (Groisman et al., 2002). However, both of these reports conflict with the determination that global translation in Xenopus extracts and embryos peaks during interphase and is significantly reduced at mitosis (Kanki and Newport, 1991). Furthermore, recent findings that cyclin levels plateau prior to CDK1 activation suggested that oscillations might not be driven by constant cyclin B synthesis (Pomerening et al., 2005). Cyclin B is essential for mitotic entry (Murray and Kirschner, 1989), and work has described a lag period before CDK1 activation that starts when a threshold level of cyclin is produced (Solomon et al., 1990). Might attenuating cyclin synthesis, rather than producing it constantly during this lag period, provide any advantage to the CDK1–anaphase-promoting complex (APC) early embryonic oscillator?
Here we investigate the pattern of cyclin B1 synthesis with respect to global translation and its impact on cell cycle oscillations in Xenopus early embryonic extracts. We find that cyclin B1 levels plateau as a result of translational repression mediated by CDK1 activity and not due to combined synthesis and proteolysis. To understand how and why this regulation occurs, we investigated the possible role of polyadenylation and the 3′ untranslated region (UTR) of the cyclin B1 mRNA and studied how increasing its levels affected cell cycle progression. Whereas polyadenylation increased levels of interphase cyclin B1 translation, neither it nor the 3′ UTR was required to reduce synthesis of the protein during mitosis. However, an excess of cyclin B1 mRNA did cause mitotic arrest due to a failure to completely degrade cyclin B1 protein. In addition, we found that the attenuation of cyclin B1 synthesis corresponds with global translation repression at M phase entry. Using a simple CDK1-APC computational model, we describe for the first time how this CDK1-mediated repression of cyclin synthesis is not only capable of driving realistic CDK1 oscillations, but also provides some design advantages over its continuously stimulated counterpart. Our results indicate that the Xenopus early embryonic cell cycle oscillator, although rapid and clock-like, is capable of tempering its own stimulus and provides a more efficient and robust control system that could be applicable to the cell cycle designs of other higher eukaryotes.
RESULTS
Cyclin B translation is attenuated before mitosis by a CDK1-dependent negative-feedback loop
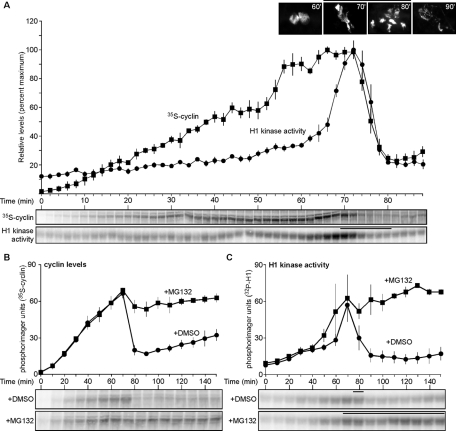
High-resolution time-course measurements of cyclin B and CDK1 activity levels throughout one cell cycle of X. laevis embryonic extract revealed that cyclin B accumulation begins early in interphase and plateaus prior to peak CDK1 activation during mitosis (Figure 1A), in agreement with previous findings (Pomerening et al., 2005). Two possible mechanisms could contribute to this behavior: either a balance between cyclin synthesis and degradation is achieved prior to complete CDK1 activation, or cyclin synthesis is impeded at that point. To distinguish between these two possibilities, cycling extract was treated with the proteasome inhibitor MG132, and levels of cyclin B and CDK1 activity were measured. Unexpectedly, despite proteasome inhibition, the amount of cyclin B produced did not increase upon mitotic onset, whereas the DMSO control exited mitosis and reinitiated new cyclin synthesis (Figure 1, B and C). No differences between cyclin levels or CDK1 activities were observed in MG132-treated extract with or without cycloheximide added, corroborating that cyclin synthesis is indeed inhibited (Supplemental Figure S1, A–D). In addition, a 35S-labeled cyclin tracer was found to be stabilized in both pre–M phase and M phase–arrested extracts treated with MG132 (Supplemental Figure S1, E and F). These results excluded the possibility that the plateau of the endogenous cyclin was caused by a balance between polyubiquitylation/degradation and cyclin synthesis. This is the first evidence that cyclin B is not continuously translated during mitosis.
FIGURE 1:
Cyclin B translation is attenuated before and during mitosis. (A) Time course of cyclin B levels and H1 kinase activities in cycling extracts. 35S-Met-labeled cyclin was precipitated using p13 agarose; H1 kinase assays were performed to measure CDK1 activity; both were subjected to SDS-PAGE separation and visualized by phosphorimaging. Sperm chromatin was added to extracts; aliquots were taken at various times and stained with DAPI. Sperm chromatin forms nuclei in interphase (60 min is shown as representative), and then nuclei undergo mitosis, as indicated by chromatin condensation and NEB (70 and 80 min). Data were shifted to align the peak H1 kinase activities of all three replicates, with two replicates shifted 2 and 10 min earlier, respectively, excluding the earliest and latest lone time points from the analysis. Quantitated data are plotted as means ± SEM (error bars; N = 3). Bars above micrographs and H1 kinase activities denote time points where NEB was observed. (B, C) Time course of cyclin B levels and H1 kinase activities for cycling extracts treated with DMSO (1%) or MG132 (1 mM; 1% final volume). Data were shifted to align the peak H1 kinase activities of all four replicates, with two replicates shifted 10 and 20 min earlier, respectively, excluding the earliest and latest lone time points from the analysis. Data were analyzed as described in A and plotted as means ± SEM (N = 4). Bars denote time points where NEB was observed. Also see Supplemental Figure S1.
Because the plateau of cyclin B accumulation was concomitant with the early gradual increase of CDK1 activity (Figure 1A), we hypothesized that there might exist a CDK1-directed negative feedback upon cyclin B synthesis. Cycling egg extract was treated with MG132 or DMSO and either roscovitine—to inhibit CDK1 activation—or ethanol as the negative control. Unlike DMSO-treated extract, which underwent two cell cycle oscillations, MG132-treated extract produced a maximum level of cyclin B that was sustained following mitotic onset (Figure 2A, left). Constant cyclin synthesis, however, occurred in roscovitine-plus-MG132–treated extract, where CDK1 activity remained low (Figure 2A, right). We next tested whether an extract could detect the presence of exogenously added cyclin and limit its production of the endogenous protein. Following the addition of 200 nM Xenopus nondegradable cyclin B1 (Δ65XCycB1)—a concentration that will not saturate CDK1 and will thus permit binding of newly synthesized cyclin—the extract produced less cyclin (Figure 2B, left), and its synthesis was impeded earlier than the controls due to the accelerated activation of CDK1 (Figure 2B, right). If there exists a negative-feedback loop on cyclin synthesis, then mimicking its effect during the window in which it is activated would neither affect the level of accumulated cyclin nor delay mitotic entry. To test this, cycloheximide (CHX) was added to cycling extract to inhibit protein synthesis at successively later points during a cycle (Figure 2C, left). CHX added at 20 min did not allow the onset of nuclear envelope breakdown (NEB; data not shown), whereas the control extract exhibited NEB at 60 min (Figure 2C, right, dark gray bar). CHX addition at 30 and 40 min delayed NEB by 10 min (Figure 2C, right, green bar) and 5 min (Figure 2C, right, blue bar), respectively. However, CHX added at 50 min contained cyclin levels similar to the control (Figure 2C, left; compare orange and dark gray histograms), and addition at this time and 55 min did not affect M phase entry (Figure 2C, right, orange and light gray bars, respectively). Similarly, peak H1 kinase activity that occurred in the untreated extract at 55 min was matched in the extract treated with CHX at 50 min. These results suggest that cyclin B synthesis is hampered ∼5–10 min prior to and during M phase. This result agreed with earlier studies that revealed a period during the cleavage of Xenopus single-cell blastomeres that was independent of protein synthesis, but no correlation with cell cycle regulation had been made (Beckhelling et al., 1999). Collectively, these results show that cyclin B is synthesized at a steady rate during early interphase, and then a CDK1-driven negative-feedback loop represses its translation near the end of interphase and before the onset of M phase.
FIGURE 2:
Inhibiting or prematurely activating CDK1 alters the level of cyclin B synthesis. (A–C) Time course of cyclin B levels (left) and H1 kinase activities (right) for cycling extracts treated with (A) DMSO (1%) and ethanol (0.64%), MG132 (1 mM) and ethanol, or MG132 (1 mM) and roscovitine (0.18 mM); (B) DMSO (1%) and buffer, MG132 (1 mM) and buffer, or MG132 (1 mM) and Δ65XCycB1 (200 nM); (C) CHX (100 μg/ml) added at different time points prior to and during mitosis in cycling egg extract. Data were shifted to align the peak H1 kinase activities of all three replicates, with one replicate shifted 5 min earlier, excluding the earliest and latest lone time points from the analysis. Data were analyzed as described in Figure 1A and plotted as means ± SEM (A, N = 3; B, N = 2; C, N = 3). Bars denote time points where NEB was observed (A–C), and histograms correspond to cyclin levels and H1 kinase activities before and after CHX addition (C); control (no addition), dark gray bars; 30 min addition, light green bars; 40 min addition, light blue bars; 50 min, orange bars; 55 min, light gray bars.
Adenylation of the cyclin B1 mRNA 3′ UTR does not alternate during CDK1 oscillations
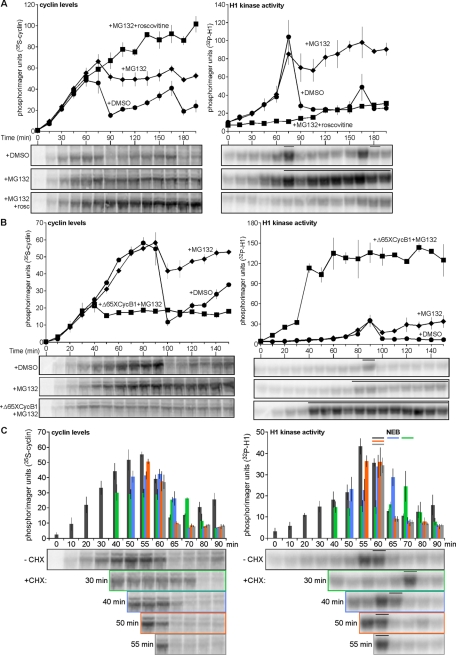
The cited data show that cyclin B production is pulse-like: it is translated starting at early interphase, and its production slows before M phase. However, previous work suggested that cyclin translation starts prior to and accelerates during mitosis as a result of polyadenylation of its mRNA (Groisman et al., 2002). To determine whether there is a connection between modification of the cyclin B1 mRNA and its translational control, we first tested the effect of in vitro–transcribed mRNA from the Xenopus cyclin B1–coding region (XCycB1cod), which was generated as described previously (Pomerening et al., 2005). Increasing doses of this mRNA were titrated into cycling egg extract and resulted in increases in the rate of cyclin accumulation and decreases in the time for mitotic onset without any adverse effects on the CDK1 oscillation (Figure 3A). Although cyclin destruction permitted a normal mitotic exit in the presence of exogenous cyclin B1 mRNA, we were curious to determine whether the absence of cyclin B1 UTRs would prevent the attenuation of its translation. This was not the case, as cycling extract supplemented with MG132 and XCycB1cod mRNA properly paused its cyclin production (Figure 3B, left), although CDK1 was activated earlier due to this addition (Figure 3B, right). These results show that the cyclin B1 UTRs are not required to attenuate cyclin B1 synthesis during early embryonic oscillations.
FIGURE 3:
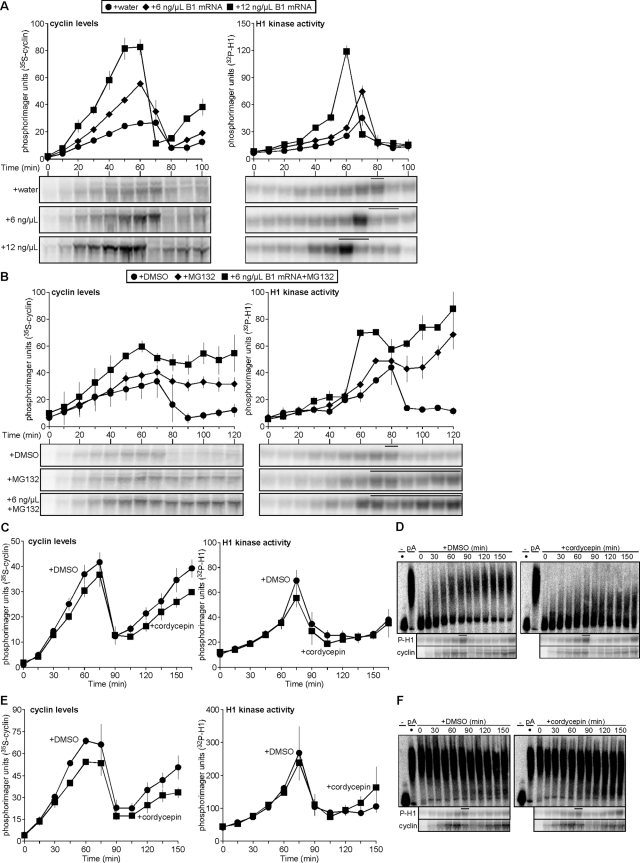
Polyadenylation of the cyclin B1 mRNA 3′ UTR does not vary with changes in cyclin B1 synthesis during cycling. (A, B) Time course of cyclin B levels (left) and H1 kinase activities (right) for cycling extracts treated with (A) water or 6 or 12 ng/μl in vitro–transcribed cyclin B1 mRNA coding region; (B) DMSO (1%), MG132 (1 mM), 6 ng/μl cyclin B1 mRNA coding region and MG132 (1 mM). For A, data were shifted to align the H1 kinase activity peaks of all three replicates, with two replicates shifted 10 min earlier, excluding the earliest and latest lone time points from the analysis; for B, one replicate was shifted 30 min earlier to align peak H1 kinase activities. Data were analyzed as described in Figure 1A and plotted as means ± SEM; A, N = 3; B, N = 2. Bars denote time points where NEB was observed. (C) Pooled data of cyclin B levels (left) and H1 kinase activities (right) plotted as means ± SEM (N = 3) for results shown in D. Data were shifted to align the H1 kinase activity peaks of all three replicates, with one replicate shifted 15 min earlier, excluding the earliest and latest lone time points from the analysis. (D) 32P-Labeled, in vitro–transcribed cyclin B1 3′ UTR was incubated in cycling extracts treated with either DMSO (1%; left) or the polyadenylation inhibitor cordycepin (1 mM; right). Dot indicates addition of nonpolyadenylated UTR. Aliquots were taken at indicated time points, and total RNAs were purified, separated on denaturing urea PAGE, and visualized by phosphorimaging. H1 kinase activities and cyclin B levels were analyzed as described in Figure 1A. Representative of one of three independent experiments is shown. (E) Pooled data of cyclin B levels (left) and H1 kinase activities (right) plotted as means ± SEM (N = 3) for results shown in F. (F) 32P-Labeled, in vitro–transcribed and polyadenylated Xenopus cyclin B1 3′ UTR incubated in extracts treatment were performed as described in D. Dot indicates addition of in vitro–polyadenylated UTR. Bars denote time points where NEB was observed. Representative of one of three independent experiments is shown. Also see Supplemental Figure S2.
Although the UTRs of Xenopus cyclin B1 are not essential for its translational attenuation, cell cycle–dependent oscillations in the adenylation state of the mRNA have been reported (Groisman et al., 2002). If alternations in adenylation of the endogenous cyclin B1 mRNA occur during the cell cycle, then changes in adenylation of the 3′ UTR should be observed. To determine whether there is a correlation between cyclin B1 mRNA adenylation and cell cycle oscillations, we measured the polyadenylation of in vitro–transcribed Xenopus cyclin B1 3′ UTR in cycling extract. We found that the 3′ UTR was polyadenylated immediately but remained modified throughout two full cell cycles; a majority of preadenylated 3′ UTR also remained modified, with some being deadenylated, and they did not increase in poly(A) tail length (Supplemental Figure S2, A and B). We next tested whether the inhibition of polyadenylation would inhibit cyclin synthesis, by observing cycling in egg extract treated with cordycepin (3′-dATP)—a poly(A) polymerase inhibitor—and then tracking the modification of either nonadenylated or polyadenylated cyclin B1 3′ UTR (Groisman et al., 2000). Neither treatment with DMSO alone nor that with cordycepin altered cyclin B synthesis (Figure 3C, left) or CDK1 oscillations (Figure 3C, right), although the bulk of nonadenylated 3′ UTR in extract treated with cordycepin was not significantly modified at any time during nearly two complete cell cycles (Figure 3D). Similarly, cordycepin treatment had no significant effect on preadenylated cyclin B1 3′ UTR (Figure 3F) and did not perturb cell cycle passage (Figure 3E). These findings supported prior studies that reported the presence of a basal deadenylation activity during a cell cycle oscillation (Paillard et al., 1996; Voeltz et al., 2001). This validates that adenylation activity during interphase is dispensable for cyclin synthesis and mitotic entry and that polyadenylation of endogenous cyclin B1 mRNA does not occur during the cell cycle after fertilization, most likely because it is already polyadenylated in the unfertilized egg (Sheets et al., 1994; Ballantyne et al., 1997; Kim and Richter, 2007; Belloc et al., 2008). According to our results, the majority of this polyadenylation would likely remain throughout early embryonic cell cycles. To confirm that polyadenylation of the unmodified cyclin B1 3′ UTR occurs independent of cell cycle oscillations, we assayed its polyadenylation in cycling egg extracts where either protein synthesis or CDK1 activation was inhibited, with both treatments preventing cell cycle progression. As indicated by H1 kinase activity, ethanol-treated control extract cycled into mitosis at 60 min, with the exogenous cyclin B1 3′ UTR being immediately and rapidly adenylated and reaching its maximal level by 60–75 min (Figure S2, C, left; and D). Neither CHX nor roscovitine treatment altered this pattern of polyadenylation, despite the inability of these extracts to cycle—the former due to inhibition of protein synthesis (Figure S2C, center), and the latter due to inhibition of CDK1 (Figure S2C, right). Taken together, these data show that cyclin B1 mRNA is polyadenylated independently of early embryonic cell cycle progression—prior to release from cytostatic factor (CSF) arrest—and that cyclin B1 3′ UTR does undergo some basal deadenylation.
Negligible cyclin B synthesis at mitosis collaborates with APC function
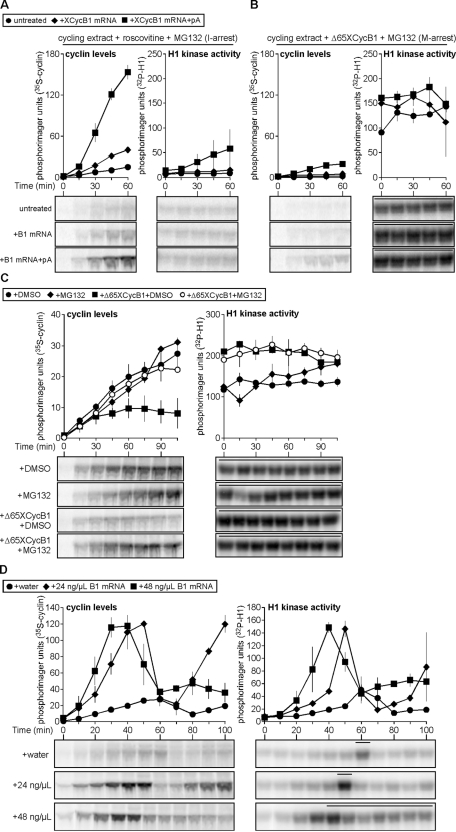
Poly(A) polymerase activity neither is necessary for cyclin translation in Xenopus early embryonic extracts nor is it required during cell cycle oscillations. However, might polyadenylation of the mRNA enhance its translation during mitotic cycles? Using MG132-treated egg extracts supplemented with nonadenylated or in vitro–polyadenylated XCycB1cod mRNA (Supplemental Figure S3), we tested the potential of the latter to increase cyclin B1 translation efficiency. Roscovitine treatment of cycling extract prevented CDK1 activation (Figure 4A, right), and adding nonadenylated mRNA or in vitro–polyadenylated mRNA permitted respective increases in cyclin B1 translation compared with the control (Figure 4A, left). Alternatively, M phase–arrested extracts maintained elevated CDK1 activity (Figure 4B, right), and adding nonadenylated or in vitro–polyadenylated message provided only slight respective increases in cyclin B translation (Figure 4B, left). These results support the idea that polyadenylation can strongly enhance translation of cyclin B1 mRNA in interphase. However, although the translation of the polyadenylated mRNA in mitotic-arrested extracts can occur (Figure 4B, left, boxes) it was only weakly enhanced compared with CDK1-inhibited interphase extracts (Figure 4A, left, boxes). Although it is clear that polyadenylated cyclin B1 mRNA can be translated more effectively, these results agree with our previous findings that cyclin translation, in general, is not optimal in M phase.
FIGURE 4:
Cyclin B translational repression is mitosis specific and collaborates with APC function. (A, B) Polyadenylation of cyclin B1 mRNA promotes translation. Water, 12 ng/μl in vitro–transcribed cyclin B1 mRNA coding region, or in vitro–polyadenylated cyclin B1 mRNA coding region were added to cycling extracts arrested in interphase (I) by roscovitine (0.18 mM) and MG132 (1 mM; A) and mitosis (M) by Δ65XCycB1 (67 nM) and MG132 (1 mM; B), respectively, in the presence of 35S-Met. Aliquots were collected at 15-min intervals and analyzed as described in Figure 1A. Cyclin B levels and H1 kinase activities are plotted as means ± SEM (N = 2). Bars denote time points where NEB was observed. (C) Cyclin B is translated in CSF-M extract. CSF-M extracts were treated with DMSO (1%) and buffer, DMSO and Δ65XCycB1 (65 nM), MG132 (1 mM) and buffer, or MG132 (1 mM) and Δ65XCycB1 (65 nM). Samples were collected at 15-min intervals and analyzed as described in Figure 1A. Quantitated data are plotted as means ± SEM (N = 2). Bars denote time points where NEB was observed. (D) Rapid production of cyclin B1 hampers its destruction and causes mitotic arrest. Time course of H1 kinase activities (right) and cyclin B levels (left) for cycling extracts treated with water, 24 or 48 ng/μl cyclin B1 mRNA coding region. Quantitated data are plotted as means ± SEM (N = 2). Bars denote time points where NEB was observed. Also see Supplemental Figures S3 and S4.
In vitro–transcribed and polyadenylated XCycB1cod mRNA was not efficiently translated in mitotic-arrested egg extract, and this raised the question of the biological context of polyadenylation itself. Endogenous cyclin B1 mRNA becomes heavily polyadenylated during Xenopus oocyte maturation and remains so in the mature egg (Sheets et al., 1994; Ballantyne et al., 1997; Kim and Richter, 2007; Belloc et al., 2008). Therefore we next inquired whether it is during this meiotic arrest in eggs that these stores of polyadenylated cyclin B1 mRNA could be more effectively translated, despite possessing high CDK1 activity. CSF egg extracts—arrested in metaphase of meiosis II—were prepared and incubated with either DMSO or MG132. CDK1 is active during CSF arrest under these conditions (Figure 4C, right), and 35S-labeled cyclin B1 was continuously generated in the presence and absence of MG132, suggesting that APC/proteasome activity is low enough to permit accumulation of newly labeled cyclin (Figure 4C, left, dots and diamonds). Treatment of the same extracts with Δ65XCycB1 increased their CDK1 activities (Figure 4C, right, boxes). This caused a decrease in the level of cyclin synthesized when proteasome activity was permitted (Figure 4C, left, boxes), whereas blocking the proteasome (Figure 4C, left, open circles) caused accumulation of cyclin that matched levels seen in extracts with unaltered CDK1 activities. This confirmed that increasing CDK1 activity can increase APC activity in CSF extracts—which had been reported previously (Hansen et al., 2007; Wu et al., 2007; Isoda et al., 2011)—but did not slow the production of cyclin B. This is the opposite of what we observed prior to and during M phase of mitotic cycles and demonstrates that CDK1 activity does not exert the same inhibition on cyclin synthesis in mature oocytes/eggs (CSF extract) that it does in embryos (cycling extract).
Although there was strong evidence supporting the hypothesis that cyclin B translation is pulse-like during early embryonic CDK1-APC oscillations, the biological relevance of this regulation remained unclear. We hypothesized that these regulated doses of cyclin B1 are a means for the early embryo to minimize the APC activity required for mitotic progression and exit. Previous work revealed that injecting embryos with 50 ng (∼100 ng/μl cytoplasm) of cyclin B2 mRNA could cause cleavage arrest (Hartley et al., 1996). This raised the following question: could this type of M phase arrest be caused by cyclin expression exceeding the level normally produced by a cyclin pulse? We explored this issue using an approach similar to that of Hartley et al. (1996) by titrating increasing doses of cyclin B1 mRNA into cycling extract. The peak level of cyclin B translated in both treatments was similar; however, the highest dose caused the most rapid rate of cyclin B1 synthesis (Figure 4D, left, boxes). This latter treatment—as scored by constant NEB after M phase onset—corresponded with reduced but sufficient levels of cyclin B and CDK1 activity to induce mitotic arrest (Figure 4D, right, boxes). Treatment of arrested extract with CHX did not release it from M phase, confirming that cyclin destruction and CDK1 inactivation were incomplete; MG132 treatment corroborated this finding and revealed that cyclin synthesis was also negligible during this arrest, since minimal additional cyclin accumulated (Supplemental Figure S4). The rapid overproduction of cyclin stimulus resulted in its ineffective proteolysis during mitosis and, coupled with continued translational repression, interrupted further cell cycle oscillations.
M phase cyclin B1 mRNA content in polysome fractions is reduced and corresponds to diminished global translation
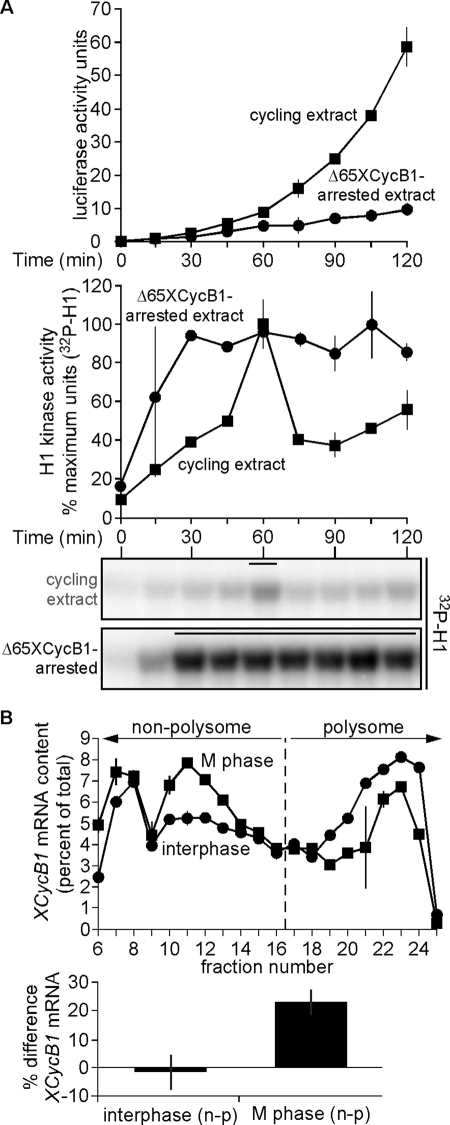
To home in on the mechanism that governs the control of cyclin B1 translation, we first tested whether this CDK1-driven negative-feedback loop has a general effect on translation. Earlier published studies described a global translational inhibition in mitotic-arrested Xenopus extracts (Kanki and Newport, 1991), and we tested this idea by adding exogenous luciferase mRNA to either buffer-treated cycling egg extract or cycling extract arrested in M phase treatment with by Δ65XCycB1. The former underwent nearly two mitotic cycles (Figure 5A, bottom), and luciferase activity increased throughout the duration of time course, corroborating that translation of the luciferase mRNA could occur (Figure 5A, top). However, Δ65XCycB1-treated extract quickly entered and then arrested in mitosis (Figure 5A, bottom), and the increase of luciferase activity was negligible (Figure 5A, top). Unlike interphase periods, M phase did not provide conditions optimal for continued protein synthesis of a general exogenous mRNA such as luciferase. To test endogenous protein synthesis on a global level, cycling egg extract treated with MG132 was labeled with 35S-Met and samples were collected at 12-min intervals up until mitosis. As reported previously (Kanki and Newport, 1991), we found that translation was substantial as interphase proceeded and then diminished at M-phase onset (Supplemental Figure S5A).
FIGURE 5:
A reduction of cyclin B1 mRNA content in polysome fractions at M phase corresponds to its diminished translation. (A) Luciferase mRNA is poorly translated in mitosis-arrested extract. In vitro–transcribed luciferase mRNA (5 ng/μl) was added to cycling extracts treated with either buffer or Δ65XCycB1 (200 nM). Samples were collected at 15-min intervals in a 96-well plate cooled on ice, and luciferase assays were then performed. H1 kinase activities were analyzed as described in Figure 1A (bottom) and plotted with luciferase signals as means ± SEM (N = 2; top), in cycling egg extract (boxes), and Δ65XCycB1-arrested extract (dots). Data were shifted to align the H1 kinase activity peaks of both replicates, with one replicate shifted 15 min earlier, excluding the earliest and latest lone time points from the analysis. Bars above H1 kinase activities denote time points where NEB was observed. (B) Top, samples of interphase extract and mitotic extract (just prior to NEB) were separated by sucrose gradient centrifugation. Total RNA from 40S, 60S, 80S, and polysome fractions was purified, and the distribution of cyclin B1 mRNA was analyzed by qRT-PCR. Left of the dashed line is defined as nonpolysome fraction (40S, 60S, and 80S) and on its right is defined as the polysome fraction. The percentage cyclin B1 mRNA in each fraction was quantitated and normalized as a percentage of total across all fractions, in duplicate. (B) Bottom, differences in the percentage of cyclin B1 mRNA in nonpolysome and polysome fractions (n − p) between interphase and mitosis is presented as mean ± SEM (N = 3). Also see Supplemental Figure S5.
If cyclin B1 synthesis is indeed controlled on a general translational level, then its mRNA might be associated with polysomes during interphase, followed by a shift to the nontranslating, nonpolysome fraction at M phase onset (White et al., 1990). To determine whether this accounts for the observed pattern of cyclin B1 synthesis during a cell cycle oscillation, a polysome profiling analysis was performed on cycling egg extract that was in interphase—when the rate of cyclin synthesis is highest—and at mitotic onset—when CDK1 is active and cyclin translation drops off (Supplemental Figure S5B, inset). As a control for resolving the different ribosomal fractions, an aliquot of interphase extract was treated with puromycin—a chemical that dissociates elongating ribosomes from their mRNA templates (Azzam and Algranati, 1973; Blower et al., 2007; Sivan et al., 2007)—and separated along with interphase and M phase samples by sucrose density gradient fractionation (Supplemental Figure S5B). Following fractionation of these samples, polysome content of the ensuing gradients was measured, and the relative distribution of cyclin B1 mRNA between the collected fractions was assayed by quantitative reverse transcription (qRT)–PCR. First, puromycin-treated extract underwent a significant shift of cyclin B1 mRNA abundance from the polysomal fraction to the nonpolysomal fraction relative to the untreated interphase extract, which had a near-equal distribution between these two fractions. This indicated that that these different translational fractions could be resolved according to ribosome content (Supplemental Figure S5C). In contrast to the nearly 50/50 distribution of cyclin B1 mRNA associated with both polysome and nonpolysome fractions in interphase extract, the RNA content shifted from the polysome to nonpolysome fraction by nearly 25% during mitosis (Figure 5B). This shift of cyclin B1 mRNA to the nonpolysome fraction is concordant with the observed decrease in cyclin B1 production upon mitotic entry and provides the first evidence that cyclin B1 synthesis is decelerated by CDK1 at the global translational level during early embryonic oscillations.
Attenuation of cyclin synthesis improves efficiency and robustness of CDK1 oscillations
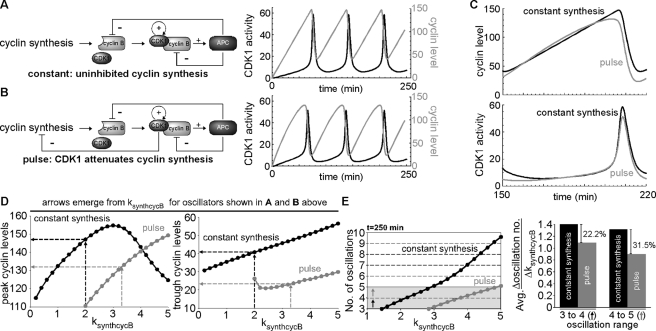
Following the discovery over 20 years ago that cyclin oscillates during embryogenesis, its constant synthesis has been the default for computational models of the Xenopus early embryonic cell cycle (Goldbeter, 1991; Goldbeter and Guilmot, 1996; Norel and Agur, 1991; Tyson, 1991; Novak and Tyson, 1993; Borisuk and Tyson, 1998; Sible and Tyson, 2007; Obeyesekere et al., 1992; Busenberg and Tang, 1994; Pomerening et al., 2005; Kim and Richter, 2007; Tsai et al., 2008; Ferrell et al., 2011). Our finding that cyclin production is attenuated prior to and during M phase raised questions about how this might affect CDK1 oscillations relative to past models driven by constant cyclin synthesis. Would this somehow be detrimental to cell cycle oscillations? Alternatively, might it confer some advantages? Because answers to these questions were not easily intuited, we set out to address them by modifying a previously published simple ordinary differential equation (ODE) model (Pomerening et al., 2005; Tsai et al., 2008; see Supplemental Materials and Methods) that included expressions for cyclin synthesis, CDK1 posttranslational modifications, and APC/cyclosome–mediated cyclin destruction. We then examined variations in cyclin production and proteolysis and also compared the sensitivities to changes in cyclin synthesis rate between the two different types of cyclin synthesis pattern.
Under constant cyclin synthesis (ksynthcycB = 2.0 nM/min; ksynthcycBoff = 0) the APC/cyclosome is responsible for removing cyclin that is complexed with CDK1 at mitotic exit, as well as new cyclin synthesized during mitosis (Figure 6A, left). These conditions generated more than three saw-toothed oscillations of cyclin B and three pulsatile oscillations of CDK1 activity in 250 min, similar to the frequency observed in cycling egg extracts (Figure 6A, right). A pulse-driven CDK1-APC oscillator—where CDK1 activity is inhibitory toward cyclin synthesis (Figure 6B, left)—requires a higher rate of synthesis (ksynthcycB = 3.3 nM/min; ksynthcycBoff = 0.2) to generate the same pattern but yields some characteristic differences from the constant-synthesis model (Figure 6B, right). The production of cyclin was blunted and its baseline reduced relative to the constant-synthesis model (Figure 6C, top), but the CDK1 activities elicited from both models remained nearly identical (Figure 6C, bottom). Attenuation of cyclin synthesis during oscillations produced 10% less cyclin input (Figure 6D, left), and this led to 43% less protein remaining after each cycle (Figure 6D, right). This pulse-driven oscillator was not only more efficient, but it also conferred decreased sensitivity to changes in frequency as cyclin synthesis rate changed (Figure 6E, left). Compared to CDK1 oscillations driven by continuous cyclin synthesis, the average change in oscillation rate per change in ksynthcycB was 22% less in the pulse-driven model between three and four oscillations and nearly one-third less from four to five oscillations (Figure 6E, right). These data supported that attenuation of cyclin stimulus not only produced comparable and realistic oscillations of CDK1 activity, but it also improved both its efficiency and robustness against changes in oscillation number as cyclin synthesis rate is varied.
FIGURE 6:
Attenuation of cyclin synthesis at M phase yields a CDK1 oscillator with some functional advantages. (A) Schematic of ODE model with constant cyclin synthesis (left); three oscillations of cyclin (gray) and CDK1 activity (black) from ODE model with constant cyclin synthesis (ksynthcycB = 0.2; ksynthcycBoff = 0; right). In this model, APC degrades all cyclin complexed with CDK1 and newly synthesized cyclin monomer. (B) Schematic of ODE model with pulses of cyclin synthesis caused by CDK1 activity repressing cyclin synthesis (left); three oscillations of cyclin (gray) and CDK1 activity (black) from ODE model with pulses of cyclin synthesis (ksynthcycB = 0.33; ksynthcycBoff = 0.2; right). In this model, CDK1 activity is inhibitory toward cyclin synthesis. Note that although the inhibitory leg of APC toward cyclin monomer shown in B is included in this computational model, it effectively disappears because no new cyclin synthesis occurs once APC is active. (C) Cyclin levels (top) and CDK1 activity (bottom) of ODE models with either constant cyclin synthesis (black) or cyclin pulses (gray) during a single oscillation (shown as the third oscillation in the simulation from 150 to 220 min to avoid minor variations in initial transients). (D) Peak cyclin levels (left) and trough cyclin levels (right) generated by ODE models with constant cyclin synthesis (black) or cyclin pulses (gray) as the parameter ksynthcycB (rate of cyclin synthesis) is varied. For consistency, peak and trough values of the limit cycle oscillations were measured. Arrows extending from the abscissa demarcate ksynthcycB values that produce a frequency of CDK1 oscillations representative of that observed in cycling egg extracts (shown in A and B, for constant synthesis and pulse, respectively); horizontal lines toward the ordinate indicate the median value required to produce the oscillations in the constant cyclin synthesis (black) and cyclin pulse (gray) models as shown in A and B, respectively. To generate oscillations in the cyclin pulse model, ksynthcycB values >1.8 were required. (E) Comparison of the sensitivity to changes in cyclin synthesis rate (ksynthcycB) between CDK1 oscillator models with constant or attenuated cyclin synthesis. Black arrow indicates changes in ksynthcycB required for an increase from three to four oscillations over a period of 250 min; gray arrow indicates changes in ksynthcycB required for an increase from four to five oscillations (left). Histogram (right) plotting differences in the average change in oscillation number over a period of 250 min per change in ksynthcycB, showing differences in sensitivity to increasing the oscillation number from three to four (left histogram pair; refer to black arrow in left plot) and from four to five (right histogram pair; refer to gray arrow in left plot).
In conclusion, our experimental data and simple computational modeling together reveal that the repression of cyclin production prior to mitosis and its destruction at mitotic exit can effectively function in tandem. Indeed, a negative-feedback loop governing cyclin production coupled to the APC-driven negative-feedback loop through CDK1 yields a more efficient and robust oscillator and would help to assure that cyclin levels are properly reset and early embryonic cell cycle oscillations are sustained at a proper rate.
DISCUSSION
A CDK1-driven negative-feedback loop represses cyclin synthesis during cell cycle oscillations
The regulatory underpinnings of the early embryonic cell cycle has been described as a “clock” being mediated by a biochemical oscillator that is autonomous and resistant to both chemical and physical perturbation (Beal and Dixon, 1975; Wasserman and Smith, 1978; Gerhart et al., 1984). Prior studies describe the constant stimulation of this oscillator with cyclin—even during mitosis (Evans et al., 1983; Groisman et al., 2002)—despite the known global repression of translation during that period (Kanki and Newport, 1991). Although this is entirely possible, it raised some questions regarding how this oscillator operates. Is there a reason to continue cyclin synthesis once a threshold level is produced? Does this system somehow benefit from constantly generating cyclin stimulus, despite the fact that newly synthesized cyclin would have to be immediately degraded during mitosis? To address these questions, we investigated the pattern of cyclin B1 synthesis with respect to global translation and its impact on cell cycle oscillations in Xenopus early embryonic extracts. We discovered that cyclin B levels plateau prior to peak CDK1 activation, and proteasome inhibition did not permit further accumulation after mitotic entry. Therefore cyclin destruction does not contribute to a flattening accumulation pattern of cyclin B1 prior to mitotic entry, and this agreed with prior work that showed APC activation is switch-like during mitosis following CDK1 activation (Georgi et al., 2002). To determine whether this slowdown in cyclin B1 accumulation is mediated by CDK1 activity, we tested whether preventing or accelerating CDK1 activation would alter the pattern of its synthesis. We found that CDK1 inhibition permitted continuous cyclin B1 accumulation, whereas early activation of CDK1 by nondegradable cyclin B1 addition prematurely halted its synthesis. It is known that CDK1 requires a threshold level of cyclin synthesis for its activation (Solomon et al., 1990), but this is the first demonstration that translation of cyclin B itself will continue if CDK1 is not activated; indeed, the oscillator is not programmed to know how much cyclin to dose in during each cycle, but CDK1 becoming active is the cue for its attenuation. This result was bolstered by experiments halting translation prior to peak CDK1 activation: adding CHX several minutes before CDK1 activation neither lessened the level of accumulated cyclin nor delayed mitotic entry in egg extract. This demonstrated that sufficient cyclin B1 is synthesized before M phase and its consequent CDK1-dependent attenuation. We then set out to determine whether the cause of this translational suppression of cyclin B1 at mitosis was either specific to its mRNA or global in nature. Levels of adenylation on the cyclin B1 3′ UTR did not alternate during oscillations, and translational repression of cyclin B1 still occurred despite the addition of exogenous mRNA lacking its UTRs. This revealed that changes in cyclin B1 synthesis during the cell cycle neither corresponded with alternating adenylation of its mRNA nor required its 3′ UTR and suggested that cyclin translation might be impeded by a general repression at mitosis. However, this disagreed with a prior study describing increased cyclin B translation during mitosis and its decrease in interphase due to alternating levels of adenylation on its mRNA (Groisman et al., 2002), and we performed experiments to address this. We found that polyadenylation of exogenous cyclin B1 mRNA greatly increased its interphase translation, but only minimally increased its translation during mitosis. The control of meiotic cyclin B translation in the unfertilized egg, however, is different from that of mitotic cycles. Polyadenylation of cyclin B1 mRNA has been reported in the unfertilized egg (Sheets et al., 1994; Ballantyne et al., 1997; Kim and Richter, 2007; Belloc et al., 2008), and we confirmed that meiotic CSF extracts are capable of continuous cyclin translation in the presence of elevated CDK1 activity, in agreement with previously published studies (Hansen et al., 2007; Wu et al., 2007; Isoda et al., 2011). Therefore, although translation of cyclin B in meiotic M phase is continuous despite CDK1 activity, it is attenuated during M phase of mitotic cycles. With no UTR of the cyclin B1 mRNA required to repress its translation during M phase, we then tested whether global translation is reduced at mitotic onset. As reported previously (Kanki and Newport, 1991), global protein synthesis is slowed at M phase, and we learned that the attenuation of cyclin B synthesis corresponds with a shift of cyclin B1 mRNA from polysome to nonpolysome fractions. Indeed, whereas the decreased translation of cyclin B prior to mitosis is mediated by CDK1 activity, this repression encompasses global protein synthesis.
Attenuation of cyclin synthesis implies an improved variation of the CDK1-APC oscillator
Unlike in eukaryotic systems such as yeast and Drosophila embryos, Xenopus embryos have apparently evolved a cell cycle oscillator capable of restraining cyclin production once a sufficient level has been generated to initiate CDK1 activation. The benefits of this regulation versus a constantly stimulated oscillator were unclear, so we used a simple ODE model to test whether there are any design advantages of a CDK1-APC system driven by CDK1-attenuated cyclin synthesis. By simply including a CDK1-mediated inhibitory term upon cyclin synthesis, we determined that this design would provide some advantages to the CDK1 oscillator. Driving CDK1 oscillations with protracted pulses of cyclin stimulus during interphase reduced the overall level of cyclin produced while making its destruction more effective during each cycle. This also lessened the sensitivity of the oscillator to changes in cyclin synthesis rate, making this cell cycle control system more robust against changes in frequency as translation rate varies compared with an oscillator driven by constant stimulus. In summary, our study reveals the novel finding that two negative-feedback loops—one restraining cyclin production through a global repression of translation, and the other promoting cyclin degradation—collaborate to provide an efficient and robust control system that delivers stimulus to and removes it from the developing early Xenopus embryo.
Although it is true that different organisms—even within their own respective developmental contexts (e.g., unfertilized and fertilized eggs)—will vary somewhat in the design and sensitivity of their cell cycle controls, our findings raise some intriguing possibilities regarding how these mechanisms evolved and diverged in higher eukaryotes. Although enforced continuous cyclin translation during mitosis does not adversely affect cell cycles in yeast (Drapkin et al., 2009) and flies (Stiffler et al., 1999), the fact that cyclin overexpression can cause mitotic arrest in a system such as Xenopus might be evidence of some plasticity in the design of the CDK1-APC oscillator. For example, cell cycles in certain organisms may require higher CDK1 activities and/or accelerated M-phase onset and could apply continuous cyclin production to accomplish this while trading off some efficiency and robustness. Alternatively, the cell cycles in the early frog embryo might require a more conservative approach to protein synthesis and cell cycle oscillations and therefore they evolved to produce the minimal amount of stimulus needed to drive its cleavages. Taken together, our results further underscore the relationship that can exist between translational and cell cycle controls. Understanding how these systems interface could be invaluable to illuminating how they evolved to function and facilitate optimized growth and development in all eukaryotes.
MATERIALS AND METHODS
Constructs
Xenopus stage VI oocytes were manually defolliculated, and total RNA was isolated using an RNeasy kit and recommended cytoplasmic protocol (Qiagen, Valencia, CA). Purified RNA served as template for cDNA synthesis using the SuperScript III RT-PCR kit (Invitrogen, Carlsbad, CA). Cyclin B1 3′ UTR was amplified from a collection of single-stranded cDNAs using Phusion polymerase and sense (5′-GCGAATTCAGGACTACGTGGCATTCCAATTG-3′) and antisense primers (5′-GCGTTAACCATGTTAAAATGAGCTTTATTAAAACC-3′), and the product was digested with EcoRI and HpaI and then cloned into pCS111, a derivative of the pCS2+ vector (Baker et al., 1999). Xenopus nondegradable cyclin B1 and cyclin B1–coding region constructs were described previously (Pomerening et al., 2005).
Purification of recombinant proteins
Baculoviruses were generated from constructs in a custom pFastBacNFLAGHIS plasmid, and recombinant proteins were overexpressed according the Bac-to-Bac Baculovirus Expression Systems Manual (Invitrogen) with minor variations. Recombinant proteins were bound in batch to anti-FLAG M2 agarose (Sigma-Aldrich, St. Louis, MO) and eluted with 0.3 mg/mL FLAG peptide (Sigma-Aldrich) in Tris-buffered saline, pH 7.5. Eluted proteins were dialyzed against two 1-l exchanges of buffer (150 mM NaCl, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 2.5 mM MgCl2, and 50 mM sucrose, pH 7.5), snap-frozen as 5-μl aliquots in liquid nitrogen, and stored at –80°C. The final concentration of each protein preparation was determined by electrophoresis, Sypro-Ruby staining (Bio-Rad, Hercules, CA), gel scanning by GelDocXR (Bio-Rad), and linear regression analysis using bovine serum albumin as a standard and QuantityOne 4.6.8 software (Bio-Rad).
Xenopus egg extract preparation, cyclin B1 precipitation, and histone H1 kinase assays
CSF extracts and cycling extracts were prepared essentially as previously described (Murray, 1991). The CSF extracts prepared from unactivated eggs in the absence of cycloheximide were incubated with either buffer or insect cell-expressed Δ65XCycB1 for 50 min at room temperature. DMSO (Sigma-Aldrich) or 1 mM MG132 (Tocris Bioscience, Ellisville, MO) was then added.
Cycling extracts used in time-course experiments were prepared from dejellied eggs treated for 2 min with 0.5 μg/ml calcium ionophore A23187 (Sigma-Aldrich) and were mixed with recombinant proteins, Xenopus cyclin B1 coding mRNA or cyclin B1 3′ UTR mRNA, MG132, cordycepin (Santa Cruz Biotechnology, Santa Cruz, CA), or protein buffer as indicated and were then incubated at room temperature. Samples of egg extract were removed at various intervals and frozen on dry ice for subsequent assays. Interphase and mitotic arrested extracts were prepared by the addition of 0.18 mM roscovitine (Cayman Chemical, Ann Arbor, MI) or Δ65XCycB1 to the cycling extracts, respectively. Cyclin B1 levels were monitored by labeling cycling egg extracts with 0.3 μCi/μl 35S-Met (PerkinElmer Life Sciences, Boston, MA) and freezing 8-μl samples on dry ice during the indicated time intervals. Samples were thawed with ice-cold EB buffer (80 mM β-glycerophosphosphate, 20 mM ethylene glycol tetraacetic acid , and 15 mM MgCl2, pH 7.3), and p13 agarose was used to precipitate Cdk1/35S-labeled cyclin complexes. Samples for histone H1 kinase assays and p13-precipitated proteins were electrophoresed through 12.5% Criterion gels (Bio-Rad) and transferred onto polyvinylidene fluoride (Millipore, Bedford, MA). Radioactive global translation assays were performed as described for cyclin B1, but total egg extract was analyzed. 32P-Labeled histone H1 and 35S-labeled cyclin or total proteins were detected by phosphorimaging and quantitated using ImageQuant software (Amersham Pharmacia Biotech, Piscataway, NJ).
To assess nuclear morphology and nuclear-envelope breakdown, demembranated sperm chromatin (prepared as described previously; Smythe and Newport, 1991) was added to extracts (1000 sperm/μl) and then stained with 4′,6-diamidino-2-phenylindole (DAPI) and analyzed by fluorescence and phase microscopy. Cell cycle phase was determined by analyzing at least five microscopic fields using a 20× objective. The criteria for M phase entry were condensed chromatin and a lack of a discernible nuclear envelope in at least 90% of the nuclei. The criterion for M phase exit was the reappearance of a smooth nuclear envelope around chromatin.
In vitro transcription
The m7G-capped Xenopus cyclin B1 RNA–coding region and cyclin B1 3′ UTR mRNAs were transcribed using a mMessage mMachine SP6 kit (Ambion, Austin, TX) with Xenopus cyclin B1-pCS111CFLAGHIS and cyclin B1 3′ UTR-pCS111CFLAGHIS templates linearized by AscI and HpaI, respectively. The m7G-capped luciferase mRNA was produced from pRL-CMV (Promega, Madison, WI) template and a mMessage mMachine T7 kit (Ambion). RNA concentration was determined by spectrophotometry, diluted to 1 mg/ml, visualized by electrophoresis through a nondenaturing 1% Tris-acetate-EDTA gel, and stored at –80°C.
In vitro and in vivo cyclin B1 3′ UTR polyadenylation, RNA purification, and RNA electrophoresis
In vitro polyadenylation was performed using Poly (A) Tailing Kit (Ambion) according to manufacturer's instructions with the following alterations: 1 μl of Escherichia coli poly(A) polymerase was added to a 50-μl reaction and incubated for a period of 20 min to obtain the desired poly(A) tail length. The reaction was stopped by the addition of an equal volume of H2O, and the RNA was extracted by LiCl precipitation (Ambion). Five micrograms of cyclin B1 coding sequence mRNA and in vitro–polyadenylated cyclin B1 coding sequence mRNA were resolved by electrophoresis through 1% agarose–3.7% formaldehyde gels and then stained with ethidium bromide. Labeling and detection of cyclin B1 3′ UTR polyadenylation in Xenopus extracts was performed as described previously, with minor modifications (Kim and Richter, 2008). Cyclin B1 3′ UTR RNA was in vitro transcribed and labeled with α-32P-uridine, then added to cycling extracts at a final concentration of 15 ng/μl. Ten-microliter samples were collected at 15-min intervals and frozen on dry ice. Total RNA was purified using TRIzol reagent (Invitrogen), separated on 6% polyacrylamide/6 M urea gels (Ambion), and visualized by phosphorimaging.
Luciferase assays
Renilla luciferase activity was measured using the Renilla Luciferase Assay System (Promega). In vitro–transcribed luciferase mRNA was added to cycling egg extract at a final concentration of 5 ng/μl. Twenty-microliter samples were collected at 15-min intervals into a 96-well plate placed on ice that contained 2 μl of a 1 mg/ml cycloheximide solution in each well. Luciferase assays were then performed as per the manufacturer's instructions.
Polysome profile analysis
Mitotic, interphase, or interphase extract (340 μl) incubated with 100 μg/ml puromycin (EMD Chemicals, Gibbstown, NJ) was mixed with an equal volume of 2× polysome buffer (50 mM KCl, 4 mM MgCl2, 4 mM HEPES, pH 7.7, cycloheximide 20 μg/ml, and 0.5% NP-40) and 80 U of murine RNase inhibitor (NEB, Ipswich, MA). Samples were centrifuged at 4°C in a refrigerated tabletop microcentrifuge at 13,000 rpm for 2 min, followed by their layering onto a 12-ml 25–56% sucrose gradient in 1× polysome buffer. Gradients were then ultracentrifuged in an SW40Ti rotor at 40,000 rpm for 160 min at 4°C without braking. The gradients were fractionated from the top in 25 fractions of equal volume while absorbance at 254 nm was recorded (GE Healthcare, Piscataway, NJ).
Determination of cyclin B1 mRNA relative abundance by qRT-PCR
Total RNA from equal volumes of each sucrose gradient fraction was isolated using GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich). The relative distributions of cyclin B1 mRNA were monitored by qRT-PCR using Brilliant II SYBR Green QRT-PCR Master Mix Kit, 1-Step (Stratagene, Santa Clara, CA). PCRs were carried out in an optical 96-well plate with an ABI StepOnePlus detection system (Applied Biosystems, Carlsbad, CA). The qRT-PCRs were carried out following the thermal profile: 50°C for 30 min and 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min. After 40 cycles, the specificity of the amplifications was tested by heating to 95°C for 1 min, 55°C for 30 s, and heating to 95°C for 30 s with a ramp speed of 0.5ºC/s, resulting in melting curves. Data were analyzed using Miner software (Zhao and Fernald, 2005). The average efficiency, E, of the cyclin B1 mRNA was used to calculate the relative transcript levels in each fraction from the equation Q = 1/(1 + E)CT, where CT is the cycle threshold. The primers used for cyclin B1 qRT-PCRs are as follows: cyclin B1 forward primer, 5′-TTCCTCCTGCTTGTCTCTCAAAATC-3′; and cyclin B1 reverse primer, 5′-CCATTTCCACAACAACATCTGACC-3′.
Ordinary differential equation modeling of the Xenopus CDK1-APC system
This model is based on that presented in Pomerening et al. (2005) but has been expanded to include an APC inactivation term (apcppase) based generally on previously published work (Lahav-Baratz et al., 1995; Wu et al., 2009) and a cyclin synthesis inhibition term (ksynthcycBoff) to model the effects of CDK1 inhibition on cyclin translation, and it has been reparametrized. The model was written for Mathematica 7.0 (Wolfram Research, Champaign, IL). Parameters were chosen by hand to yield reasonably realistic oscillations whether driven by constant or attenuated cyclin stimulus. The change that yields constant cyclin synthesis is setting ksynthcycBoff = 0; to produce a pulse-like behavior, set ksynthcycBoff = 0.2. Equations and parameters for the model are included in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank the Adam Zlotnick and Cheng Kao labs for providing technical assistance, the Indiana University Center for Genomics and Bioinformatics for facilities and assistance with qRT-PCR procedures, David Hendrickson for advice regarding sucrose gradients and qRT-PCR procedures, Srividhya Jeyaraman for advice regarding the modeling, and members of the Pomerening lab for critical review of the manuscript. J.R.P. is a Pew Scholar in the Biomedical Sciences and is supported by a grant from the National Institutes of Health (R01GM086526).
Abbreviations used:
- APC
anaphase-promoting complex
- CDK1
cyclin-dependent kinase 1
- CHX
cycloheximide
- CSF
cytostatic factor
- DAPI
4′,6-diamidino-2-phenylindole
- Δ65XCycB1
Xenopus nondegradable cyclin B1 protein
- NEB
nuclear envelope breakdown
- ODE
ordinary differential equation
- UTR
untranslated region
- XCycB1cod
Xenopus cyclin B1–coding region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0768) on November 30, 2011.
REFERENCES
- Azzam ME, Algranati ID. Mechanism of puromycin action: fate of ribosomes after release of nascent protein chains from polysomes. Proc Natl Acad Sci USA. 1973;70:3866–3869. doi: 10.1073/pnas.70.12.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JC, Beddington RS, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 1999;13:3149–3159. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne S, Daniel DL Jr, Wickens M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol Biol Cell. 1997;8:1633–1648. doi: 10.1091/mbc.8.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal CM, Dixon KE. Effect of UV on cleavage of Xenopus laevis. J Exp Zool. 1975;192:277–283. doi: 10.1002/jez.1401920218. [DOI] [PubMed] [Google Scholar]
- Beckhelling C, Penny C, Clyde S, Ford C. Timing of calcium and protein synthesis requirements for the first mitotic cell cycle in fertilised Xenopus eggs. J Cell Sci. 1999;112:3975–3984. doi: 10.1242/jcs.112.22.3975. [DOI] [PubMed] [Google Scholar]
- Belloc E, Pique M, Mendez R. Sequential waves of polyadenylation and deadenylation define a translation circuit that drives meiotic progression. Biochem Soc Trans. 2008;36:665–670. doi: 10.1042/BST0360665. [DOI] [PubMed] [Google Scholar]
- Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau AM, Sonenberg N. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J Biol Chem. 1987;262:11134–11139. [PubMed] [Google Scholar]
- Borisuk MT, Tyson JJ. Bifurcation analysis of a model of mitotic control in frog eggs. J Theor Biol. 1998;195:69–85. doi: 10.1006/jtbi.1998.0781. [DOI] [PubMed] [Google Scholar]
- Busenberg S, Tang B. Mathematical models of the early embryonic cell cycle: the role of MPF activation and cyclin degradation. J Math Biol. 1994;32:573–596. doi: 10.1007/BF00573462. [DOI] [PubMed] [Google Scholar]
- Creanor J, Mitchison JM. Patterns of protein synthesis during the cell cycle of the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1982;58:263–285. doi: 10.1242/jcs.58.1.263. [DOI] [PubMed] [Google Scholar]
- Datta B, Datta R, Mukherjee S, Zhang Z. Increased phosphorylation of eukaryotic initiation factor 2alpha at the G2/M boundary in human osteosarcoma cells correlates with deglycosylation of p67 and a decreased rate of protein synthesis. Exp Cell Res. 1999;250:223–230. doi: 10.1006/excr.1999.4508. [DOI] [PubMed] [Google Scholar]
- Drapkin BJ, Lu Y, Procko AL, Timney BL, Cross FR. Analysis of the mitotic exit control system using locked levels of stable mitotic cyclin. Mol Syst Biol. 2009;5:328. doi: 10.1038/msb.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Fan H, Penman S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr, Tsai TY, Yang Q. Modeling the cell cycle: why do certain circuits oscillate? Cell. 2011;144:874–885. doi: 10.1016/j.cell.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Georgi AB, Stukenberg PT, Kirschner MW. Timing of events in mitosis. Curr Biol. 2002;12:105–114. doi: 10.1016/s0960-9822(01)00662-5. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Wu M, Kirschner M. Cell cycle dynamics of an M-phase-specific cytoplasmic factor in Xenopus laevis oocytes and eggs. J Cell Biol. 1984;98:1247–1255. doi: 10.1083/jcb.98.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A. A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase. Proc Natl Acad Sci USA. 1991;88:9107–9111. doi: 10.1073/pnas.88.20.9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Guilmot JM. Arresting the mitotic oscillator and the control of cell proliferation: insights from a cascade model for cdc2 kinase activation. Experientia. 1996;52:212–216. doi: 10.1007/BF01920708. [DOI] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Pomerening JR, Summers MK, Miller JJ, Ferrell JE Jr, Jackson PK. Emi2 at the crossroads: where CSF meets MPF. Cell Cycle. 2007;6:732–738. doi: 10.4161/cc.6.6.3937. [DOI] [PubMed] [Google Scholar]
- Hartley RS, Rempel RE, Maller JL. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- Isoda M, et al. Dynamic regulation of Emi2 by Emi2-bound Cdk1/Plk1/CK1 and PP2A-B56 in meiotic arrest of Xenopus eggs. Dev Cell. 2011;21:506–519. doi: 10.1016/j.devcel.2011.06.029. [DOI] [PubMed] [Google Scholar]
- Kanki JP, Newport JW. The cell cycle dependence of protein synthesis during Xenopus laevis development. Dev Biol. 1991;146:198–213. doi: 10.1016/0012-1606(91)90460-k. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. Measuring CPEB-mediated cytoplasmic polyadenylation-deadenylation in Xenopus laevis oocytes and egg extracts. Methods Enzymol. 2008;448:119–138. doi: 10.1016/S0076-6879(08)02607-4. [DOI] [PubMed] [Google Scholar]
- Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton M, Cormier P, Belle R, Mulner-Lorillon O, Morales J. Translational control during mitosis. Biochimie. 2005;87:805–811. doi: 10.1016/j.biochi.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Malureanu L, Jeganathan KB, Jin F, Baker DJ, van Ree JH, Gullon O, Chen Z, Henley JR, van Deursen JM. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J Cell Biol. 2010;191:313–329. doi: 10.1083/jcb.201003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Norel R, Agur Z. A model for the adjustment of the mitotic clock by cyclin and MPF levels. Science. 1991;251:1076–1078. doi: 10.1126/science.1825521. [DOI] [PubMed] [Google Scholar]
- Novak B, Tyson JJ. Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J Cell Sci. 1993;106:1153–1168. doi: 10.1242/jcs.106.4.1153. [DOI] [PubMed] [Google Scholar]
- Obeyesekere MN, Tucker SL, Zimmerman SO. Mathematical models for the cellular concentrations of cyclin and MPF. Biochem Biophys Res Commun. 1992;184:782–789. doi: 10.1016/0006-291x(92)90658-8. [DOI] [PubMed] [Google Scholar]
- Paillard L, Legagneux V, Osborne HB. Poly(A) metabolism in Xenopus laevis embryos: substrate-specific and default poly(A) nuclease activities are mediated by two distinct complexes. Biochimie. 1996;78:399–407. doi: 10.1016/0300-9084(96)84746-8. [DOI] [PubMed] [Google Scholar]
- Pomerening JR, Kim SY, Ferrell JE Jr. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–578. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Sonenberg N. Cell-cycle-dependent translational control. Curr Opin Genet Dev. 2001;11:13–18. doi: 10.1016/s0959-437x(00)00150-7. [DOI] [PubMed] [Google Scholar]
- Salb JM, Marcus PI. Translational inhibition in mitotic HeLa cells. Proc Natl Acad Sci USA. 1965;54:1353–1358. doi: 10.1073/pnas.54.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Sible JC, Tyson JJ. Mathematical modeling as a tool for investigating cell cycle control networks. Methods. 2007;41:238–247. doi: 10.1016/j.ymeth.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Aviner R, Elroy-Stein O. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J Biol Chem. 2011;286:27927–27935. doi: 10.1074/jbc.M111.255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Stiffler LA, Ji JY, Trautmann S, Trusty C, Schubiger G. Cyclin A and B functions in the early Drosophila embryo. Development. 1999;126:5505–5513. doi: 10.1242/dev.126.23.5505. [DOI] [PubMed] [Google Scholar]
- Tsai TY, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE Jr. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 2008;321:126–129. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ. Modeling the cell division cycle: cdc2 and cyclin interactions. Proc Natl Acad Sci USA. 1991;88:7328–7332. doi: 10.1073/pnas.88.16.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Orr-Weaver TL. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev Cell. 2007;12:157–166. doi: 10.1016/j.devcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WJ, Smith LD. The cyclic behavior of a cytoplasmic factor controlling nuclear membrane breakdown. J Cell Biol. 1978;78:R15–R22. doi: 10.1083/jcb.78.1.r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MW, Degnin C, Hill J, Morris DR. Specific regulation by endogenous polyamines of translational initiation of S-adenosylmethionine decarboxylase mRNA in Swiss 3T3 fibroblasts. Biochem J. 1990;268:657–660. doi: 10.1042/bj2680657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Guo JY, Tang W, Yang CS, Freel CD, Chen C, Nairn AC, Kornbluth S. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol. 2009;11:644–651. doi: 10.1038/ncb1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, et al. A role for Cdc2- and PP2A-mediated regulation of Emi2 in the maintenance of CSF arrest. Curr Biol. 2007;17:213–224. doi: 10.1016/j.cub.2006.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.