ABSTRACT
Little is known about the mechanisms of gene targeting within the nucleus and its effect on gene expression, but most studies have concluded that genes located near the nuclear periphery are silenced by heterochromatin. In contrast, we found that early herpes simplex virus (HSV) genome complexes localize near the nuclear lamina and that this localization is associated with reduced heterochromatin on the viral genome and increased viral immediate-early (IE) gene transcription. In this study, we examined the mechanism of this effect and found that input virion transactivator protein, virion protein 16 (VP16), targets sites adjacent to the nuclear lamina and is required for targeting of the HSV genome to the nuclear lamina, exclusion of heterochromatin from viral replication compartments, and reduction of heterochromatin on the viral genome. Because cells infected with the VP16 mutant virus in1814 showed a phenotype similar to that of lamin A/C−/− cells infected with wild-type virus, we hypothesized that the nuclear lamina is required for VP16 activator complex formation. In lamin A/C−/− mouse embryo fibroblasts, VP16 and Oct-1 showed reduced association with the viral IE gene promoters, the levels of VP16 and HCF-1 stably associated with the nucleus were lower than in wild-type cells, and the association of VP16 with HCF-1 was also greatly reduced. These results show that the nuclear lamina is required for stable nuclear localization and formation of the VP16 activator complex and provide evidence for the nuclear lamina being the site of assembly of the VP16 activator complex.
IMPORTANCE
The targeting of chromosomes in the cell nucleus is thought to be important in the regulation of expression of genes on the chromosomes. The major documented effect of intranuclear targeting has been silencing of chromosomes at sites near the nuclear periphery. In this study, we show that targeting of the herpes simplex virus DNA genome to the nuclear periphery promotes formation of transcriptional activator complexes on the viral genome, demonstrating that the nuclear periphery also has sites for activation of transcription. These results highlight the importance of the nuclear lamina, the structure that lines the inner nuclear membrane, in both transcriptional activation and repression. Future studies defining the molecular structures of these two types of nuclear sites should define new levels of gene regulation.
Introduction
Eukaryotic gene transcription is regulated at many levels, including initiation of transcription by RNA polymerase II, binding of repressors and activators, chromatin structure, and on a larger scale, targeting of the genes to specific domains within the nucleus. Much is known about the processes of transcriptional initiation and regulation by activators, repressors, and chromatin, but little is known about the role of intranuclear positioning in the regulation of transcription. The eukaryotic nucleus is organized into structural domains including the nuclear envelope and lamina, nucleoplasm, and nucleolus. Localization of genes and chromosomes to the nuclear periphery has traditionally been associated with gene silencing (reviewed in references 1 and 2). Several lines of evidence support this conclusion. Major sites of heterochromatin are located near the nuclear periphery (3, 4), and gene-poor chromosomal regions are located near the nuclear periphery (5–8). Selection of genomic sequences attached to lamin B by the DamID approach has shown that gene-poor and heterochromatin regions are associated with the nuclear lamina (9). Furthermore, genes move away from the nuclear periphery coincident with activation of transcription (10–13). Active alleles of serum-activated genes are located in the nuclear interior (14), and cDNAs from hematopoietic cells hybridize with the nuclear interior (15). Tethering genes at the nuclear periphery silences many, but not all, genes (16–18). However, some peripheral sites may not be silencing sites, because lamin A/C-rich microdomains have been associated with euchromatin and active genes (19).
In contrast to the bulk of the evidence for peripheral targeting and silencing, we found that lamin A, a major component of the nuclear lamina, was required for targeting of the herpes simplex virus (HSV) genome to the periphery of the nucleus at early times postinfection, which correlated with reduced heterochromatin on viral genes and increased viral immediate-early (IE) gene expression (20). Therefore, in the HSV system, targeting of the viral genome to the nuclear periphery is linked to removal of heterochromatin and activation of IE gene expression. HSV infection of mammalian cells provides a good model system for the study of nuclear compartmentalization, because its genome is found initially in genome complexes and small replication compartments near the nuclear periphery (20–22). HSV gene expression occurs in a cascade, with the expression of IE genes activating early (E) gene expression, which subsequently activates viral DNA replication and late (L) gene expression (23). Viral DNA replication and late gene transcription take place in intranuclear domains called replication compartments (21, 24).
The critical issue of how lamin A/C promotes viral IE gene transcription, the initial process affected in the lamin A/C−/− cells, remained unresolved. HSV IE gene expression is transactivated by the HSV virion protein 16 (VP16), a tegument protein that assembles into a transactivator complex with two host proteins, host cell factor 1 (HCF-1) (25–28) and the octamer binding transcription factor 1 (Oct-1) (29–32). After VP16 loads onto IE gene promoters, it recruits general transcription factors, RNA polymerase II, and chromatin-remodeling enzymes to IE promoters (33–35) through its acidic domain, and HCF-1 recruits the Set1 histone methyl transferase to IE gene promoters for the euchromatic histone H3 lysine 4 (H3K4) methylation modification and the LSD1 demethylase to remove the heterochromatic H3K9 methylation modification (36, 37). As a result, the histones associated with IE gene promoters contain euchromatic histone modifications that correspond to active transcription of IE genes (38, 39), although some of the recruited chromatin-modifying enzymes are not essential for active IE gene expression (40, 41). In this study, we have investigated the mechanism(s) by which the nuclear lamina promotes the transcription and expression of viral IE genes through the targeting of the genome to the nuclear periphery.
RESULTS
Targeting of HSV genomes and early replication complexes to the nuclear lamina requires functional HSV VP16.
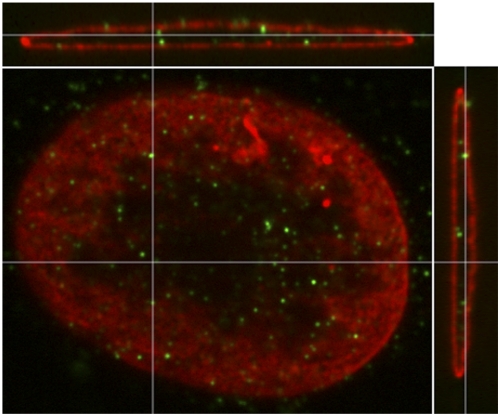
We have shown that in lamin A/C−/− cells, HSV genome complexes fail to target to the nuclear periphery, heterochromatin accumulates on viral DNA, and viral gene expression is reduced (20). These results demonstrated a linkage between targeting of HSV genome complexes to the nuclear lamina and viral gene expression. The first detectable effect on gene expression was on IE gene products (20), whose expression is promoted by the VP16 activator complex. To determine where the initial genome complexes involving VP16 localize in the infected-cell nucleus, we infected murine embryonic fibroblasts (MEFs) with the herpes simplex virus 1 (HSV-1) DG-1, which expresses a VP16-green fluorescent protein (GFP) fusion protein (42), at a multiplicitiy of infection (MOI) of 100 in the presence of cycloheximide throughout infection to restrict the detection of VP16 to input virion protein. At 3 h postinfection (hpi), we fixed the infected cells and immunostained with a GFP-specific antibody to aid detection of the low levels of input VP16 protein and with a lamin B1 antibody to detect the lamina, and the immunostained cells were analyzed by three-dimensional (3D) confocal microscopy. We observed that input VP16-GFP localized to punctate sites near the nuclear periphery at these early times postinfection (Fig. 1). In these cells, the input viral genomes in the nucleus are templates for IE gene transcription; therefore, these results suggested that the VP16 transactivator complex promotes IE gene transcription at the nuclear periphery.
FIG 1 .
Localization of input virion VP16-GFP in infected cells. Lmna+/+ MEFs were infected with HSV-1 DG1 virus at an MOI of 100 in the presence of cycloheximide (100 µg/ml) and fixed at 3 hpi. The cells were stained with antibodies specific for GFP (green) and lamin B1 (red) and imaged in 3D on a confocal microscope. A single xy confocal plane near the top of the nucleus is shown in the center along with xz and yz cross-sectional views adjacent to it. The cross-hairs show the planes of the other images.
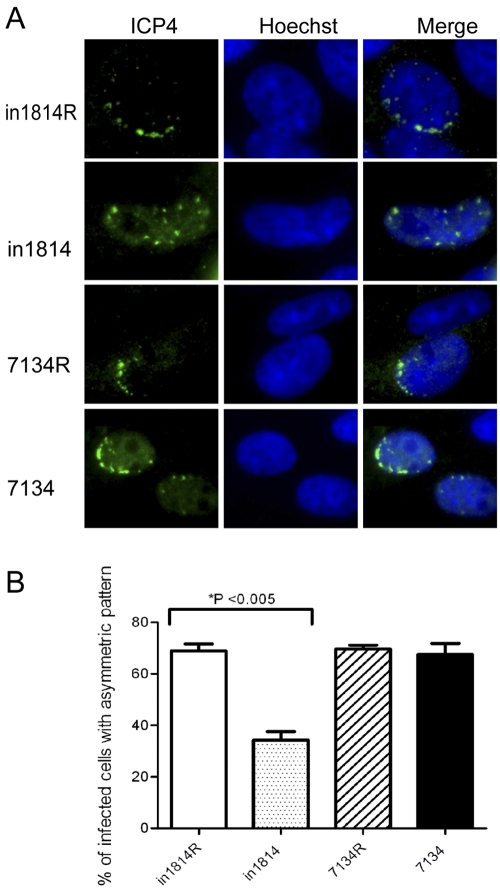
We then tested whether VP16 played a role in genome targeting to the nuclear periphery by studying cells infected with the HSV-1 mutant virus in1814, which carries a gene encoding a mutant VP16 molecule defective for binding to HCF-1 and Oct-1 (43), and its rescued virus in1814R. For a control, we studied the HSV-1 7134 ICP0-null mutant virus, and the 7134R rescued virus (44), because ICP0 also promotes euchromatin on viral lytic gene promoters (45). We first assessed targeting of genome complexes by immunofluorescence examination of Vero cells at the edge of a developing plaque, under which conditions genome complexes are initially localized at the inner edge of the nucleus proximal to the center of the plaque (20, 22, 46). In cultures infected with the in1814R or 7134R rescued virus with a small number of PFU of virus, approximately 70% of the infected cells (n = 100) at the edge of plaques contained small replication compartments along one edge of the nucleus, as detected by ICP4 immunofluorescence (Fig. 2). In contrast, only 35% of cells infected with the VP16 mutant in1814 displayed an asymmetric distribution of replication compartments (P < 0.005) (Fig. 2B). Infection with the 7134 ICP0 gene null mutant virus resulted in 70% of cells displaying an asymmetric distribution of replication compartments, similar to infection with the rescued virus 7134R (Fig. 2B). Similar results were also observed with permissive U2OS cells infected with 7134 virus (results not shown). These results argued that VP16, but not ICP0, plays a role in the targeting and recruitment or the stable association of HSV genome complexes with the nuclear lamina under these experimental conditions.
FIG 2 .
VP16 is required for HSV genome targeting to the nuclear lamina. Vero cells were infected with the in1814 VP16 mutant virus (0.05 PFU/cell), the 7134 ICP0-null virus (0.05 PFU/cell), or the respective rescued viruses (0.005 PFU/cell) so as to obtain approximately 15 to 30 plaques per coverslip. The cells were fixed at 36 hpi and stained with an antibody specific for ICP4 (green) as a marker of genome complexes and Hoechst to stain the nuclei. (A) Images of representative cells. (B) Quantification of distributions of ICP4 genome complexes. Nuclei (n = 100) of cells around plaques that contained small ICP4 foci were scored according to whether the ICP4 foci were distributed along one side of the nucleus (asymmetric) or throughout the nucleus (symmetric). The data shown are mean values plus standard deviations (error bars) from three experiments. Values that are significantly different (P < 0.005) using a paired Student’s t test are indicated.
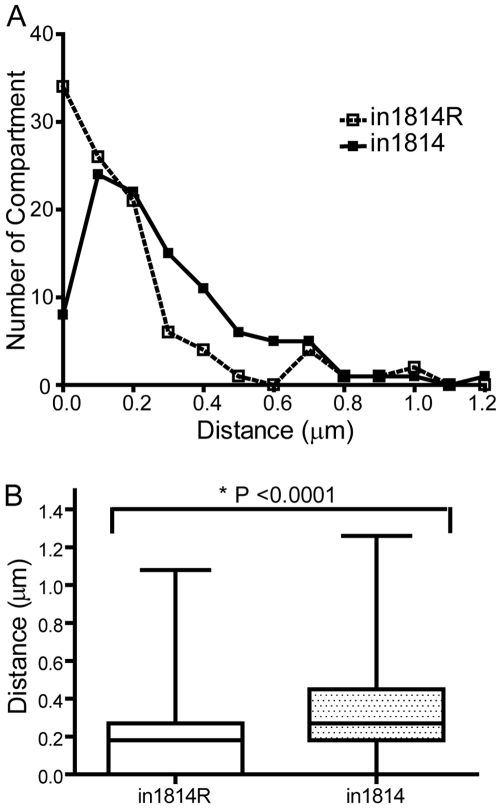
We also analyzed the localization of genome complexes by measuring the distance of small replication compartments from the nuclear lamina at early times postinfection using confocal microscopy. We infected HeLa cells with in1814 or in1814R virus, fixed the cells at 4 hpi, and stained them with antibodies specific for ICP8 and lamin B1 to visualize early replication compartments and the nuclear lamina, respectively. To determine the distance between a replication compartment and the nuclear lamina, we measured the distance in three orthogonal planes (xy, xz, and yz), for individual replication compartments (n = 100). The shortest distance measured from the three orthogonal views was used as the distance between the replication compartment and lamina. In in1814R virus-infected cells, many of the replication compartments were adjacent to the lamina, whereas in in1814 virus-infected cells, fewer compartments were adjacent to the lamina and tended to be located further away from the lamina (Fig. 3A). Overall, replication compartments were significantly closer to the nuclear lamina in in1814R-infected cells (median, 0.18 µm) than in in1814-infected cells (median, 0.27 µm) (P <0.0001) (Fig. 3B). Taken together, the results from the two experimental assays argued for a role for VP16 or its associated components in targeting of genome complexes and replication compartments or association with sites near the nuclear lamina during the early stages of viral infection.
FIG 3 .
Quantification of replication compartment-lamina distance during in1814 and in1814R infection. HeLa cells were infected at an MOI of 1 with either the in1814 VP16 mutant virus or the in1814R rescued virus, fixed at 4 hpi, stained with antibodies to ICP8 and lamin B, and imaged in 3D on a confocal microscope. The distances of individual replication compartments (n = 100) from the lamina were measured in the xy, xz, and yz planes using Slidebook software, and the shortest distance was selected to represent the compartment-lamina distance. (A) Distribution of distances of replication compartments from the lamina. (B) Compartment-lamina distances (µm) plotted as a box-and-whiskers graph. The upper and lower lines of the boxes represent the 75th and 25th percentile values, and the whiskers represent the maximal and minimal values. Values that are significantly different (P < 0.0001) using the nonparametric Mann-Whitney rank sum test are indicated.
Heterochromatin exclusion from HSV-1 replication compartments requires functional VP16.
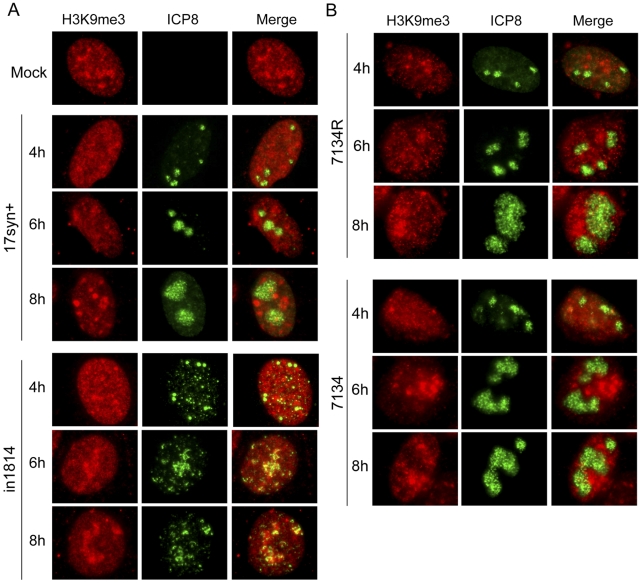
VP16 and its associated host proteins recruit histone-modifying enzymes (37, 47) and promote euchromatin on IE gene promoters (35, 37). We have shown that lamin A/C is required for targeting of the HSV genome to the nuclear periphery and for reduced levels of heterochromatin on the ICP4 IE gene promoter (20). These results are evidence that viral DNA targeted to the nuclear lamina is protected from chromatin silencing. Because VP16 increased the efficiency in targeting of early replication complexes to the nuclear periphery, we tested whether it also played a role in reducing heterochromatin association with the viral genome. We infected HeLa cells with wild-type (WT) 17syn+ virus, the in1814 VP16 mutant virus, or the in1814R rescued virus, fixed the cells at intervals between 4 and 8 hpi, and stained them with antibodies specific for the heterochromatin marker histone H3 lysine 9 trimethyl (H3K9me3) and for the HSV replication compartment marker ICP8. In WT virus-infected cells (17syn+; Fig. 4A) or in in1814R rescued virus-infected cells (not shown), heterochromatin was excluded from replication compartments. In contrast, replication compartments in cells infected with the VP16 mutant in1814 virus appeared smaller and frequently colocalized with the heterochromatin marker (Fig. 4A). These results argued that VP16 is important for heterochromatin exclusion from replication compartments. However, the observed phenotype with the VP16 mutant virus could also be an indirect result of reduced levels of ICP0 due to defective VP16 transactivator function for IE gene expression (45). We tested this possibility by infecting HeLa cells with the HSV-1 7134 ICP0-null mutant virus or the 7134R rescued virus, under the same conditions as those used in Fig. 4A. Cells were fixed at 4 to 8 hpi and stained with antibodies specific for ICP8 and the heterochromatin marker H3K9me3. Mature replication compartments in ICP0-null virus-infected cells excluded heterochromatin as efficiently as cells infected with the rescued virus (Fig. 4B). These results are evidence that under these experimental conditions, VP16 promotes heterochromatin exclusion from replication compartments.
FIG 4 .
Exclusion of heterochromatin from HSV-1 replication compartments requires VP16 but not ICP0. (A) HeLa cells were infected with wild-type 17syn+ virus or the in1814 VP16 mutant virus at an MOI of 1 from 4 to 8 hpi. The cells were then stained with antibodies for the replication compartment marker ICP8 (green) and the heterochromatin marker H3K9me3 (red). (B) HeLa cells were infected with the 7134 ICP0-null mutant virus or the 7134R rescued virus at an MOI of 1 and then stained as described above for panel A.
Heterochromatin exclusion from the HSV-1 IE promoters requires VP16.
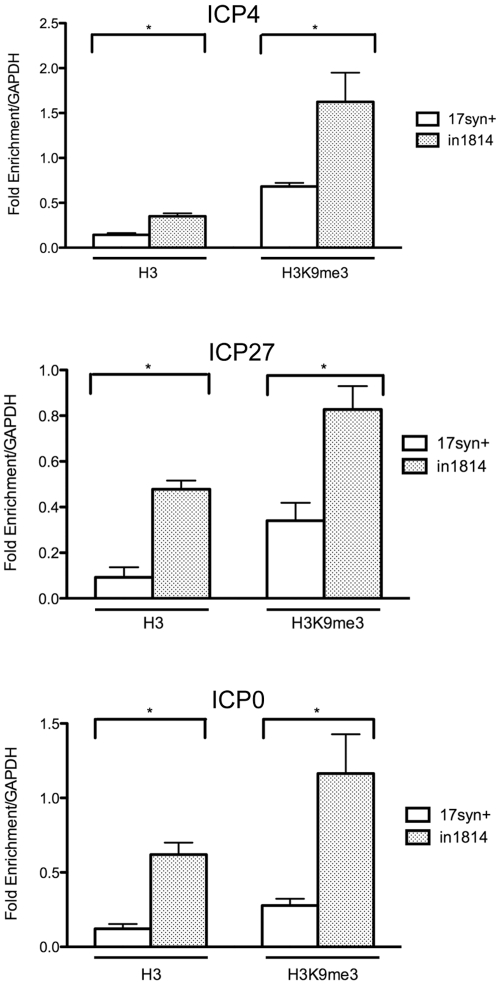
To directly test the association of heterochromatin with viral DNA, we performed chromatin immunoprecipitation (ChIP). We infected HeLa cells with the WT virus, 17syn+, or the VP16 mutant virus, in1814, at an MOI of 1 and prepared chromatin extracts at 4 hpi. Antibodies specific for H3K9me3 and histone H3 were used to immunoprecipitate heterochromatin and total chromatin, respectively. The levels of immunoprecipitated DNA were determined by real-time (RT) PCR analysis using primers for the ICP4 gene transcriptional start site, the ICP27 gene transcription start site, the ICP0 gene promoter, and a cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) pseudogene (Table 1). Consistent with previous results (35), we observed an increased association of histone H3 with the ICP4 gene (2.8-fold), ICP27 gene (5-fold), and ICP0 gene promoters (5-fold) during infection with in1814 virus (Fig. 5). We also observed increased levels of the heterochromatin marker H3K9me3 on the ICP4 (3-fold), ICP27 (2.5-fold), and ICP0 (4.2-fold) promoters in in1814-infected cells as compared with WT virus-infected cells (Fig. 5). To determine the efficiency of H3K9me3 ChIP among each sample set, we used satellite 2 (Sat2) sequences as a positive cellular control for H3K9me3 enrichment, because these satellite sequences are heavily associated with heterochromatin. There was no significant difference in H3K9me3 enrichment on the Sat2 sequences between the WT virus- and VP16 mutant virus-infected cells (results not shown). Therefore, VP16 or its associated components promote the reduction of histone H3 and heterochromatin on HSV IE gene promoters at early times during lytic infection.
TABLE 1 .
Primers used for quantitative PCR analysis
| Gene | Primer directiona | Primer sequence (5’–3’) |
|---|---|---|
| ICP0 promoterb | F | TAACTTATACCCCACGCCTTTC |
| R | TCCGGTATGGTAATGAGTTTC | |
| ICP4 promoterc | F | CGCATGGCATCTCATTACCG |
| R | TAGCATGCGGAACGGAAGC | |
| ICP4 transcriptional start sited | F | GCCGGGGCGCTGCTTGTTCTCC |
| R | CGTCCGCCGTCGCAGCCGTATC | |
| ICP27 transcriptional start sitec | F | GCCACGTGTAGCCTGGATCCC |
| R | CGGGGGTGGATACGCTGGCT | |
| TK promoter | F | CCGGAGGCGCGAGGGACTGC |
| R | CAACGGGCCACGGGGATGAAGC | |
| GAPDHc | F | TTCGACAGTCAGCCGCATCTT |
| R | CAGGCGCCCAATACGACCAAA |
FIG 5 .
Increased association of chromatin at IE gene promoters in the absence of functional VP16. HeLa cells were infected with either the wild-type 17syn+ virus or the VP16 mutant virus in1814 at an MOI of 1 and fixed at 4 hpi. ChIP was performed using antibodies specific for histone H3 or H3K9me3 or as a control, normal rabbit IgG. The immunoprecipitated DNA fragments were quantified by real-time PCR with primers specific for the ICP4, ICP27, and ICP0 genes. Values were normalized to GAPDH to determine fold enrichment. The data shown are mean values plus standard deviations from three experiments. Values that are significantly different (P < 0.05) using a paired Student’s t test are indicated by an asterisk and brackets.
Lamin A/C is required for efficient association of VP16 and Oct-1 transcription factor with IE gene promoters.
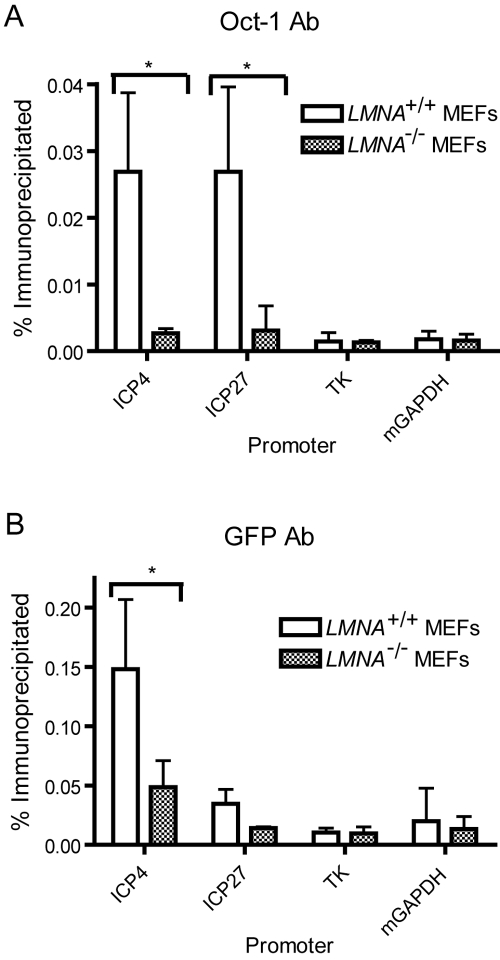
Because infection of lamin A/C−/− cells and infection with the VP16 mutant virus both resulted in defective targeting of HSV genome complexes to the nuclear periphery and increased heterochromatin association with HSV IE gene promoters early in infection, we hypothesized that the two phenotypes might be related. We hypothesized that lamin A/C is required for efficient VP16 activator complex formation, which in turn is required to facilitate IE gene expression. To test this hypothesis, we examined the association of VP16 and Oct-1 with HSV IE gene promoters in Lmna−/− and Lmna+/+ MEFs. We infected cells with the HSV-1 DG1 virus and prepared chromatin extracts at 2 hpi for ChIP analysis using antibodies specific for GFP or Oct-1. Real-time PCR was performed with primers specific for the promoters of the IE ICP4 and ICP27 genes, to which the Oct-1/HCF-1/VP16 complex is known to bind (29). Primers specific for the E thymidine kinase (TK) gene were used as a negative control, because E genes lack VP16 binding sites. We observed 10-fold and 8.7-fold increases in Oct-1 association with the ICP4 (P < 0.05) and ICP27 (P < 0.05) promoters, respectively, in Lmna+/+ MEFs compared to Lmna−/− MEFs (Fig. 6A). Furthermore, we observed 3-fold and 2.4-fold increases in VP16-GFP association with the ICP4 (P < 0.05) and ICP27 (P = 0.12) promoters, respectively, in Lmna+/+ MEFs compared to Lmna−/− MEFs (Fig. 6B). There was minimal enrichment of Oct-1 or VP16-GFP at the TK and GAPDH promoters in both cell lines. Therefore, in the absence of lamin A/C, the VP16 activator complex was not efficiently assembled at IE gene promoters.
FIG 6 .
Decreased association of VP16-GFP and Oct-1 at IE promoters in the absence of lamin A/C. Lmna+/+ and Lmna−/− MEFs were infected with HSV-1 DG1 virus at an MOI of 10 and fixed at 2 hpi. Cell lysates were analyzed by ChIP with antibodies (Ab) specific for Oct-1 (A) or GFP (B). The levels of total and immunoprecipitated DNA were quantified by real-time PCR with primers specific for the IE ICP4 and ICP27 gene promoters. The viral TK and cellular GAPDH promoters were used as controls. The results shown are mean values plus standard deviations from three independent experiments. Values that are significantly different (P < 0.05) using a paired Student’s t test are indicated by an asterisk and brackets. mGAPDH, mouse GAPDH.
Lamin A/C is required for stable nuclear localization of HCF-1 and VP16 and their association into a complex.
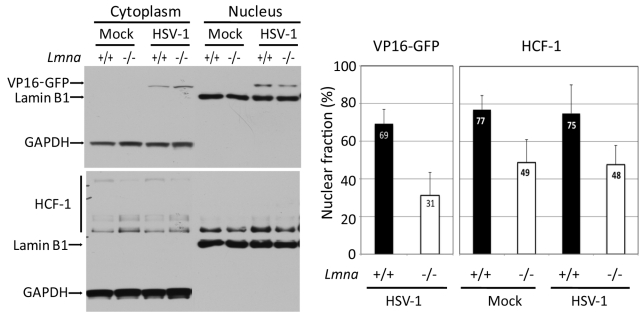
The reduced levels of VP16 associated with viral DNA in lamin A/C−/− cells could be due to reduced levels of VP16 in the nucleus, defective activator complex assembly, or reduced ability of the complex to bind to viral DNA. To determine whether lamin A/C regulated the nuclear localization of VP16, we performed subcellular fractionation of Lmna+/+ and Lmna−/− MEFs. We infected the two types of cells with the HSV-1 DG1 virus at an MOI of 50 in the presence of cycloheximide to analyze input VP16. Mock-infected cells were used as a control. We harvested the cells at 3 hpi and prepared cytoplasmic and nuclear fractions. VP16-GFP and HCF-1 proteins were detected by Western blotting (Fig. 7, left panel). In the Lmna+/+ MEFs, 69% of the VP16-GFP was in the nuclear fraction, while only 31% was associated with the nuclear fraction in Lmna−/− MEFs (Fig. 7, right) (P < 0.05 using the Wilcoxon signed-rank test). HCF-1 also showed reduced association with the nucleus with 77% in the Lmna+/+ MEF nuclear fraction compared to 49% observed in nuclear fractions of mock-infected Lmna−/− MEFs (P < 0.05). HSV-1 infection did not affect the distributions of HCF-1. These results demonstrated that lamin A/C is required for the nuclear localization of HCF-1 and VP16 and/or their stable association with the nucleus.
FIG 7 .
Decreased nuclear association of VP16-GFP and HCF-1 in the absence of lamin A/C. (Left) Lmna+/+ and Lmna−/− MEFs were infected with the HSV-1 DG1 virus at an MOI of 50 in the presence of cycloheximide (100 µg/ml), harvested at 3 hpi, and fractionated into cytoplasmic and nuclear fractions. Cytoplasmic and nuclear fractions were loaded at a 1:2 ratio onto an SDS-polyacrylamide gel, and the proteins were resolved in the gel. VP16-GFP and HCF-1 were detected by Western blotting with GFP- and HCF-1-specific antibodies, respectively. GAPDH and lamin B1 were detected as fractionation and loading controls (left). (Right) The percentages of VP16-GFP and HCF-1 in the nuclear fractions were quantified using ImageJ software. Histograms represent the mean values and standard deviations from at least five independent experiments.
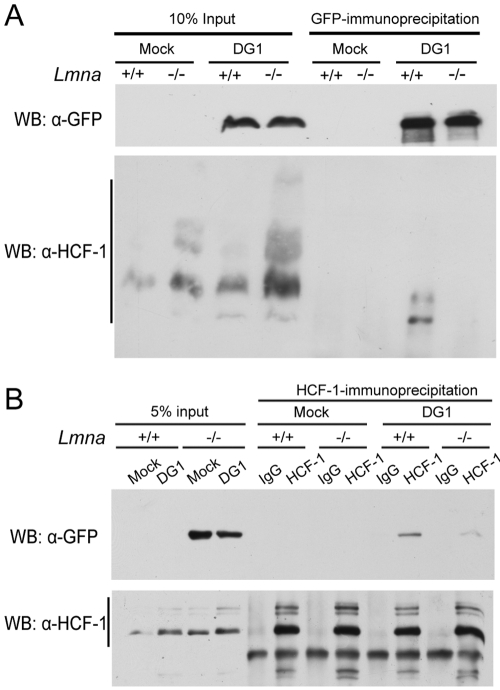
To determine whether VP16 activation complex assembly was impaired in the absence of lamin A/C, we analyzed VP16–HCF-1 association by immunoprecipitation from Lmna+/+ and Lmna−/− MEFs. To examine input VP16, we infected cells with HSV-1 DG1 virus at an MOI of 100 and harvested the cells at 2 hpi. VP16 was derived completely from input virus under these conditions (results not shown). Because of the differences in cell localization described above, we used total cell lysates for immunoprecipitation. Immunoprecipitation of VP16-GFP from Lmna+/+ cell lysates using a GFP antibody resulted in coimmunoprecipitation with HCF-1 (Fig. 8A). In contrast, HCF-1 did not efficiently coimmunoprecipitate with VP16-GFP from Lmna−/− cell lysates. Similar amounts of input and immunoprecipitated VP16-GFP in both cell lines suggested that these results were not due to differences in VP16-GFP levels (Fig. 8A). HCF-1 did not detectably coimmunoprecipitate from the mock-infected cells or IgG control samples (not shown), indicating that the coimmunoprecipitation was specific for VP16-GFP. To further confirm these coimmunoprecipitation results, we performed immunoprecipitations with an HCF-1 antibody. The HCF-1 antibody immunoprecipitated similar amounts of HCF-1 from both Lmna+/+ and Lmna−/− cell total cell lysates, and infection did not affect these levels (Fig. 8B). As observed with the GFP antibody, there was significantly less VP16-GFP coimmunprecipitated from infected Lmna−/− cell lysates than from infected Lmna+/+ cell lysates (Fig. 8B). These results provided evidence that in the absence of lamin A/C, there was reduced association of VP16-GFP with HCF-1.
FIG 8 .
Reduced association of HCF-1 and VP16-GFP in the absence of lamin A/C. Lmna+/+ and Lmna−/− MEFs were infected with HSV-1 DG1 virus at an MOI of 100 and harvested at 2 hpi for immunoprecipitation with antibodies specific for GFP (A) or HCF-1 (B). Rabbit IgG was used as a control. Immunoprecipitated proteins were resolved by SDS-PAGE, and VP16-GFP and HCF-1 were detected by GFP- and HCF-1-specific antibodies, respectively. WB, Western blotting; α-GFP, anti-GFP antibody; α-HCF-1, anti-HCF-1 antibody.
DISCUSSION
We had shown previously that lamin A is needed for targeting of HSV genome complexes to the periphery of the nucleus, reduction of heterochromatin association with IE gene promoters, and stimulation of viral IE gene expression (20). Those results contrasted with the dogma that peripheral nuclear targeting silences genes through heterochromatin association or targeting of genes or chromosomes to heterochromatin domains. In this study, we found that the mechanism for our observed effects is that lamin A/C promotes the stable accumulation or retention of VP16 and HCF-1 with the nucleus and the assembly of the VP16 activator complex on IE genes, thereby promoting viral IE gene transcription. Because input virion VP16 targeted sites adjacent to the nuclear lamina, we hypothesize that the VP16 activator complex is assembled and associates with viral IE gene promoters at sites adjacent to or connected with the nuclear lamina. These results point out the potential role of specific sites on the nuclear lamina for assembly of transcriptional activator complexes, while other sites serve as sites for assembly of silencing complexes.
Role of the nuclear lamina in assembly of the transactivation complex.
We observed that, in the absence of lamin A/C, VP16 and HCF-1 associated less well with the nucleus and formed activator complexes less efficiently. The reduction in nuclear association was less than the reduction in activator complex formation, so we infer that the localization defect may contribute to but does not fully explain the defect in assembly of the activator complex. The primary defect in assembly of the activator complex on IE genes may be the association of Oct-1 with IE promoters, because VP16 and HCF-1 association is dependent on Oct-1 binding (48).
The reduced association of HCF-1 and VP16 in the nuclei of lamin A/C−/− MEFs argues that their nuclear association is at least in part due to tethering to molecules associated with the nuclear lamina. Alternatively, the nuclear lamina may play a role in regulating the nuclear export of HCF-1 by the hematopoietic PBX-interacting protein (HPIP) cellular export factor (49). Although there is no evidence for HCF-1 or VP16 interactions with nuclear lamina components, another member of the activator complex, Oct-1, has been shown to colocalize with lamin B (50, 51). However, in these systems, Oct-1 associated with lamin B seems to be sequestered so that it cannot bind to DNA and activate gene expression. Thus, in lamin B knockout cells, Oct-1 is released from the nuclear lamina and moves inward and is free to bind to cognate sequences and activate specific promoters. In the case of HSV, VP16 and HCF-1 may bind to Oct-1 associated with the lamina and free it to bind to other sites nearby where the complex can bind to viral IE gene promoters. It is interesting to note that HCF-1 is found in the cytoplasm of sensory neurons, thought to result from retention at the Golgi apparatus (52, 53).
The requirement for the lamina in assembly of VP16 activator complexes and the localization of input virion VP16 to the sites on the nuclear lamina further support the idea that the activator complex is assembled at sites on the nuclear lamina. An interesting question is whether the activator complex or viral DNA is primarily targeted to the nuclear lamina sites. A corollary question is whether viral DNA that lacks Oct-1 binding sites will target the nuclear lamina without VP16 activator complex bound to it. It is conceivable that all incoming DNA such as transfected or viral DNA is shunted to the nuclear lamina where it is normally silenced by association with heterochromatin. HSV DNA may bypass this by association with the VP16 activation complex bound to alternative sites on the nuclear lamina.
Implications for genome targeting.
In contrast to most cellular genes at the nuclear periphery that are associated with heterochromatin and transcriptionally inactive, we have found that the targeting of the input HSV genomes to the nuclear periphery, an association that requires lamin A/C and the viral protein VP16, is linked to transcriptional activation of the viral IE genes and reduction in heterochromatin association with the viral genome. Our results argue that lamina subdomains can either silence genes through heterochromatin association or serve to activate gene transcription through euchromatin association. This was first raised as a general concept by Shimi et al. (19) who reported microdomains enriched for lamin A or lamin B that were associated with actively transcribing genes or inactive genes, respectively. Our work provides specific examples of genes that are activated by association with the nuclear periphery, likely in association with molecules associated with the nuclear lamina. It will be important to define the molecules associated with the nuclear lamina and with which the VP16 activator complex or viral genome interact that facilitate assembly of the complex on viral IE promoters. Further studies are needed to confirm that viral DNA is located in the sites to which input VP16 is localized and to identify the cellular factors and gene products needed for targeting of VP16 and viral DNA to these sites.
It is interesting to note that some of the gene constructs that resisted silencing when tethered to the nuclear lamina were based on the human cytomegalovirus immediate-early enhancer promoter (18). Therefore, it is conceivable that viral immediate-early promoters have evolved to evade chromatin-silencing mechanisms at the periphery of the nucleus when the viral genomes invade the nucleus.
Implications for viral latent infection.
These results may also have implications for the mechanisms by which HSV undergoes a latent infection in neurons. Sensory neurons may be deficient for a component(s) associated with the nuclear lamina, which is needed for assembly of the VP16 activator complex. It is known that HCF-1 is localized in the cytoplasm in sensory neurons, and there is evidence that this is due to retention at the Golgi apparatus (26). Given our results, it is conceivable that a component of the nuclear lamina that normally tethers HCF-1 in the nucleus is missing in sensory neurons or that the nuclear lamina regulates the export of HCF-1 from the nucleus by the HPIP protein (49). Our initial results indicate that murine trigeminal ganglia that are latently infected and express the viral latency-associated transcript (LAT) do contain lamin A (L. Chang and D. M. Knipe, unpublished data) , but other components might be missing or altered in the neurons. Once the molecules that bridge the viral genome complexes to the nuclear periphery are identified, these would be obvious candidates for molecules that might be altered or missing in the nuclei of sensory neurons.
Our results document the importance of intranuclear nuclear targeting of the HSV genome during lytic infection, and this may extend to latent infection as well. These results raise a potential new model for HSV lytic versus latent infection mechanisms in which intranuclear targeting of the viral genome determines the fate of the viral genome. VP16 and its associated proteins target the incoming viral genome to sites on the nuclear lamina where the activator complex can form on the viral DNA, euchromatin is assembled, and IE gene transcription can ensue. In contrast, in neurons, VP16 and HCF-1 are not available to target the viral genome to these sites, but other factors such as CCCTC binding factor (CTCF) which are known to affect genome targeting in the nucleus (54), could target the viral genome to sites on the nuclear lamina or elsewhere in the nucleus where heterochromatin silencing take place, resulting in a latent infection. In addition, this may integrate with transcription of LAT that may target the viral genome to Polycomb bodies in the nucleus where heterochromatin is assembled on viral lytic genes (55–57).
Viruses have often provided very sensitive probes of the mechanisms by which their host cells function. These studies of HSV infection showing the importance of intranuclear targeting of the viral genome for initiation of viral gene transcription raise the concept of gene regulation mechanisms that involve the targeting of genes to specific sites in the nucleus where preformed protein complexes are located or where new complexes are formed on the genes that either activate or silence the genes. This concept differs from the view that genes and binding factors diffuse in solution and find each other randomly. These studies of the HSV genome provide insight into this little-studied area of gene regulation in mammalian cells and highlight the need for further investigation of the role of intranuclear targeting of other specific viral and cellular genes.
MATERIALS AND METHODS
Cells and viruses.
HeLa, Vero, and U2OS cells were obtained from the American Type Culture Collection (Manassas, VA). Immortalized Lmna−/− murine embryonic fibroblasts (MEFs) and litter-matched Lmna+/+ control MEFs (58) were provided by Brian Kennedy, Buck Institute for Research on Aging. Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco) supplemented with 5% fetal bovine serum (FBS) plus 5% bovine calf serum (BCS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. The wild-type strain of HSV-1 (17syn+) (59) used in this study was grown and titrated on Vero cells. The HSV-1 in1814 VP16 insertion mutant and in1814R rescued viruses (43) were provided by Chris M. Preston and grown and titrated on U2OS cells. The VP16-GFP-tagged HSV-1 DG1 virus (42) was grown and titrated on Vero cells. HSV-1 KOS 7134 virus, which has a lacZ expression cassette in place of the ICP0 gene and the 7134R rescued viruses (44) were originally provided by Priscilla Schaffer, and the titers of the viruses were determined on U2OS cells.
Virus infections.
HeLa cells, Lmna+/+ MEFs, or Lmna−/− MEFs were seeded 24 h prior to infection. Virus was diluted in phosphate-buffered saline (PBS) containing 0.1% glucose (wt/vol) and 1% heat-inactivated calf serum and applied to cells for 1 h at 37°C. One hour after the addition of viral inoculum, the cells were washed three times for 30 s each time with an acid wash buffer (135 mM NaCl, 10 mM KCl, 40 mM citric acid [pH 3]) and then washed with DMEM before incubation in DMEM with 1% FBS at 37°C for the indicated time period.
Immunofluorescence microscopy.
HeLa cells, Lmna+/+ MEFs, or Lmna−/− MEFs were seeded at 1 × 105 cells/well on glass coverslips in 24-well plates 24 h prior to infection. The cells were processed for indirect immunofluorescence as described previously (60). The primary antibodies used were histone H3K9me3 (Abcam), HSV-1 ICP4 58S (N. DeLuca), GFP (Clontech), lamin B1 (Abcam), or HSV-1 ICP8 mouse monoclonal 39S (61). Secondary antibodies conjugated to Alexa Fluor 594 and 488 dyes and Prolong gold antifade mounting reagent were obtained from Molecular Probes Inc.
Wide-field images of cells were acquired on a Zeiss Axioplan 2 microscope with a Plan Apochromat 63× 1.4-numerical-aperture (1.4-NA) objective lens, a Photometrics CoolSNAP HQ2 charge-coupled device (CCD) camera, and the Zeiss AxioVision 4 image acquisition software. Three-dimensional confocal images of cells were captured on a spinning disk confocal imaging system consisting of a Zeiss Axiovert 200M microscope, a Plan Apochromat 63× 1.4-NA objective lens, a PerkinElmer Yokogawa spinning disk confocal head, a Roper Scientific, Cascade electron microscope (EM)-CCD camera, and SlideBook 4.2 image acquisition and analysis software (Intelligent Imaging Innovations).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (20). Briefly, HeLa cells were seeded at 3 × 106 cells per 100-mm dish 24 h prior to infection. Cells were infected at an MOI of 1 with the WT parental strain 17syn+ or the VP16 mutant virus in1814. Chromatin samples were incubated with 1.5 µg of anti-histone H3 IgG (Abcam), 0.9 µg anti-histone H3K9me3 IgG (Abcam), or equivalent amounts of rabbit immunoglobulin G (Millipore) as the negative control. ChIP assays for Oct-1 and VP16-GFP were performed by the method of Malhas et al. (51) using antibodies from Santa Cruz Biotechnology and Abcam, respectively.
Real-time PCR.
Real-time PCR was performed using the Power SYBR Green PCR master mix and a Prism 7300 sequence detection system (Applied Biosystems) as previously described (20). The primers used in this study are shown in Table 1. The percent immunoprecipitated values were determined by subtracting the normal rabbit IgG control values from the enriched antibody immunoprecipitation (IP) values and dividing by the input DNA. The fold enrichment of viral DNA immunoprecipitated compared to the input sample was normalized to the fraction of cellular GAPDH DNA precipitated in the same reaction.
IP.
Lmna +/+ and Lmna−/− MEFs were seeded at ~1 × 107 cells in 150-mm dishes 24 h prior to infection. MEFs were infected at an MOI of 100 with the HSV-1 DG1 virus. At 2 hpi, the cells were washed twice with cold PBS on ice. The cells were resuspended in 0.5 ml of IP buffer (120 mM potassium acetate, 20 mM Tris acetate [pH 7.9], 5 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.1% Nonidet P-40, 1 mM leupeptin, 1 mM aprotinin, and 1 Complete protease inhibitor cocktail tablet [Roche]) as previously described (62). Lysates were incubated with 1.5 µl of the GFP rabbit polyclonal antibody (Abcam), the HCF-1 rabbit polyclonal antibody (Bethyl Lab) or normal rabbit IgG (Millipore). Beads containing immune complexes were boiled in Laemmli buffer, and proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cell fractionation.
Lmna+/+ and Lmna−/− MEFs were infected with the HSV-1 DG1 virus at an MOI of 50 in the presence of cycloheximide (100 µg/ml) and fixed at 3 hpi. Cell fractionation was performed as described previously (63). Briefly, the cells were washed with PBS twice, swelled in reticulocyte standard buffer (RSB) (10 mM Tris hydrochloride [pH 7.6], 10 mM NaCl, 1.5 mM MgCl2) for 5 min on ice. The cells were disrupted with 40 to 50 strokes with a Dounce homogenizer, and 0.2 volume of 60% sucrose in RSB was added. The cytoplasmic fraction was transferred to a new tube after centrifugation at 3,000 rpm for 5 min. The crude nuclear fraction pellet was washed with 1 ml of RSB containing 10% sucrose and 0.5% NP-40. The nuclear fraction pellet was recovered by centrifugation. The pellet was lysed in standard radioimmunoprecipitation assay (RIPA) buffer. Protease inhibitor (Roche) was added to all the buffers during the fractionation.
SDS-PAGE and Western blotting.
Proteins in the IPs were resolved in 4 to 12% polyacrylamide gradient gels (Invitrogen) and then transferred to a polyvinylidene difluoride membrane (PerkinElmer Life Sciences). The anti-HCF-1 (Bethyl Lab) and anti-GFP (Clontech) antibodies were used at 1:10,000 dilutions.
ACKNOWLEDGMENTS
We thank Thomas Kristie for providing the HCF-1 antibody reagent and Chris Preston for providing the VP16 mutant virus in1814 and in1814R rescued virus. We also thank Anna Cliffe for assistance with the chromatin immunoprecipitation assays.
This work was funded by National Institutes of Health (NIH) grant AI 063106 to D.M.K. We also thank the New England Regional Center of Excellence for Biodefense and Emerging Infectious Disease imaging facility at the Immune Disease Institute, which was funded by NIH grant AI057159 for use of the spinning disk confocal microscope.
Footnotes
Citation Silva L, et al. 2012. Roles of the nuclear lamina in stable nuclear association and assembly of a herpesviral transactivator complex on viral immediate-early genes. mBio 3(1):e00300-11. doi:10.1128/mBio.00300-11.
REFERENCES
- 1. Deniaud E, Bickmore WA. 2009. Transcription and the nuclear periphery: edge of darkness? Curr. Opin. Genet. Dev. 19:187–191 [DOI] [PubMed] [Google Scholar]
- 2. Meister P, Mango SE, Gasser SM. 2011. Locking the genome: nuclear organization and cell fate. Curr. Opin. Genet. Dev. 21:167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilchrist S, Gilbert N, Perry P, Bickmore WA. 2004. Nuclear organization of centromeric domains is not perturbed by inhibition of histone deacetylases. Chromosome Res. 12:505–516 [DOI] [PubMed] [Google Scholar]
- 4. Weierich C, et al. 2003. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 11:485–502 [DOI] [PubMed] [Google Scholar]
- 5. Boyle S, et al. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219 [DOI] [PubMed] [Google Scholar]
- 6. Croft JA, et al. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer R, et al. 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 6:44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolzer A, et al. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3:e157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guelen L, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951 [DOI] [PubMed] [Google Scholar]
- 10. Pickersgill H, et al. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38:1005–1014 [DOI] [PubMed] [Google Scholar]
- 11. Kosak ST, et al. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296:158–162 [DOI] [PubMed] [Google Scholar]
- 12. Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. 2004. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur. J. Immunol. 34:3604–3613 [DOI] [PubMed] [Google Scholar]
- 13. Williams RR, et al. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119:132–140 [DOI] [PubMed] [Google Scholar]
- 14. Levsky JM, et al. 2007. The spatial order of transcription in mammalian cells. J. Cell. Biochem. 102:609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kosak ST, et al. 2007. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 5:e309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finlan LE, et al. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4:e1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452:243–247 [DOI] [PubMed] [Google Scholar]
- 18. Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180:51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shimi T, et al. 2008. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22:3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silva L, Cliffe A, Chang L, Knipe DM. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 4:e1000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Bruyn Kops A, Knipe DM. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857–868 [DOI] [PubMed] [Google Scholar]
- 22. Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex viruses, p 2501–2602 In Knipe DM, Howley PM, Fields virology, 7th ed Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 24. Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868 [DOI] [PubMed] [Google Scholar]
- 25. Gerster T, Roeder RG. 1988. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc. Natl. Acad. Sci. U. S. A. 85:6347–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kristie TM, LeBowitz JH, Sharp PA. 1989. The octamer-binding proteins form multi-protein—DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 8:4229–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilson AC, LaMarco K, Peterson MG, Herr W. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115–125 [DOI] [PubMed] [Google Scholar]
- 28. Xiao P, Capone JP. 1990. A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein-DNA complex assembly with Oct-1. Mol. Cell. Biol. 10:4974–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kristie TM, Roizman B. 1987. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc. Natl. Acad. Sci. U. S. A. 84:71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McKnight JL, Kristie TM, Roizman B. 1987. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1 infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. U. S. A. 84:7061–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Hare P, Goding CR. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52:435–445 [DOI] [PubMed] [Google Scholar]
- 32. Preston CM, Frame MC, Campbell ME. 1988. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell 52:425–434 [DOI] [PubMed] [Google Scholar]
- 33. Neely KE, et al. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649–655 [DOI] [PubMed] [Google Scholar]
- 34. Utley RT, et al. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498–502 [DOI] [PubMed] [Google Scholar]
- 35. Herrera FJ, Triezenberg SJ. 2004. Vp16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 78:9689–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15:1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kent JR, et al. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78:10178–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang J, et al. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 80:5740–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kutluay SB, DeVos SL, Klomp JE, Triezenberg SJ. 2009. Transcriptional coactivators are not required for herpes simplex virus type 1 immediate-early gene expression in vitro. J. Virol. 83:3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kutluay SB, Triezenberg SJ. 2009. Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection. J. Virol. 83:5835–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ottosen S, et al. 2006. Phosphorylation of the VP16 transcriptional activator protein during herpes simplex virus infection and mutational analysis of putative phosphorylation sites. Virology 345:468–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ace CI, McKee TA, Ryan JM, Cameron JM, Preston CM. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cai WZ, Schaffer PA. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor TJ. 2002. Intranuclear localization of the herpes simplex virus ICP8 protein. Dissertation Harvard University, Cambridge, MA [Google Scholar]
- 47. Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kristie TM, Sharp PA. 1990. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 4:2383–2396 [DOI] [PubMed] [Google Scholar]
- 49. Mahajan SS, Little MM, Vazquez R, Wilson AC. 2002. Interaction of HCF-1 with a cellular nuclear export factor. J. Biol. Chem. 277:44292–44299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Imai S, et al. 1997. Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol. Biol. Cell 8:2407–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malhas AN, Lee CF, Vaux DJ. 2009. Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol. 184:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kristie TM, Vogel JL, Sears AE. 1999. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl. Acad. Sci. U. S. A. 96:1229–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kolb G, Kristie TM. 2008. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J. Virol. 82:9555–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13:291–298 [DOI] [PubMed] [Google Scholar]
- 55. Wang QY, et al. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83:8182–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83:8173–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sullivan T, et al. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown SM, Ritchie DA, Subak-Sharpe JH. 1973. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18:329–346 [DOI] [PubMed] [Google Scholar]
- 60. Simpson-Holley M, Baines J, Roller R, Knipe DM. 2004. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Showalter SD, Zweig M, Hampar B. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Knipe DM, Spang AE. 1982. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J. Virol. 43:314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]