Dictyostelium NE81 is the first protein found in a lower eukaryote with properties justifying its denomination as a lamin-like protein. Knockout and overexpression mutants revealed an important role for NE81 in nuclear integrity, chromatin organization, and mechanical stability of cells.
Abstract
Lamins build the nuclear lamina and are required for chromatin organization, gene expression, cell cycle progression, and mechanical stabilization. Despite these universal functions, lamins have so far been found only in metazoans. We have identified protein NE81 in Dictyostelium, which has properties that justify its denomination as a lamin-like protein in a lower eukaryote. This is based on its primary structure, subcellular localization, and regulation during mitosis, and its requirement of the C-terminal CaaX box as a posttranslational processing signal for proper localization. Our knockout and overexpression mutants revealed an important role for NE81 in nuclear integrity, chromatin organization, and mechanical stability of cells. All our results are in agreement with a role for NE81 in formation of a nuclear lamina. This function is corroborated by localization of Dictyostelium NE81 at the nuclear envelope in human cells. The discovery of a lamin-like protein in a unicellular organism is not only intriguing in light of evolution, it may also provide a simple experimental platform for studies of the molecular basis of laminopathies.
INTRODUCTION
The nuclear envelope consists of an inner and outer nuclear membrane. Proteins of the inner membrane interact with a filamentous protein network called the nuclear lamina. Its main structural components are specialized intermediate filament (IF) proteins called lamins. Owing to their presence in even the most primitive metazoans, such as Hydra, lamins are considered the most ancestral IF proteins. There are no IF proteins at all in plants, fungi, and protozoa. Cytoplasmic IF proteins appear to be absent in many arthropods, although a cytosolic IF protein, isomin, has very recently been characterized in primitive insects (Herrmann and Strelkov, 2011; Mencarelli et al., 2011). While mammals express four lamin isoforms, lamins A, C, B1, and B2, most invertebrates possess only one lamin isoform belonging to the B type. B-type lamins are ubiquitously expressed and are essential for cell viability, while A-type lamins are restricted to differentiated cells.
During interphase, lamins form a network of organized filaments underneath the inner nuclear membrane (Goldberg et al., 2008; Herrmann et al., 2009). On its nuclear face, the lamin network is associated with heterochromatin, histones, transcriptional regulators, and chromatin modifiers. On its reverse face, it is coupled to transmembrane proteins of the nuclear envelope, which in turn are linked to all cytosolic cytoskeletal systems. This has been discovered to be crucial for the mechanical stability of the whole cell (Dahl et al., 2004; Friedl et al., 2011). However, bearing in mind the diversity of lamin interactions with chromatin and other proteins, it is not surprising that lamins serve more functions than just providing mechanical support and a structural barrier between two compartments. They are also involved in chromatin organization, epigenetic regulation, transcription, and mitosis (Andres and Gonzalez, 2009). This became evident when researchers elucidated the molecular defects underlying several devastating, inheritable diseases, including muscular dystrophy, cardiomyopathy, partial lipodystrophy, and progeria (Worman et al., 2009). These diseases are known as laminopathies, because they are usually caused by mutations in the lamin A gene or in genes encoding lamin-binding or lamin-processing proteins, which are required for proper assembly of the lamin-based nucleoskeleton.
A-type and B-type lamins are translated as proproteins, undergoing several processing steps to yield the mature protein. First, they are prenylated by a farnesyl transferase at the cysteine residue of the C-terminal CaaX box (C = cysteine, a = aliphatic amino acid, X = any amino acid). In a further step, a specific protease removes the three C-terminal amino acids (= aaX), either by prenyl protein peptidase, in the cases of prelamin A, B1, and B2, and also by ZMPSTE24, in the case of prelamin A. After C-terminal cleavage, the farnesylated cysteine residue is methylated by isoprenylcysteine methyl transferase. While this is the last processing step in the case of B-type lamins, prelamin A is cleaved once again by ZMPSTE24, which removes the last 15 amino acids, including the farnesyl anchor.
Fluorescence recovery after photobleaching (FRAP) experiments have revealed that lamins integrated into the nuclear lamina show little or no mobility (Broers et al., 1999). Closer analyses using FRAP revealed that mature lamin B1 already begins to assemble at the nuclear envelope in late mitosis, while the majority of lamin A is only integrated into the stable nuclear lamina in late G1 (Moir et al., 2000). On entry into mitosis, lamin phosphorylation by CDK1 causes disassembly of the nuclear lamina, which in turn is a prerequisite for nuclear envelope breakdown (NEBD) in higher cells (Foisner, 2003). In many fungi and lower eukaryotes, such as Dictyostelium discoideum amoebae, there is no NEBD and chromosome segregation takes place within an intact or sometimes fenestrated nuclear envelope.
Recent studies in Dictyostelium have brought many insights into chromatin dynamics, particularly in the areas of transcriptional activity and histone modification (reviewed by Stevense et al., 2011). Yet relatively little is known about the structural organization of the Dictyostelium nuclear envelope. A nuclear lamina has not yet been described in Dictyostelium. With the exception of the nuclear pore complex proteins Nup43, interaptin, and Sun1, nothing is known about proteins associated with the nuclear membranes (Xiong et al., 2008; Schulz et al., 2009a). Sun1 is present in both nuclear membranes, and is a key component of the physical connection between centrosomes and centromeres, spanning both nuclear membranes and persisting throughout interphase (Schulz et al., 2009a). As in all unicellular organisms, no intermediate filaments, and thus no lamins, have been identified in the Dictyostelium genome to date. Yet Dictyostelids represent an interesting order of organisms. Under certain environmental conditions, these usually unicellular, vegetatively growing amoebae are capable of forming a multicellular entity (Kessin, 2001). From an evolutionary point of view, they are positioned between protists, which lack lamins, and metazoans, which possess a nuclear lamina composed of lamins. We have now identified the first lamin-like protein in D. discoideum. In this work, we demonstrate the functional similarity of NE81 to lamins. In the future, Dictyostelium, a genetically accessible and well-established model organism, may provide a simple experimental platform that could help us to understand the mechanisms underlying human laminopathies.
RESULTS
In a recent attempt to identify novel centrosomal components through mass spectrometric analysis of the proteome of isolated Dictyostelium centrosomes (Reinders et al., 2006), we have identified not only novel centrosomal components, but also several proteins allocated to centrosome-associated structures (Schulz et al., 2009b). One of these proteins, encoded by gene DDB_G0289429, is localized at the nuclear envelope in interphase cells when expressed as a green fluorescent protein (GFP) fusion protein. Thus the protein was called NE81, according to its calculated molecular mass of 81 kDa. Furthermore, NE81 copurified with isolated centrosomes. This accidental finding was not investigated further in our earlier study. In this paper, we present a functional characterization of the NE81 protein. Our results strongly suggest that NE81 represents an evolutionary precursor of metazoan lamins.
The primary structure of NE81 is reminiscent of lamins
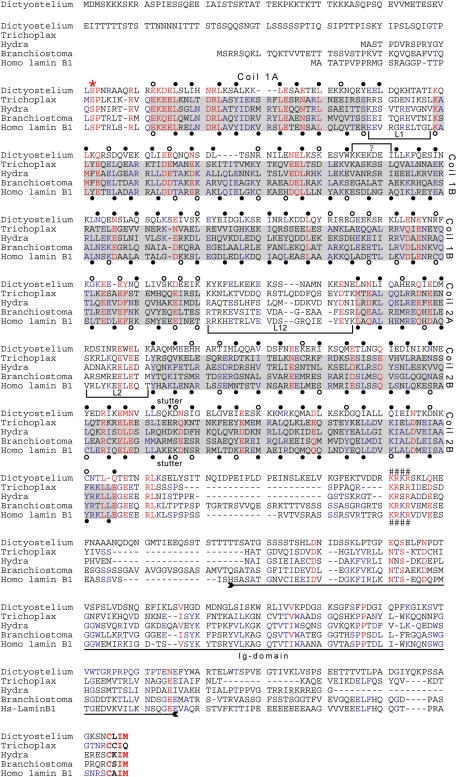
NE81 consists of 716 amino acid residues. Sequence analysis revealed a similarity to nuclear lamins, with which it shares a predicted central coiled-coil domain flanked by more or less globular head and tail domains. With a predicted length of 370 amino acids, the coiled-coil region is comparable in size with the corresponding region of nuclear lamins of metazoans. Figure 1 shows a protein sequence alignment of Dictyostelium NE81 with four bona fide lamins from plakozoans, hydrozoans, cephalochordates, and humans. The coiled-coil region of lamins is characterized by four α-helical rods named coils 1A, 1B, 2A, and 2B, which are interrupted by short linker sequences called L1, L12, and L2 (Herrmann and Aebi, 2004). Within the NE81 sequence, assignment is straightforward for coils 1A, 2A, and 2B. A small linker (marked by a bracketed “?” in Figure 1) must be introduced to match the heptad pattern of the lamins. Moreover, at the end of the putative coil 1B, a large number of unfavorable residues are found in a and d positions (open circles). As in the lamin sequences, a stutter has been introduced in coil 2B of NE81. This discontinuity in the heptad pattern at this position is a characteristic feature of IF proteins (arrows below and above the sequences in Figure 1). Note, however, that in contrast with bona fide lamin sequences, the same number of favorable amino acid residues are found in NE81 in the a and d positions, even without a stutter. One of the hallmarks of IF proteins is the highly conserved sequences of the N- and C-termini of their rod domains and, to a lesser extent, the conserved sequences at the beginning of coil 1B. At the end of the NE81 rod region, similarity to the IF consensus sequence (YRKLLEGEEeRLr/kL/I) is only very weak. The immunoglobulin (Ig) domain within the tail is a shared characteristic of most lamins. The alignment highlights only few amino acid identities in this region of NE81, giving no clear hints as to whether or not this region actually represents an Ig-type folding. As in the canonical lamins, the coiled-coil region of NE81 is directly preceded by a CDK1 phosphorylation consensus sequence (SPNR; the consensus is S/T-P-X-R/K) and is followed by an evolutionarily conserved, basic nuclear localization sequence (KRKR), and the sequence carries a CaaX box for isoprenylation (CLIM) at its C-terminal end. The KRKR motif is conserved among lamins, and its requirement for nuclear localization has been proven by Mattout et al. (2007). The methionine at the X-position of the CaaX box is also typical for lamins and indicates farnesylation instead of geranylgeranylation (Casey and Seabra, 1996). NE81 is conserved among the other sequenced Dictyostelids, Dictyostelium purpureum, Dictyostelium fasciculatum, and Polysphondylium pallidum, with amino acid sequence identities ranging from 36% (P. pallidum) to 57% (D. purpureum; sequence data kindly provided by G. Glöckner, IGB, Berlin). These sequence similarities strongly suggested NE81 be classified as a lamin-like protein with a clear evolutionary relationship to IFs. We set out to corroborate this idea with experimental data.
FIGURE 1:
Protein sequence alignment of four bona fide lamins with Dictyostelium NE81. The lamin sequences of the plakozoon Trichoplax adhaerens, the hydrozoon Hydra magnipapillata, the cephalochordate Branchiostoma floridae, and the human lamin B1 were aligned with Dictyostelium NE81 using MultAlin software (Corpet, 1988). The sequence of the CaaX-encoding exon of Hydra was manually aligned to highlight the presence of the CaaX motif in all five sequences. The sequences of Trichoplax, Hydra, and Branchiostoma were deduced from genomic sequences (Peter and Stick, unpublished data); the human (AAC37575) and Dictyostelium (this study) sequences were deduced from cDNA sequences. Coiled-coil regions are shaded in gray, and linker regions separating the coils are marked by brackets. Shading is restricted to bona fide lamin sequences, to avoid biased interpretation. Heptad repeats of the human B1 are marked at the first and fourth position (a and d positions) below the sequence. Filled circles indicate hydrophobic and uncharged amino acid residues favorable for coiled-coil formation (L, I, V, A, M, N, Q, Y, F, W); open circles indicate less favorable amino acid residues (E, D, K, R, H, S, T, C, G). The discontinuity in the heptad pattern in the second half of coil 2B (stutter) is indicated by an arrow above and below the Dictyostelium and B1 sequences, respectively. Guided by the lamin heptad pattern, tentative heptad positions were assigned to the Dictyostelium NE81 sequence (above the sequence). A small linker, marked by a bracket with a ?, had to be introduced into the NE81 sequence to match the heptad pattern of the lamins. The region of the Ig domain within the tail of the lamin sequences is indicated by a double-headed arrow. Three sequence motifs characteristic for lamins are highlighted: a CDK1 phosphorylation consensus sequence upstream of the rod domain (*), a putative nuclear localization sequence (####), and the C-terminal CaaX box (bold letters).
NE81 is associated with the nuclear envelope during the entire cell cycle
If NE81 were a lamin-like protein, it should associate with the inner membrane of the nuclear envelope. To investigate this by immuno-electron microscopy (immuno-EM), we generated a specific polyclonal antiserum against recombinant NE81, expressed as an MBP-NE81 fusion protein in Escherichia coli (Figure 2A). In immuno-EM of isolated nuclei, the vast majority of gold particles associated with the inner nuclear membrane (Figure 2B). This was even more evident in regions in which the outer nuclear membrane peeled away from the inner nuclear membrane during specimen preparation, and gold particles were exclusively located at the inner membrane. Thus, the predicted nuclear localization signal (NLS) of NE81 appears to be functional and indicates NE81 is a nuclear protein associated with the inner nuclear membrane. The functionality of the NLS was also shown by fusion of the NLS-encoding sequence to GFP-CP224ΔC. This N-terminal fragment of the XMAP215-family protein CP224 normally localizes to the cytosol (Hestermann and Gräf, 2004). After introduction of the NLS sequence from NE81 (RKRKRSK), the GFP-NLS-CP224ΔC fusion protein was found exclusively in the nucleus (Supplemental Figure S1).
FIGURE 2:
NE81 is associated with the inner nuclear envelope. (A) Immunoblot of nuclear proteins stained with anti-NE81 to show the specificity of the antibody. (B) Immunogold EM of isolated nuclei. Inner nuclear membrane association is particularly evident in regions in which the outer nuclear membrane is ripped off (upper edge in B). Scale bar: 0.5 μm.
Anti-NE81 antibodies were also used for immunofluorescence microscopy to study the behavior of the endogenous protein during mitosis. While NE81 was evenly distributed over the entire nuclear envelope during interphase, we observed an uneven distribution of NE81 at the nuclear envelope during mitosis, with a reduced concentration of the protein in the equatorial plane. This was evident at the metaphase–anaphase transition, and more obviously in telophase, in which NE81 was almost absent between the chromosome masses of the two daughter nuclei (arrowheads in Figure 3). The same mitotic localization was also shown by the GFP-NE81 fusion protein (Figure S2). From our earlier investigations of GFP-Sun1 cells (Schulz et al., 2009a), we know that the central spindle connecting the two daughter nuclei is still associated with the nuclear envelope at this stage of mitosis. Therefore the absence of NE81 in this region indicates a regulatory process, and the protein is likely to be more mobile at this stage than during interphase.
FIGURE 3:
Cell cycle–dependent localization of endogenous NE81. Immunofluorescence microscopy of wild-type cells fixed with glutaraldehyde and stained with anti-NE81/anti–rabbit-Alexa Fluor 488 and anti–tubulin YL1/2 (Wehland and Willingham, 1983)/anti–rat-Alexa Fluor 568. Mitotic stages are indicated at the top. The merged panel shows GFP fluorescence in green, tubulin in red, and nuclei in blue. The bottom panel shows GFP-NE81 fluorescence only. Arrowheads point at regions with reduced amounts of NE81. Scale bar: 2 μm.
Mobility of NE81 at the nuclear envelope is regulated during mitosis
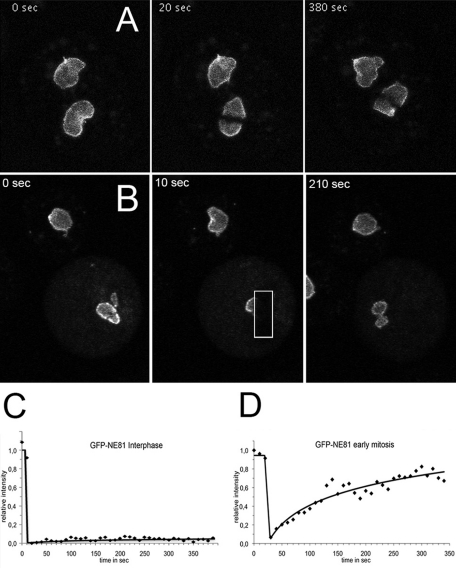
If NE81 were a major constituent of a nuclear lamina, we would not expect considerable turnover of the protein at the nuclear envelope of interphase nuclei, but we would anticipate increased mobility during mitosis to allow constriction of the nuclear envelope during karyokinesis. Indeed, our FRAP experiments clearly showed that GFP-NE81 is almost immobile at the nuclear envelope during interphase (Figure 4A and Supplemental Movie S1). Within the monitored time period of 390 s, there was no noteworthy recovery of the bleaching process, suggesting that NE81 is stably associated with the nuclear envelope, and there is little turnover during interphase. In contrast, during mitosis, prior to the metaphase–anaphase transition, GFP-NE81 exhibited considerable mobility, and reappeared in the bleached region of interest with a half-time of recovery of approximately 100 s (Figure 4B and Movie S2). The window of time of high GFP-NE81 mobility appears to be quite narrow, since we no longer observed a noteworthy mobility in early telophase (Movie S3).
FIGURE 4:
GFP-NE81 is virtually immobile within the nuclear envelope during interphase but mobile during mitosis. FRAP experiments in a three-dimensional time series recorded on a confocal laser-scanning microscope. The bleached region is marked by the white rectangle. Time-lapse acquisition rate was 6 stacks/min (at a frame rate of 5 frames/s); maximum intensity projection of 5 slices per image stack. (A and C) Bleach during interphase. (B and D) Bleach during metaphase. (C and D) Corrected FRAP curves of representative experiments (n = 15 in C; n = 4 in D).
Proper localization of NE81 at the nuclear envelope requires the prenylation signal
As in case of lamins, we expected the C-terminal CaaX box to serve as a signal for protein processing and targeting, including isoprenylation, proteolytic cleavage, and isoprenylcysteine methylation. Therefore we expressed a truncated GFP-NE81 fusion protein in which the CaaX box was deleted (GFP-NE81ΔCLIM) both in the control strain AX2, for analyses of fixed cells, and in a cherry-H2B strain with red fluorescent chromatin, for live-cell studies. During interphase, GFP-NE81ΔCLIM was no longer clearly associated with the nuclear membrane but accumulated in discrete clusters. These clusters were always clearly located inside the nucleus (Figure 5 and Movie S4). They were often found close to the nuclear envelope. GFP-NE81ΔCLIM was completely absent from the cytosol. The localization of GFP-NE81ΔCLIM clusters in the nucleus clearly showed that the CaaX box is functional and required for proper localization of the fusion protein to the nuclear envelope. These results confirm that NE81 is an inner nuclear protein.
FIGURE 5:
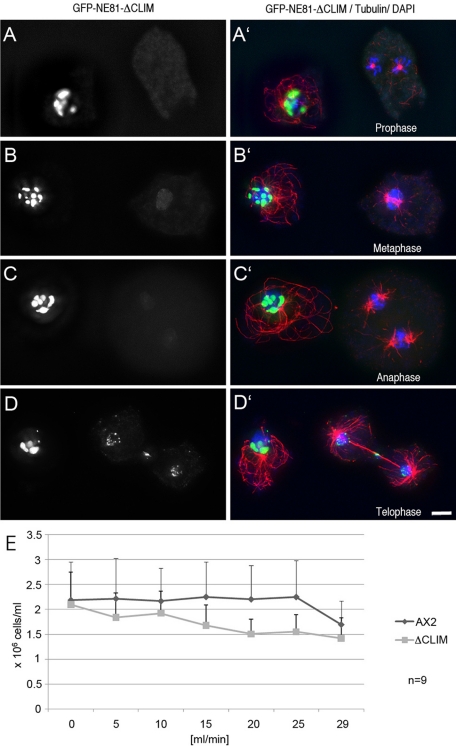
GFP-NE81ΔCLIM reversibly forms clusters within the nucleus and causes increased mechanosensitivity. Immunofluorescence microscopy of GFP-NE81ΔCLIM cells fixed with glutaraldehyde and stained with anti-tubulin YL1/2/anti–rat-Alexa Fluor 568 (A–D). Mitotic stages are indicated in (A′–D′) below the respective mitotic cells. The merged panel (A′–D′) shows GFP fluorescence in green, tubulin in red, and DAPI-stained nuclei in blue. The left panel (A–D) shows GFP fluorescence only. Bar: 2 μm. (E) AX2 control cells and GFP-NE81ΔCLIM cells were passed through polycarbonate filters (12-μm pore size) at the flow rate given at the x-axis. The number of surviving cells was counted and is given on the y-axis. Mean values ± SD are given.
Formation of GFP-NE81ΔCLIM clusters is cell cycle dependent
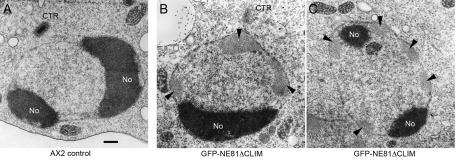
Analysis of fixed cells suggested that GFP-NE81ΔCLIM clusters dissolve at the G2–M transition, causing the fusion protein to diffuse across the cell through the permeable nuclear envelope. This was concluded from the absence of GFP-NE81ΔCLIM clusters from prophase to early telophase, and the appearance of a weak green fluorescent background in the nucleus and the cytosol (cells on the right in Figure 5, A–C), which was absent during interphase (cells on the left in Figure 5, A–D). Live-cell imaging of GFP-NE81ΔCLIM/cherry-H2B cells clearly confirmed these observations, since GFP-NE81ΔCLIM clusters suddenly disappeared at the onset of mitosis, and started to reappear at the chromatin masses and the central spindle only in telophase (Movie S4). GFP-NE81ΔCLIM clustering is reversible and may be regulated by a mitotic signal. Moreover, this behavior argues against GFP-NE81ΔCLIM clusters representing unstructured aggregates of improperly folded protein. This view is also corroborated by EM of GFP-NE81ΔCLIM clusters (Figure 6). In ultrathin sections, these clusters appeared as spongy structures of intermediate electron density. They were mostly located in the nuclear periphery at the nuclear envelope. This may be explained by their association with farnesylated endogenous NE81. The appearance of GFP-NE81ΔCLIM clusters as seen using EM clearly distinguishes them from protein aggregates, which are usually quite electron dense (Kopito, 2000). Based on these observations, we believe that NE81 possesses an inherent capacity to reversibly form polymeric structures. The dissociation of GFP-NE81ΔCLIM clusters at the onset of mitosis may be triggered through phosphorylation of serine residue 122 within the predicted CDK1 phosphorylation site. To corroborate this idea, we created a Dictyostelium strain expressing point-mutated GFP-NE81ΔCLIM-S122A, in which Ser-122 was mutated to alanine. The S122A version also formed green fluorescent clusters within the nucleus, but unlike GFP-NE81ΔCLIM clusters, they persisted throughout mitosis (Figure 7). This behavior strongly suggests that dynamic formation and dissociation of GFP-NE81ΔCLIM clusters is regulated by phosphorylation at Ser-122.
FIGURE 6:
GFP-NE81ΔCLIM forms spongy clusters of intermediate electron density within the nucleus. Ultrathin section through the nucleus of control cells (A) and GFP-NE81ΔCLIM cells (B and C). Cells were processed for EM essentially as described in (Ueda et al., 1999). Cells on coverslips were fixed for 30 min with 1% glutaraldehyde in PHEM buffer 12 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 1.6 mM ethylene glycol tetraacetic acid, 1 mM MgCl2) supplemented with 0.5% Triton X-100. Coverslips were washed three times for 5 min each time with 50 mM Na-cacodylate buffer (pH 7.0) and postfixed in cacodylate-buffered 1% OsO4 for 30 min. After dehydration in a graded ethanol series and acetone, specimens were embedded in Spurr's resin. Ultrathin sections were further processed as described in Materials and Methods. GFP-NE81ΔCLIM clusters are indicated by arrowheads. No, nucleolus; CTR centrosome. Scale bar: 0.5 μm.
FIGURE 7:
Nuclear GFP-NE81ΔCLIM-S122A clusters persist during mitosis. Immunofluorescence microscopy of GFP-NE81ΔCLIM-S122A cells fixed with glutaraldehyde and stained with anti–tubulin YL1/2/anti–rat-Alexa Fluor 568 and DAPI (A–E). Single fluorescence channels are given on top and mitotic stages are indicated in (A–E). The merged panel shows GFP fluorescence in green, tubulin in red and nuclei in blue (A′′′–E′′′). Scale bar: 2 μm.
NE81 is required for mechanical stability of cells
On fivefold overexpression of the fusion protein GFP-NE81ΔCLIM, cells were unable to grow in shaking culture, while they grew normally in adherent culture. This suggested an increased sensitivity of GFP-NE81ΔCLIM cells to mechanical stress. This notion was confirmed in an assay in which GFP-NE81ΔCLIM cells and control cells were passed through polycarbonate filters with an exact pore size of 12 µm at defined speed using an adjustable pumping device. As control cells, GFP-NE81ΔCLIM cells usually had a size well below 10 μm and showed no multiple nuclei; the cell survival rate after this treatment was a good measure for their resistance against mechanical shear forces. At a flow rate of 20 ml/min, only approximately two-thirds of GFP-NE81ΔCLIM cells survived this treatment, while control cells were still unaffected (Figure 5E). This increased mechanosensitivity of GFP-NE81ΔCLIM cells is in agreement with a role of an NE81-based nucleoskeleton as a mechanical support and thus substantiates a further lamin-like function of NE81. GFP-NE81ΔCLIM expression may exert this dominant-negative effect through copolymerization with endogenous NE81. Recruitment of endogenous NE81 into the GFP-NE81ΔCLIM clusters may result in an insufficient amount of functional NE81 at the nuclear envelope. The NE81/GFP-NE81ΔCLIM copolymer may still be anchored to the membrane through prenylation of endogenous NE81, which explains the occurrence of the majority of clusters in vicinity of the nuclear envelope.
Overexpression and knockout of NE81 reveals lamin-like functions
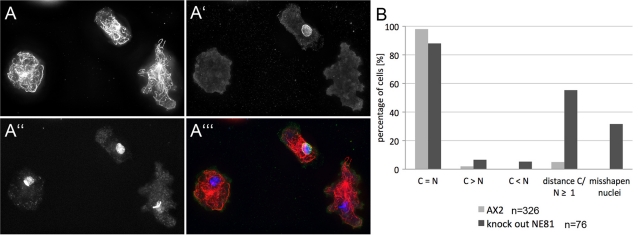
High mechanosensitivity of GFP-NE81ΔCLIM cells was not the only indication that NE81 could function in a lamin-like manner. When Dictyostelium GFP-NE81 cells were analyzed soon after transformation (i.e., after they had reached a total number of ∼2 × 106 cells), about half of the cells showed aberrant phenotypes, including misshapen nuclei with obviously disorganized chromatin (Figure 8). These GFP-NE81 cells were obviously unable to survive and disappeared after several passages, due to selective pressure against these phenotypes. In surviving cells, the expression level of GFP-NE81 was attenuated (to ∼3.2-fold overexpression based on Western blot densitometry), and nuclei were characterized only by a green fluorescent nuclear envelope, but otherwise displayed normal morphology (Figure S2). We also constructed a knockout plasmid to disrupt the NE81 gene using homologous recombination. We obtained a mixed cell population of null mutants and cells obviously carrying an ectopic integration of the blasticidin resistance gene in five independent transformations. While null mutants showed no NE81 protein expression in immunofluorescence microscopy, the latter displayed anti-NE81–labeled nuclear envelopes (Figures 9A′ and S3B). Unfortunately, we were not able to clone these NE81 knockout cells. In mixed cultures, as obtained after transformation, knockout cells were rapidly overgrown by the healthier cells carrying the ectopic integration of the knockout vector. All attempts to obtain a pure strain by cloning on bacterial lawns failed. In 4′,6-diamidino-2-phenylindole (DAPI) staining, cells not expressing NE81 were characterized by misshapen nuclei with irregularly condensed chromatin (32%), increased distance of centrosomes from nuclei (55%), supernumerary centrosomes (7%), and missing centrosomes (5%; Figure 9B; compare cells with and without anti-NE81 staining in Figures 9, A–A′′′, and S3, A–D). Taken together, the phenotypes observed upon NE81 overexpression and in cells lacking NE81 indicate that NE81 shares many functions attributed to lamins in metazoan cells, including maintenance of nuclear architecture, chromatin integrity, and cross-talk of nuclear structures with cytosolic structures, such as the centrosome and microtubules.
FIGURE 8:
GFP-NE81 overexpression results in misshapen nuclei with disorganized chromatin. Immunofluorescence microscopy of GFP-NE81 cells shortly after transformation. Two examples are shown. Cells were fixed with glutaraldehyde and stained with anti–tubulin YL1/2/anti–rat-Alexa Fluor 568. The merged panel shows GFP fluorescence in green, tubulin in red, and DAPI-stained nuclei in blue. Scale bar: 2 μm.
FIGURE 9:
Knockout of NE81 results in irregularly condensed chromatin, centrosomal detachment from nuclei, and altered centrosome numbers. Immunofluorescence microscopy of NE81 knockout cells shortly after transformation. (A–A′′′) Cells were fixed with glutaraldehyde and stained with anti–tubulin YL1/2/anti–rat-Alexa Fluor 568 (A), anti-NE81/anti–rabbit-Alexa Fluor 488 (A′), and DAPI (A′′). A merged image in shown in (A′′′). See Figure S3 for more examples. (B) Phenotypic quantification in cells without anti-NE81 labeling. Percentages are given for cells with equal numbers of centrosomes and nuclei (C = N), supernumerary centrosomes (C > N), missing centrosomes (C < N), centrosome/nucleus distance of more than 1 μm, and misshapen nuclei. Scale bar: 2 μm.
NE81 localizes to the nuclear envelope in mammalian cells
Finally, we wanted to assess whether NE81 is also capable of localizing to the nuclear envelope in mammalian cells. We transiently expressed a codon-optimized version of NE81 as a GFP fusion protein in HeLa cells. Fluorescence microscopy revealed that GFP-NE81 clearly colocalized with lamin B1 at the nuclear envelope in these cells. This is another strong indication that NE81 functions as a lamin-like protein and is associated with the inner nuclear membrane (Figure 10).
FIGURE 10:
Colocalization of GFP-NE81 and lamin B1 at the nuclear envelope in HeLa cells. Immunofluorescence microscopy 24 h after transformation showing GFP-NE81 fluorescence (A, green in C) and lamin B1 (B, red in C). Formaldehyde fixation stained with anti–lamin B1 (Abcam, Cambridge, UK) and anti–rabbit-Alexa Fluor 568 and DAPI (blue in C). A merged image is shown in (C) and a phase-contrast image in (D). Scale bar: 10 μm.
DISCUSSION
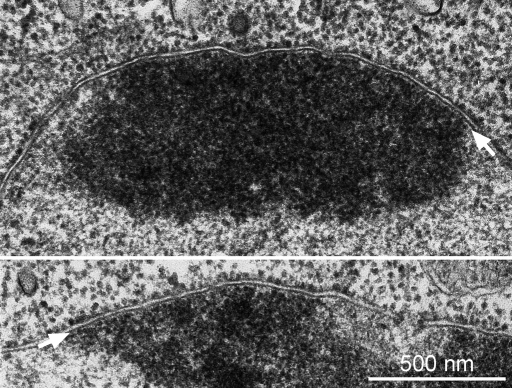
The existence of a nuclear lamina in Dictyostelium amoebae was already postulated in the late 1970s, when as yet unpublished electron micrographs of Dictyostelium cells revealed a thin, continuous, low-electron-density layer between the inner nuclear membrane and the chromatin (Figure 11). Its appearance was very reminiscent of the so-called fibrous lamina detected in ultrathin sections of animal cells in the 1960s (Fawcett, 1966). Today this layer is known as the lamin-based nuclear lamina. Yet the molecular correlate to a nuclear lamina in Dictyostelium was completely unknown. All our data presented in this work strongly suggest that NE81 is the first lamin-like protein to be characterized in a nonmetazoan organism. NE81 is associated almost exclusively with the inner nuclear envelope. Its predicted structural organization—a 370–amino acid, coiled-coil domain, whose α-helical pattern shares similarities with IFs; a head domain that may serve as a CDK1 target; and a tail domain with an NLS and a CaaX box as a putative farnesylation signal—is very reminiscent of lamins. Aside from lamins, Drosophila kugelkern is the only other known inner nuclear protein containing a CaaX box (Melcer and Gruenbaum, 2006). Kugelkern is also involved in formation of the nuclear lamina, but is clearly distinguished from lamins by its much shorter α-helical, coiled-coil region. Thus NE81 is much more likely to represent a distant relative of lamins than of kugelkern. Database analysis revealed that the genes encoding all proteins known to be required for CaaX-box processing, that is, farnesyl transferase (fntA/fntB), ZMPSTE24 endopeptidase (zmpste24), and isoprenylcysteine methylase (icmA) are represented in the Dictyostelium genome, suggesting that NE81 is processed in the same manner as lamins. Indeed, we could clearly demonstrate that the CaaX box of NE81 is functional and is, as in lamins, required for proper protein targeting to the nuclear envelope. Deletion of the C-terminal CaaX box in GFP-NE81 abolished proper localization of the protein to the nuclear envelope. Instead, it resulted in clustering of the GFP-NE81ΔCLIM fusion protein inside the nucleus. The spongy appearance of GFP-NE81ΔCLIM clusters, and their relatively low electron density in EM images, indicated that these clusters were not aggregates of unfolded, overexpressed protein. This view was corroborated by their intriguing dynamic behavior, which was observed by live-cell imaging of mitotic cells in which clusters dissolved at the onset of mitosis and reappeared in telophase. Such a cell cycle–dependent dynamic behavior was already expected from the uneven distribution of the endogenous protein in fixed, mitotic cells. Here NE81 was almost absent from the equatorial zone of the nuclear envelope during metaphase and, later, from the constriction zone of the nuclear envelope during karyokinesis. This already suggested regulation and some mobility of NE81 during mitosis, which was proven by FRAP experiments. While GFP-NE81 does not exhibit considerable mobility at the nuclear envelope during interphase, as was expected for a structural component of a putative nuclear lamina, it is highly mobile between the onset of mitosis and early telophase. It is tempting to speculate that the mitotic signal triggering both increased mobility of GFP-NE81 and disassembly of GFP-NE81ΔCLIM clusters is phosphorylation by cyclin-dependent kinase 1 (CDK1). First, NE81 contains a CDK1 phosphorylation consensus sequence at the same position as in nuclear lamins of metazoans. Second, the time course of GFP-NE81ΔCLIM cluster disassembly/assembly and high mobility of GFP-NE81 matches exactly the time course of activity of CDK1. We suggest that GFP-NE81ΔCLIM clusters are a result of an inherent capacity of NE81 to form high-order polymers, or paracrystals, and that this capability could be regulated by phosphorylation. According to our hypothesis, endogenous NE81 becomes isoprenylated and anchored to the inner nuclear envelope, which favors polymerization in two dimensions. By contrast, in the absence of the CaaX box, NE81 cannot be isoprenylated and does not attach properly to the inner nuclear membrane, which causes polymerization to proceed in three dimensions within the nuclear matrix. Depolymerization may be triggered by phosphorylation through CDK1. In the case of endogenous NE81 or GFP-NE81, which may be still anchored to the inner nuclear envelope, this results only in increased two-dimensional mobility, while in case of GFP-NE81ΔCLIM, it causes diffusion of the unanchored protein across the cell through open nuclear pores. This is another parallel to nuclear lamins, whose phosphorylation by CDK1 is a prerequisite for NEBD. At first view, such regulation is unexpected in an organism without NEBD. However, even in these cells, the nuclear envelope needs to be rendered more flexible during karyokinesis, at least in the equatorial plane, to allow tapering of the nuclear envelope between the two newly forming daughter nuclei. Such an increased flexibility could be a consequence of NE81 phosphorylation by CDK1.
FIGURE 11:
Dictyostelium nuclei possess a continuous, low-electron-density layer between the inner nuclear membrane and the chromatin. Ultrathin sections of D. discoideum amoebae. Cells grown in suspension were fixed for 45 min with 1% glutaraldehyde in 50 mM cacodylate (pH 7.2). After being washed, cells were embedded in 2% agar, postfixed with 1% osmium tetroxide in cacodylate buffer for 1h at 4°C, washed with H2O, dehydrated with ethanol—including block staining with 2% uranyl acetate at the 70% ethanol step—and embedded in Epon. The white arrow points at the low-electron-density layer between the inner nuclear membrane and the chromatin. Two examples are shown. Scale bar: 0.5 μm.
Distribution and dynamics of NE81 at the nuclear envelope may influence the mechanical properties not only of the nucleus, but also of the whole cell. In metazoan cells, the nuclear lamina is indirectly connected to all cytosolic cytoskeletal elements through linker of nucleoskeleton and cytoskeleton complexes, which consist of Sun-family proteins in the inner nuclear membrane interacting with so-called Klarsicht, ANC-1, Syne homology (KASH)-domain proteins in the outer nuclear membrane (Stewart-Hutchinson et al., 2008). Owing to these interconnections with the cytoskeleton, the nuclear lamina is thought to serve as a “molecular shock absorber” (Dahl et al., 2004) in mechanically stressed cells. The important role of the nuclear lamina and the SUN/KASH complexes in this respect has recently been proven in embryonic fibroblasts (Stewart-Hutchinson et al., 2008). The increased mechanosensitivity of GFP-NE81ΔCLIM cells is a strong indication that NE81 plays the role of lamins in Dictyostelium and is a major constituent of the putative nuclear lamina. Although the existence of such a nuclear lamina remains to be shown by EM analyses, there are several good arguments for Dictyostelium cells requiring a nuclear lamina. First, we should expect that even unicellular organisms require proteins that serve the function of lamins as a scaffold for proteins regulating chromatin organization, DNA synthesis, transcription, DNA damage response, cell cycle progression, and cell migration (Andres and Gonzalez, 2009). Second, in contrast with protists or fungi, in which no lamin-like proteins have been identified to date, Dictyostelium cells contain no specific structures to enhance mechanical membrane rigidity, such as a pellicula or a chitin-based cell wall, respectively, even though this free-living amoeba must resist mechanical challenges posed by environmental conditions or its rapid, amoeboid cell motility. Although cross-reactivity of lamin-specific antibodies has suggested the existence of lamins in Saccharomyces, Tetrahymena, Physarum, and dinoflagellates, no putative lamin has been identified at the molecular level in these organisms (Melcer et al., 2007). In plants and some unicellular organisms, such as Trypanosoma, other filamentous coiled-coil proteins may functionally replace lamin-like proteins. All these described proteins are also located at the nuclear periphery, but they are clearly unrelated to lamins from an evolutionary point of view (Harder et al., 2000; Rout and Field, 2001; Gindullis et al., 2002; Mans et al., 2004; Picchi et al., 2011).
In Dictyostelium, we have identified a nuclear protein NE81 associated with the inner nuclear envelope representing the first known lamin-like protein in a nonmetazoan organism. Yet we prefer to classify NE81 as an IF-like protein, rather than a real IF protein. Despite the same overall length of the α-helical, coiled-coil region, the strictly conserved pattern of rod and linker domains in the α-helical, coiled-coil region of IF proteins, and the typical signatures of coil 1A and coil 2B are missing in NE81. NE81 resembles lamins not only with regard to protein sequence features and its localization at the nuclear envelope, which is observed even in mammalian cells after heterologous expression, but it also possesses lamin-like functions, such as maintenance of nuclear architecture, chromatin integrity, centrosome/nucleus attachment, and mechanical stabilization of the cell. Since Dictyostelium cells are positioned at the border of multicellularity, further functional characterization of NE81 and its binding partners is of outstanding interest for the evolution of eukaryotic cellular architecture and chromatin function. Furthermore, since the enzymatic machinery for processing of lamin-like proteins appears to be conserved in Dictyostelium, we see a chance that this simple model organism could be useful for investigations of the basic processes in the pathogenesis of laminopathies.
MATERIALS AND METHODS
Vector constructions and expression of recombinant NE81 for immunizations
The GFP-NE81 fusion plasmid pIS191 containing the complete coding sequence of DDB_G0289429 (Schulz et al., 2009b) was used as a starting point for all further constructs. For expression of NE81 in E. coli, the complete coding sequence was cloned into pMALc2 (NEB, Frankfurt, Germany). Protein expression at room temperature and purification by amylose affinity chromatography was performed according to the manufacturer's instructions. The fusion protein was used for custom immunization of two rabbits (BioGenes, Berlin, Germany). Antisera were affinity-purified according to the manufacturer's instructions (GE Healthcare, München, Germany) using NHS-activated Sepharose with coupled MBP-NE81.
To generate the GFP-NE81ΔCLIM vector, pIS191 was used as a template for PCR amplification of the complete NE81 sequence, excluding the C-terminal CLIM motif, using SalI-linker primer TAAATTGTCGACTAATGGATATGTCAAAAAAGAAAAGTAAAC-3′ and BamHI linker primer 5′-GCGCGGATCCTTAATTTGATTTACCAGCTGAAGAAGG-3′. The PCR product was cloned into the N-terminal GFP-fusion vector pIS77 (Schulz et al., 2009a) to yield pAK35 (G418 resistance).
The GFP-NE81ΔCLIM-S122A vector was generated after PCR amplification of two fragments of the NE81 sequence. The following primer combinations were used: fragment1 (385 base pairs): SalI-linker primer (see above), 5′-GTTGAGCTGCTCTATTTGGTGCTAATGGTG-3′; fragment2 (1808 base pairs): 5′-CACAAATAGGTACACCATTAGCACCAAATAG-3′; BamHI-linker primer; the exchanged bases yielding the S122A point mutation are underlined. The amplified fragments were used as templates for an overlap extension PCR using the SalI- and BamHI-linker primers. The PCR product was cloned into the N-terminal GFP-fusion vector pIS77 (Schulz et al., 2009a) to yield pPB14-29.
To generate the GFP-NLS-CP224ΔC, two complementary oligonucleotides were annealed (5′-TAAATTGTCGACAAC GTAAGAGAAAGAGATCAAAAGAGCTCAATTTA-3′, 5′-TAAATTGAGCTCTTTTGATCTCTTTCTCTTA CGTTGTCGACAATTTA-3′) and cloned with SalI and SacI into the GFP-CP224ΔC vector (Hestermann and Gräf, 2004) to yield pPB45-7.
The homologous recombination vector, containing a blasticidin S resistance cassette flanked by genomic fragments of the NE81 gene, was constructed in pLPBLP (Faix et al., 2004). Suitable linker primers were used to amplify genomic Dictyostelium DNA fragments by PCR. The 5′ SaII/HindIII fragment consisted of 820 base pairs of 5′ noncoding sequence starting at base position −852, and the 3′ PstI/BamHI fragment comprised 679 base pairs of coding sequence ending at base position 2353 of the genomic sequence. The resulting knockout plasmid (pAK43) was digested with PvuII prior to transformation into Dictyostelium cells.
For expression in human cells, the AT-rich NE81 sequence was codon-optimized for expression in mammalian cells by gene synthesis (DNA2.0, Menlo Park, CA) and cloned into pEGFP-C2 (BD Biosciences, Heidelberg, Germany).
Microscopy
Light microscopy and image processing of fixed samples were conducted as described recently on a Zeiss CellObserver HS system equipped with PlanApo 1.4/100× objective, an Axiocam MRm Rev. 3 CCD camera, a piezo stage, and the Axiovision 4.7 fast-iterative deconvolution software package (Schulz et al., 2009a). Maximum intensity projections of deconvolved image stacks (focus step size = 0.25 μm) and centrosome/nucleus distances were calculated with Axiovision. The same system was used for wide-field, live-cell imaging using a LCI PlanNeofluar 1.3/63× objective (Schulz et al., 2009a). FRAP experiments were performed on a Zeiss LSM710 laser-scanning confocal microscope equipped with a EC PlanNeofluar 1.3/40× objective as reported recently (Samereier et al., 2010). Live cells were prepared in glass bottom Petri dishes (Fluorodish, WPI, Berlin, Germany) and flattened by agar overlay (Fukui et al., 1987).
Image stacks for confocal live-cell imaging consisted of 5 frames, recorded at a focus step size of 0.8 μm at a rate 5 frames/s. Stacks were recorded every 5 s. A square region of interest covering < 10% of the area of the cell was bleached with the 488-nm Ar laser line in 40 iterations at top speed and maximum laser intensity. Fluorescence intensities during recovery of the moving region of interest were measured with ImageJ within a 4 × 4 pixel square area (0.13 μm2). As a reference for constitutive bleaching during image recording, fluorescence intensities of comparable regions in unbleached cells were measured. Background intensity was set to zero.
EM of isolated nucleus/centrosome complexes was performed as recently published (Schulz et al., 2009a). For immuno-EM, nuclei were fixed with glutaraldehyde, labeled with anti-NE81 and Nanogold-conjugated anti-rabbit Fab′ fragments (Aurion, Wageningen, Netherlands), silver-enhanced, osmicated, and finally embedded in Spurr's resin. Ultrathin sections stained with Uranyl acetate and lead citrate were viewed on a Philips CM100 electron microscope.
Mechanosensitivity measurements
AX2 control cells and GFP-NE81ΔCLIM cells were passed through polycarbonate filters with a diameter of 25 mm and a uniform pore size of 12 μm (Nuclepore; Whatman, Dassel, Germany) using syringes mounted in a Harvard Apparatus, PHD 2000 Infuse/Withdraw pump device (Harvard Apparatus, Holliston, MA). The flow rate was set between 0 and 29 ml/min. Cells in the filtrate were analyzed for survival and counted in an improved Neubauer chamber.
Other methods
Dictyostelium cells (strain AX2) were grown and transformed as described earlier (Gräf et al., 1998, 2000). HeLa cells were grown in low-glucose medium (DMEM with 10% fetal calf serum and 1 mM l-glutamate). The day before transfection, cells were seeded on coverslips coated with poly-l-lysine or collagen. For transient transfection, the polymer-based DNA transfection reagent jetPEI was used according to the manufacturer's instructions (Polyplus Peqlab, Erlangen, Germany). After a minimum of 24 h from transfection, expression of reporter genes was investigated by light microscopy. SDS gel electrophoresis and Western blotting were carried out according to Gräf et al. (1998). Over- and underexpression levels of DdNE81 estimated from immunoblot band intensities and centrosomal GFP fluorescence in microscopic images were evaluated with ImageJ.
Supplementary Material
Acknowledgments
We thank Kirsten Krüger for technical assistance and Alexandra Lepier for critically reading the manuscript. This work was supported by Deutsche Forschungsgemeinschaft grants GR1642/3-1 and GR1642/4-1.
Abbreviations used:
- CDK1
cyclin-dependent kinase 1
- DAPI
4′,6-diamidino-2-phenylindole
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- IF
intermediate filament
- Ig
immunoglobulin
- immuno-EM
immuno-electron microscopy
- KASH
Klarsicht, ANC-1, Syne homology
- NEBD
nuclear envelope breakdown
- NLS
nuclear localization signal
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0595) on November 16, 2011.
REFERENCES
- Andres V, Gonzalez JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187:945–957. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, Ramaekers FC. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Foisner R. Cell cycle dynamics of the nuclear envelope. ScientificWorldJournal. 2003;3:1–20. doi: 10.1100/tsw.2003.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Yumura S, Yumura TK. Agar-overlay immunofluorescence: high resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol. 1987;28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- Gindullis F, Rose A, Patel S, Meier I. Four signature motifs define the first class of structurally related large coiled-coil proteins in plants. BMC Genomics. 2002;3:9. doi: 10.1186/1471-2164-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Fiserova J, Huttenlauch I, Stick R. A new model for nuclear lamina organization. Biochem Soc Trans. 2008;36:1339–1343. doi: 10.1042/BST0361339. [DOI] [PubMed] [Google Scholar]
- Gräf R, Daunderer C, Schliwa M. Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J Cell Sci. 2000;113:1747–1758. doi: 10.1242/jcs.113.10.1747. [DOI] [PubMed] [Google Scholar]
- Gräf R, Euteneuer U, Ueda M, Schliwa M. Isolation of nucleation-competent centrosomes from Dictyostelium discoideum. Eur J Cell Biol. 1998;76:167–175. doi: 10.1016/S0171-9335(98)80031-9. [DOI] [PubMed] [Google Scholar]
- Harder PA, Silverstein RA, Meier I. Conservation of matrix attachment region-binding filament-like protein 1 among higher plants. Plant Physiol. 2000;122:225–234. doi: 10.1104/pp.122.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Strelkov SV. History and phylogeny of intermediate filaments: now in insects. BMC Biol. 2011;9:16. doi: 10.1186/1741-7007-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest. 2009;119:1772–1783. doi: 10.1172/JCI38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hestermann A, Gräf R. The XMAP215-family protein DdCP224 is required for cortical interactions of microtubules. BMC Cell Biol. 2004;5:24. doi: 10.1186/1471-2121-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge, UK: Cambridge University Press; 2001. [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- Melcer S, Gruenbaum Y. Nuclear morphology: when round kernels do the Charleston. Curr Biol. 2006;16:R195–R197. doi: 10.1016/j.cub.2006.02.032. [DOI] [PubMed] [Google Scholar]
- Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res. 2007;313:2157–2166. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Mencarelli C, Ciolfi S, Caroti D, Lupetti P, Dallai R. Isomin: a novel cytoplasmic intermediate filament protein from an arthropod species. BMC Biol. 2011;9:17. doi: 10.1186/1741-7007-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picchi GF, Ferreira AM, Souza FS, Lourenco EE, Arauco PR, Lorusso A, Bordignon J, Krieger MA, Goldenberg S, Fragoso SP. Trypanosoma cruzi: Identification of DNA targets of the nuclear periphery coiled-coil protein TcNUP-1. Exp Parasitol. 2011;127:147–152. doi: 10.1016/j.exppara.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Reinders Y, Schulz I, Gräf R, Sickmann A. Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J Proteome Res. 2006;5:589–598. doi: 10.1021/pr050350q. [DOI] [PubMed] [Google Scholar]
- Rout MP, Field MC. Isolation and characterization of subnuclear compartments from Trypanosoma brucei. Identification of a major repetitive nuclear lamina component. J Biol Chem. 2001;276:38261–38271. doi: 10.1074/jbc.M104024200. [DOI] [PubMed] [Google Scholar]
- Samereier M, Meyer I, Koonce MP, Gräf R. Live cell-imaging techniques for analyses of microtubules in Dictyostelium. Methods Cell Biol. 2010;97:341–357. doi: 10.1016/S0091-679X(10)97018-0. [DOI] [PubMed] [Google Scholar]
- Schulz I, Baumann O, Samereier M, Zoglmeier C, Gräf R. Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur J Cell Biol. 2009a;88:621–638. doi: 10.1016/j.ejcb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Schulz I, Erle A, Gräf R, Kruger A, Lohmeier H, Putzler S, Samereier M, Weidenthaler S. Identification and cell cycle-dependent localization of nine novel, genuine centrosomal components in Dictyostelium discoideum. Cell Motil Cytoskeleton. 2009b;66:915–928. doi: 10.1002/cm.20384. [DOI] [PubMed] [Google Scholar]
- Stevense M, Chubb JR, Muramoto T. Nuclear organization and transcriptional dynamics in Dictyostelium. Dev Growth Differ. 2011;53:576–586. doi: 10.1111/j.1440-169X.2011.01271.x. [DOI] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Schliwa M, Euteneuer U. Unusual centrosome cycle in Dictyostelium: correlation of dynamic behavior and structural changes. Mol Biol Cell. 1999;10:151–160. doi: 10.1091/mbc.10.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehland J, Willingham MC. A rat monoclonal antibody reacting specifically with the tyrosylated form of alpha-tubulinII. Effects on cell movement, organization of microtubules, and intermediate filaments, and arrangement of Golgi elements. J Cell Biol. 1983;97:1476–1490. doi: 10.1083/jcb.97.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, Vogel A, Gassen B, Noegel AA. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.