Abstract
Background
Rhamnolipids are potent biosurfactants with high potential for industrial applications. However, rhamnolipids are currently produced with the opportunistic pathogen Pseudomonas aeruginosa during growth on hydrophobic substrates such as plant oils. The heterologous production of rhamnolipids entails two essential advantages: Disconnecting the rhamnolipid biosynthesis from the complex quorum sensing regulation and the opportunity of avoiding pathogenic production strains, in particular P. aeruginosa. In addition, separation of rhamnolipids from fatty acids is difficult and hence costly.
Results
Here, the metabolic engineering of a rhamnolipid producing Pseudomonas putida KT2440, a strain certified as safety strain using glucose as carbon source to avoid cumbersome product purification, is reported. Notably, P. putida KT2440 features almost no changes in growth rate and lag-phase in the presence of high concentrations of rhamnolipids (> 90 g/L) in contrast to the industrially important bacteria Bacillus subtilis, Corynebacterium glutamicum, and Escherichia coli. P. putida KT2440 expressing the rhlAB-genes from P. aeruginosa PAO1 produces mono-rhamnolipids of P. aeruginosa PAO1 type (mainly C10:C10). The metabolic network was optimized in silico for rhamnolipid synthesis from glucose. In addition, a first genetic optimization, the removal of polyhydroxyalkanoate formation as competing pathway, was implemented. The final strain had production rates in the range of P. aeruginosa PAO1 at yields of about 0.15 g/gglucose corresponding to 32% of the theoretical optimum. What's more, rhamnolipid production was independent from biomass formation, a trait that can be exploited for high rhamnolipid production without high biomass formation.
Conclusions
A functional alternative to the pathogenic rhamnolipid producer P. aeruginosa was constructed and characterized. P. putida KT24C1 pVLT31_rhlAB featured the highest yield and titer reported from heterologous rhamnolipid producers with glucose as carbon source. Notably, rhamnolipid production was uncoupled from biomass formation, which allows optimal distribution of resources towards rhamnolipid synthesis. The results are discussed in the context of rational strain engineering by using the concepts of synthetic biology like chassis cells and orthogonality, thereby avoiding the complex regulatory programs of rhamnolipid production existing in the natural producer P. aeruginosa.
Keywords: flux analysis, quantitative physiology, metabolic network, biodetergent, non-pathogenic Pseudomonas, biosurfactants, rhamnolipids, off-gas analysis, 13C labeling, BlueSens
Background
Rhamnolipids are biosurfactants with high industrial potential. The possible applications are manifold, as reviewed by Lang and Wullbrandt [1] and Maier and Soberón-Chávez [2]. For example, rhamnolipids can be used as detergents in washing agents due to their tensio-active properties. They can also be used as emulsifiers in the cosmetic and food industry [3,4]. Particularly, they present a good alternative to chemically synthesized detergents, because they are biodegradable, show novel properties like antimicrobial effects [5-7], and are produced from renewable resources.
Rhamnolipids are mainly produced by bacteria of the genus Pseudomonas; with most reports focusing on strains of the opportunistic pathogen P. aeruginosa. Rhamnolipids are composed of one or two hydrophobic β-hydroxy fatty acids, which are linked through a β-glycosidic bond to one or two rhamnose molecules [8,9] forming the hydrophilic moiety. According to the number of rhamnose moieties, mono- and di-rhamnolipids are differentiated. The fatty acids alkyl chain length in P. aeruginosa can vary from C8 to C14 [10]; the most abundant species contains two β-hydroxy fatty acids with C10 chains [8]. The alkyl chains can also contain up to two unsaturated C-C bonds [5].
The rhamnose moiety is synthesized from glucose-6-phosphat in five consecutive enzymatic steps [11]. The β-hydroxy fatty acids in turn originate from fatty acid de novo synthesis [12]. Both metabolic pathways exist in many Gram-negative bacteria; besides de novo fatty acid synthesis, the rhamnose synthesis pathway provides building blocks for lipopolysaccharides of the outer membrane of these organisms [13,14].
The synthesis pathway of rhamnolipids consists of three dedicated enzymatic reactions. In the first step two activated β-hydroxy fatty acids are linked by RhlA, the 3-hydroxyacyl-ACP (acyl carrier protein):3-hydroxyacyl-ACP O-3-hydroxyacyltransferase, to a dimer, called 3-(3-hydroxyalkanoyloxy)alkanoate (HAA) [12,15]. Mono-rhamnolipids are produced by RhlB, the rhamnosyltransferase I, by the condensation of HAA and dTDP-L-rhamnose [16,17]. The rhamnosyltransferase II (RhlC) adds another rhamnose moiety to the mono-rhamnolipid resulting in a di-rhamnolipid [17] (Figure 1). Notably, RhlG, a β-hydroxyacyl-ACP:CoA (coenzyme A) transacylase, previously associated with rhamnolipid synthesis, is not required for rhamnolipid synthesis in vitro [12,18]. The genes rhlA and rhlB are organized in a single operon, while rhlC is localized in another region of the P. aeruginosa genome and forms an operon with a gene of unknown function.
Figure 1.
Rhamnolipid biosynthesis pathway in P. putida. All enzymatic steps required for the synthesis of essential precursors including host-intrinsic enzymes and exogenous RhlA and RhlB are shown. *) Annotated in P. putida as XanA [31].
In P. aeruginosa the expression of all three genes involved in rhamnolipid synthesis is regulated by two quorum sensing systems and is active when P. aeruginosa is cultivated under phosphate or nitrogen limiting conditions [6,19-21]. The σ54 factor is responsible for the expression of rhlAB under these conditions [22]. LasI/R and RhlI/R are also regulating transcription of many virulence factors [23].
P. aeruginosa has the highest rate of rhamnolipid production on plant oils as sole energy and carbon source [6,24]. The use of plant oils in industrial rhamnolipid production is a major drawback, as rhamnolipids are strong emulsifiers requiring elaborate and expensive methods for product purification, including organic solvent intensive extraction [20,25].
There are however approaches to produce rhamnolipids in bacteria other than P. aeruginosa by heterologous gene expression. Recombinant rhamnolipid production generally enables industrial important advantages. They facilitate both disconnecting biosynthesis from the complex quorum sensing regulation and the opportunity of avoiding pathogenic production strains, in particular P. aeruginosa. A topical review on rhamnolipid production by genetic engineering strategies using P. aeruginosa rhl-genes is given by Toribio et al. [26]. For example, Ochsner et al. [27] examined rhamnolipid synthesis by recombinant P. fluorescens, P. putida, P. oleovorans, and E. coli, which were equipped with the rhlAB operon. Recombinant strains of P. fluorescens were able to produce 0.25 g/L, those of P. putida 0.6 g/L rhamnolipid. Active rhamnosyltransferase was also synthesized in E. coli and P. oleovorans, but no rhamnolipids were formed. Wang et al. [28] reported that heterologous expression of the rhlAB genes in the E. coli strains BL21 and TnERAB led to the formation of rhamnolipids. Cabrera-Valladares et al. [29] also examined the heterologous production of mono-rhamnolipids in E. coli W3110. They constructed an E. coli strain that expressed the P. aeruginosa operons rhlAB and rmlBDAC. The latter encodes the dTDP-L-rhamnose synthesis pathway. With glucose as sole carbon source, they achieved a concentration of 120 mg/L mono-rhamnolipid and were able to show that recombinant production of mono-rhamnolipid in E. coli is limited by the availability of dTDP-L-rhamnose. Cha et al. [30] examined the option of expressing rhlAB from P. aeruginosa in P. putida. They were able to produce up to 7.3 g/L rhamnolipid with a recombinant P. putida strain, utilizing soybean oil as substrate. Another study with soybean oil as carbon source was carried out by Wang et al. [28]. They inserted the rhlAB genes into P. aeruginosa PEER02 through transposon-mediated chromosome integration; the resulting strain produced 1.8 g/L rhamnolipid. Cabrera-Valladares et al. [29] also used a hydrophobic substance (oleic acid) as substrate and detected 52 mg/L rhamnolipid with recombinant E. coli HB101.
In the present study, we designed, constructed, and analyzed a mono-rhamnolipid producing P. putida KT2440 that overcomes the outlined limitations of the natural producer P. aeruginosa. The constructed strain is devoid of pathogenicity factors [31] and complex cellular regulation, and uses glucose as carbon source thereby avoiding extensive downstream processing. The results are discussed in the context of different approaches of heterologous rhamnolipid production.
Results
High rhamnolipid resistance as prerequisite for the production host
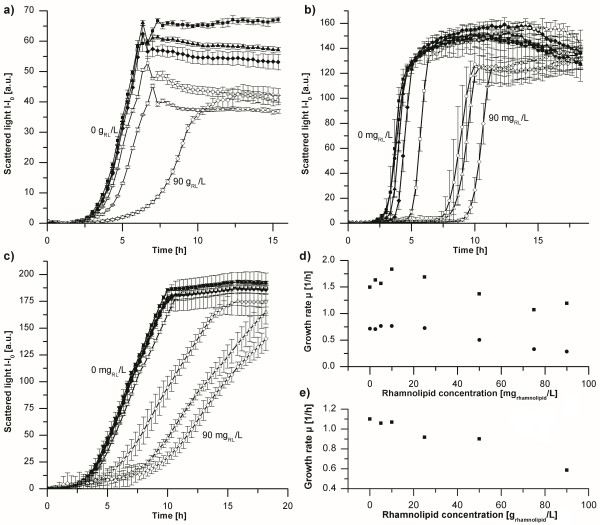
The non-pathogenic host for rhamnolipid production from glucose has to withstand high rhamnolipid concentrations to sustain industrially relevant production titers. The industrial workhorses E. coli, B. subtilis, and C. glutamicum, as well as the closely related, but non-pathogenic Pseudomonas, P. putida were tested for resistance against di-rhamnolipids. Using a microbioreactor system, the growth of the species was monitored (Figure 2) in the presence of up to 90 g/L di-rhamnolipid (purity of 95%).
Figure 2.
Toxicity studies with di-rhamnolipids using different bacteria. (a) Inhibiting effect of di-rhamnolipids on E. coli. Di-rhamnolipid concentrations tested were 0 g/L (■), 5 g/L (▲), 10 g/L (♦), 25 g/L (□), 50 g/L (○), and 90 g/L (◊). (b) Inhibiting effect of di-rhamnolipids on B. subtilis. Di-rhamnolipid concentrations tested were 0 mg/L (■), 2.5 mg/L (●), 5 mg/L (▲), 10 mg/L (♦), 25 mg/L (□), 50 mg/L (○), 75 mg/L (Δ), and 90 mg/L (◊). (c) Inhibiting effect of di-rhamnolipids on C. glutamicum. Di-rhamnolipid concentrations tested were 0 mg/L (■), 2.5 mg/L (●), 5 mg/L (▲), 10 mg/L (♦), 25 mg/L (□), 50 mg/L (○), 75 mg/L (Δ), and 90 mg/L (◊). (d) Growth rates resulting from toxicity experiments with B. subtilis (■) and C. glutamicum (●). (e) Growth rates resulting from toxicity experiments with E. coli.
The effect of rhamnolipids on Gram-positive C. glutamicum is dramatic, where concentrations of less than 100 mg reduce the rate of growth by 60% (Figure 2d). B. subtilis on the other hand, being Gram-positive as well, features only slightly decreased growth rates, while the duration of the lag-phase increases significantly in the presence of rhamnolipids (Figure 2b). B. subtilis secretes lipases, which might be able to disassemble di-rhamnolipid, by splitting off the fatty acids from the sugar molecules, reducing the toxic effect significantly. Then, B. subtilis would be able to grow unaffected by the rhamnolipids derived residues, explaining the elongated lag-phase and the almost unimpaired growth rates. Possible candidates that might extracellularly digest rhamnolipids are the enzymes lipoyl synthase (EC 2.8.1.8) and triacylglycerol lipase (EC 3.1.1.3), being products of lipA and lipB, respectively [32].
In contrast, Gram-negative species appear to be less affected and can grow in the presence of highly concentrated rhamnolipids. At a concentration of 90 g/L di-rhamnolipid E. coli only grows with half the growth rate it features when growing in absence of rhamnolipids (Figure 2e). Importantly, P. putida KT2440, certified as a safety strain [31,33] showed little change in growth rate in dependence of rhamnolipid concentrations as high as 90 g/L (data not shown). As this strain is closely related to P. aeruginosa, contains both necessary pathways for rhamnolipid precursor synthesis (i.e., de novo lipid synthesis and creation of activated rhamnose) [31], and grows with a very high rate on glucose [34], P. putida KT2440 was chosen as host for rhamnolipid production using glucose as carbon source.
Blueprint of an optimal metabolic network for rhamnolipid production
Having the chassis, the design of a metabolic network with high capacity for rhamnolipid synthesis in P. putida, using flux balance analysis with the rate of rhamnolipid production as linear programming objective, was in focus. The constraints of the metabolic network were, besides its structure, the substrate uptake rate, the rate of biomass formation, and the energetic demand for cell maintenance.
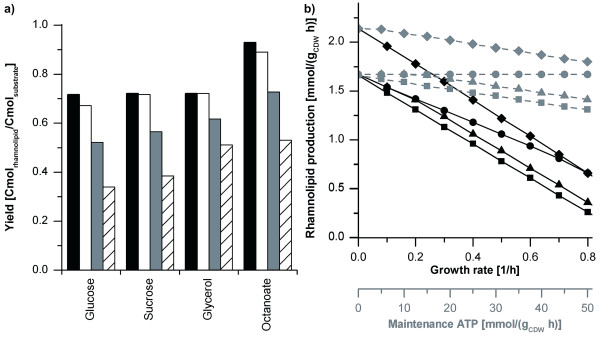
The theoretically achievable yields of rhamnolipids on industrial relevant substrates (glucose, sucrose, glycerol, and fatty acids (here as example octanoate)) were estimated. The computational results indicate that cell growth should be minimized to achieve high rhamnolipid yields (Figure 3). In simulations without growth and no or low cell maintenance, rhamnolipid yields varied only slightly between the two sugars and glycerol. The choice of substrate had an effect during growth or high cell maintenance metabolism. Specifically, sucrose and glycerol simulations were superior to simulations with glucose, as the glucose ABC-transporter requires one molecule of ATP per glucose molecule transported, while glycerol is transported via an ion channel by diffusion [31,35]. P. putida does not feature a sucrose uptake system, which therefore has to be established by simulating a sucrose porin channel [36] present in some P. syringae strains [37,38]. According adaptations of the computational metabolic network yielded in a non-cellular energy consuming sucrose uptake. In subsequently carried out simulations, octanoate enabled the highest yield (Figure 3b). Assuming the presence of an ACP-ligase [39], octanoate requires only activation by CoA/ACP to form HAA (Figure 4), thereby avoiding metabolically expensive de novo lipid synthesis. The rhamnose moiety requires β-oxidation and gluconeogenic reactions.
Figure 3.
Theoretical yields in rhamnolipid production with P. putida under different conditions. (a) Rhamnolipid yields on alternative carbon substrates. The yields were calculated for zero growth (black bars), zero growth and 30 mmol ATP/(gCDW h) maintenance metabolism (white bars), growth at a rate of 0.4 1/h with 30 mmol ATP/(gCDW h) maintenance metabolism (gray bars), and a growth rate of 0.8 1/h with 50 mmol/(gCDW h) maintenance metabolism (shaded bars). The substrate uptake rates were constrained to 120 mCmol/(gCDW h). (b) Rhamnolipid production capacity depending on biomass formation and maintenance metabolism for alternative carbon substrates. The course of rhamnolipid production for uptake of glucose (■), glycerol (●), sucrose (▲) and octanoate (♦) are displayed. The black curves display rhamnolipid production in dependence on the rate of growth; grey dashed curves display rhamnolipid production in dependence of maintenance metabolism.
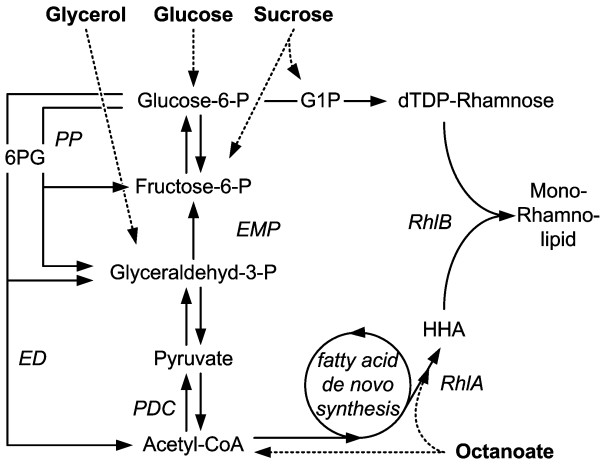
Figure 4.
Scheme of utilized pathways during rhamnolipid production by P. putida. ED, Entner-Doudoroff pathway; PP, pentose phosphate pathway; PDC, pyruvate decarboxylation; G1P, glucose-1-phosphate; HAA, β-D-(β-D-hydroxyalkanoyloxy)alkanoic acid; RhlA, 3-hydroxyacyl-ACP:3-hydroxyacyl-ACP O-3-hydroxyacyltransferase; RhlB, rhamnosyltransferase I.
Notably, in simulations with low growth rates and maintenance metabolism, rhamnolipid yield on glycerol equaled yields on sugars; with higher growth rate and maintenance metabolism, the yield on glycerol equaled the yield on octanoate. The observation that a more reduced carbon substrate is not beneficial for rhamnolipid production by non-growing cells suggests that under these conditions a carbon limitation exists, while production by growing cells can be energy limited. Glycerol feeds into the central carbon metabolism at the level of glyceraldehyde-3-phosphate and thus does not utilize the pentose phosphate (PP) pathway, which wastes carbon via CO2 production (Figure 4). Glucose and sucrose enter the central carbon metabolism via the Entner-Doudoroff (ED) pathway. To supply the cell with the necessary energy for maintenance (via redox cofactor synthesis), the PP pathway was active, hence wasting carbon via CO2 formation. In scenarios with very high energy demand (i.e., high growth rates and/or high maintenance metabolism), full oxidation via acetyl-CoA and the tricarboxylic acid (TCA) cycle was observed, again resulting in CO2 formation and concomitant lower rhamnolipid yield. The theoretically high rhamnolipid yield on octanoate is a result of the omitted reaction from pyruvate to acetyl-CoA (via the pyruvate dehydrogenase).
In summary, the carbon substrates used for rhamnolipid production by P. aeruginosa are theoretically ideal for achieving high yields of product. However, the extensive product purification from a second, hydrophobic phase is not desirable. Therefore, the industrially important carbon source glucose is preferred. It is desirable to produce rhamnolipids from glucose with a host that has low maintenance requirements and forms no side-products. Ideally, such a host allows production during non-growth conditions, to maximize the yield of product on substrate.
Having the carbon substrate defined, subsequent simulations focused on a metabolic network that is optimal for rhamnolipid production from glucose. Possible reaction candidates include glucose uptake and glucose catabolism. The earlier via a newly introduced phosphotransferase system to improve the stoichiometry of glucose uptake and the latter via an artificial Embden-Meyerhof-Parnas (EMP) pathway by introducing a phosphofructokinase to improve ATP generation by substrate level phosphorylation, respectively. Notably, the improvements of the tested scenarios were minor (below 1% of additional rhamnolipid) again highlighting that carbon and not energy availability determines the yield of rhamnolipid production. Hence, these optimizations were not considered as valuable targets for the improvement of rhamnolipid production. Instead, removing enzymatic reactions that are not necessary for rhamnolipid production, but potentially waste carbon moved into focus. As already mentioned, no side products from central carbon metabolism (e.g., carboxylic or amino acids) were observed in the growth medium. Therefore, the avoidance of biomass components that are not necessary for survival and production are the only means for metabolic optimization. One such target is the storage polymer polyhydroxyalkanoate (PHA). In P. putida, PHA-synthesis directly competes with the production of the rhamnolipid precursor HAA (Figure 1). Eliminating this reaction in vivo should lead to enhanced rhamnolipid yields. Choi et al. [40] recently showed in P. aeruginosa PA14 and PAO1 ΔphaC1 mutants that indeed PHA-production was lower. Again, after reaching a defined concentration of cells in the media, which suits rhamnolipid production best, biomass formation should be deactivated or avoided to enable optimal carbon exploitation.
Rhamnolipid production from glucose by non-pathogenic P. putida
To enable the production of rhamnolipids in P. putida, the essential genes for rhamnolipid production, located in P. aeruginosa on the rhlAB operon (encoding RhlA and RhlB) were introduced. The resulting recombinant strain P. putida KT2440 pVLT33_rhlAB produced mono-rhamnolipids (Figure 5). In comparison, P. aeruginosa PAO1 produced mono- and di-rhamnolipids (Figure 5, lane 2). First quantification of rhamnolipids indicated up to 0.35 g/L rhamnolipid in the P. aeruginosa culture, and about 0.22 g/L mono-rhamnolipid in the culture of the newly engineered P. putida KT2440 pVLT33_rhlAB.
Figure 5.
Thin layer chromatography of rhamnolipids. The sample of P. aeruginosa PAO1 (lane 2), grown in PPGAS-medium at 37°C, contains mono- and di-rhamnolipid as does the commercial rhamnolipid (JBR425, Jeneil Biosurfactant Co.) (lane 1). P. putida, expressing the rhlAB operon of P. aeruginosa from the plasmid pVLT33_rhlAB, cultivated in glucose containing LB-medium produced mono-rhamnolipid (lane 4). Empty vector control pVLT33 (lane 3). The band located above the di-rhamnolipids is IPTG (i.e., lanes 3 and 4).
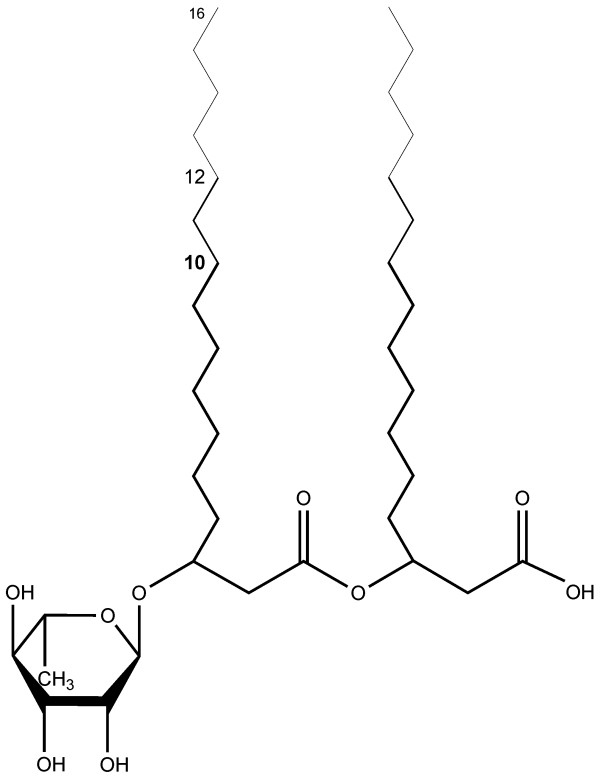
The product spectrum of P. putida KT2440 pVLT33_rhlAB was investigated by HPLC-ESI-MS. The results illustrated that the new strain produces rhamnolipids with fatty acids featuring chain lengths between C8 and C12 in different combinations and in addition very low amounts of rhamnolipids with C14 and C16 chains. Furthermore, some of the alkyl chains contained one unsaturation. The most abundant rhamnolipid species contained two fatty acids with C10 chains (Figure 6). This was also the case for rhamnolipids that contained only one β-hydroxy fatty acid chain. Although P. aeruginosa and P. putida KT2440 pVLT33_rhlAB produced rhamnolipids with different number of rhamnose residues, the produced rhamnolipids consisted of alkyl chains with the same length. Hence, the production of rhamnolipid with similar composition to the rhamnolipid of P. aeruginosa (rhlC addition leads to di-rhamnolipid synthesis) using glucose as carbon source was achieved.
Figure 6.
Molecular structure of mono-rhamnolipids from recombinant P. putida. The major compound (C10:C10) is indicated by thick black lines, while minor compounds are indicated by thin lines.
Rhamnolipid synthesis by a PHA-negative mutant
While it was shown that the engineered P. putida KT2440 pVLT33_rhlAB produced rhamnolipids, the yield on glucose was significantly below the theoretical limit of 0.72 Cmolrhamnolipid/Cmolglucose (Figure 3). The identified PHA production pathway was chosen as potentially valuable target. P. putida uses β-hydroxyacyl-ACP from de novo fatty acid synthesis as precursor for PHA storage [41-43]. The β-hydroxyacyl-ACP is converted to β-hydroxyacyl-CoA by PhaG β-hydroxyacyl-ACP:CoA transacylase [44]. PhaC1 poly(3-hydroxyalkanoic acid) synthase 1 catalyzes the reaction leading to PHA [45]. As these reactions compete with formation of hydroxyalkanoyl-alkanoates in the rhamnolipid synthesis pathway, the use of a PhaC1-negative strain was chosen as first optimization target. P. putida KT42C1 (ΔphaC1::Kmr) [41], a derivative of P. putida KT2442 [46], was tested as a host for rhamnolipid production. P. putida KT2442 in turn is a spontaneous rifampicin resistant mutant of P. putida KT2440. As the knockout was produced using a kanamycin resistance cassette, the rhlAB operon was subcloned into the plasmid pVLT31, a pVLT33 derivative carrying a tetracycline resistance. The new strain P. putida KT42C1 pVLT31_rhlAB produced up to 1.5 g/L of rhamnolipid, about seven times more than the original strain. The product profile of P. putida KT42C1 pVLT31_rhlAB revealed that not only mono-rhamnolipids, but also up to 20% of the free hydroxy fatty acid was produced (Figure 7). To avoid wasting of resources the formation of the hydroxy fatty acids should be minimized. Therefore, future experiments concentrate on the two alternative reactions forming the free fatty acids: de novo synthesis or rhamnolipid degradation. Also conceivable would be the knocking out of the gene for the second poly(3-hydroxyalkanoic acid)synthase phaC2, which features minor PHA-producing activities [41].
Figure 7.
Percentage distribution of compounds gained by hydrophobic adsorption from the supernatant of a fermentation of P. putida KT42C1 pVLT31_rhlAB.
Uncoupling growth and rhamnolipid production: towards minimal biomass formation
Ideally, to achieve optimal product yields, high production rates without biomass formation are desirable. During the fermentation, the growth rate of P. putida KT42C1 pVLT31_rhlAB declined (Figure 8). This growth behavior on lysogeny broth (LB) medium was described for E. coli and is explained by multiauxic growth due to sequential compound uptake [47,48]. The kinetics underlying growth of P. putida can best be described by a logistic growth formulation (Equations (1) to (3)).
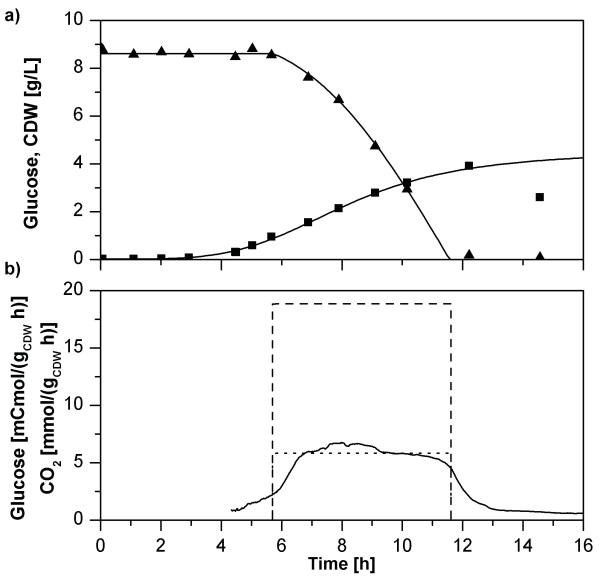
Figure 8.
Uncoupling of rhamnolipid production and cell growth of P. putida in a 50 mL baffled flask. (a) Fermentation characteristics including cell growth (■) and course of rhamnolipid (▲) and glucose (♦) concentrations and their respective fitted courses. CDW, cell dry weight. (b) Specific rates resulting from the fitted experimental data. The black line represents the course of the growth rate, while the dashed line and the dotted line show the specific glucose uptake rate and the specific rhamnolipid production rate respectively.
Although counterintuitive, changing growth rates did not result in changing neither glucose uptake nor rhamnolipid production rates (Figure 8). Importantly, the constant rhamnolipid formation rate suggests that the recombinant regulation of the rhamnolipid synthesis operon cannot be influenced by the host and hence is truly orthogonal during the growth and production phases. Hence, rhamnolipids are produced from cells that grow minimally or not at all, which opens opportunities for long-term stable production with high product yields (Figure 3). This is especially true, if glucose is the carbon source for rhamnolipid production, while the components of the LB medium are precursors for biomass formation.
To elucidate if glucose primarily serves as substrate for rhamnolipid production by P. putida KT42C1 pVLT31_rhlAB the use of glucose versus the use of alternative carbon sources was discriminated using uniformly labeled 13C6-glucose in combination with appropriate analytics. A new custom-made off-gas sensor (BlueSens GmbH, Herten, Germany) allowed the simultaneous quantification of 12CO2 and 13CO2 concentrations. Assuming that glucose is the carbon source for rhamnolipid production, the conversion of pyruvate to acetyl-CoA (the monomer of fatty acid polymerization) releases 13CO2.
The production of 13CO2 did not agree with time-invariant glucose uptake and rhamnolipid production rates, but rather a distinct and abrupt start of glucose metabolization followed by a constant 13CO2 production of 0.21-0.27 mmol/(gCDW h) (CDW: cell dry weight). The specific production rate of 13CO2 remained constant until glucose was depleted (Figure 9). The growth and glucose uptake kinetics suggest that glucose metabolism is suppressed by LB constituents, followed by glucose catabolism after these constituents are depleted, i.e., multiauxic growth [49]. Rhamnolipid production on glucose-complemented LB medium can be divided into two phases, of which the first consists of rapid growth on the most favored carbon substrates contained in the LB medium, while the second phase is characterized by a decreasing growth rate at constant specific glucose uptake and rhamnolipid production rates, i.e., by gradual uncoupling of biomass and rhamnolipid production.
Figure 9.
Kinetics of rhamnolipid-production in P. putida in a stirred 300 mL reactor. (a) Development of biomass (■) and glucose (▲) concentrations. The experimental data is depicted by symbols, while the lines present the fits using Equations (1) to (3). CDW, cell dry weight. (b) Specific rates characterizing rhamnolipid production in P. putida. The solid line presents the measured 13CO2 production rate originating from 13C-labeled glucose. The dashed line represents the glucose uptake rate and was calculated from the fitted glucose concentration curve. The dotted line shows the rate of CO2 evolution, produced in the rhamnolipid synthesis pathway. The curve was calculated from the experimentally determined yield of rhamnolipid on glucose and the estimated rate of glucose consumption.
The new off-gas sensor from BlueSens allowed to rationalize these observed fermentation kinetics and provided valuable insights into metabolic network operation, which is of paramount importance for the development of rhamnolipid production by P. putida KT42C1 pVLT31_rhlAB under non-growth conditions.
Discussion
Comparison to P. aeruginosa wild type strains
Until now, rhamnolipid production mainly was carried out by P. aeruginosa. In this work, a functional substitution to the opportunistic pathogenic, nosocomial P. aeruginosa, a non-pathogenic P. putida was developed. The engineered strain has several advantages when compared to rhamnolipid production with P. aeruginosa. We were thus for instance able, to circumvent the complex quorum sensing regulation in P. aeruginosa.
The achieved rhamnolipid production rate in the carried out experiments amounted to two thirds of the rate observed in optimized fermentations with P. aeruginosa (Table 1). The obtained carbon yield also was in the same range. For industrial production of rhamnolipids the yield is an important parameter, as it determines the expenses for the substrate. The rate of rhamnolipid production might be raised by increasing activated rhamnose availability as observed earlier [29]. Nevertheless, the obtained rhamnolipid titer in the medium is still far from the almost 40 g/L rhamnolipid reported with P. aeruginosa [20], which can be explained by the low biomass concentration in the described experiments. Fortunately, P. putida is suited for high cell density fermentations. While Kim et al. achieved 100 gCDW/L with P. putida BM01 [50], even higher concentrations were obtained with P. putida KT2442 [51]. In another study, with P. putida KT2440, cell concentrations of up to 62 gCDW/L were achieved [52].
Table 1.
Recombinant rhamnolipid production
| Organism | Medium | Substrates | Cell Dry Weight | Carbon Yield1 | Maximal Titer | Process Time | RL-Production Rate | Space-Time Yield | Specific RL-Production Rate2 | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Substance | [g/L] | [gCDW/L] | [Cmolrhamnolipid/Cmolsubstrate] | [gRL/L] | [h] | [mgRL/(Lh)] | [mgRL/(Lh)] | [g/(gCDW h)] | |||
| P. aeruginosa PAO1 | MS | Sunflower Oil | 250 | 16.3 | 0.07 | 39.00 | 90 | 16.67 | 433.33 | 0.027 | Müller et al. 2010b [20] |
| P. putida KCTC 1067 | MS | Soybean Oil | 20 | 3.2 | 0.17 | 7.30 | 72 | 3.90 | 101.39 | 0.031 | Cha et al. 2008 [30] |
| P. aeruginosa PEER02 | MS | Soybean Oil | 20 | not given | 0.04 | 1.82 | 96 | 0.73 | 18.95 | not calculable | Wang et al. 2007 [28] |
| E. coli HB101 | LB | Oleic Acid | 4 | not given | 0.01 | 0.05 | 24 | 0.08 | 2.18 | not calculable | Cabrera-Valladares et al. 2006 [29] |
| P. putida KT42C1 | LB | Glucose | 10 | 2.0 | 0.23 | 1.50 | 42 | 1.37 | 35.71 | 0.018 | This study |
| P. aeruginosa PG201 | GS | Glycerol | 20 | not given | 0.17 | 2.20 | 168 | 0.50 | 13.10 | not calculable | Ochsner et al. 1995 [27] |
| E. coli TnERAB | LB | Glucose | 4 | not given | 0.07 | 0.18 | 24 | 0.28 | 7.30 | not calculable | Wang et al. 2007 [28] |
| P. aeruginosa PEER02 | MS | Glucose | 20 | not given | 0.06 | 0.79 | 96 | 0.31 | 8.18 | not calculable | Wang et al. 2007 [28] |
| P. putida KT2442 | GS | Glycerol | 20 | 1.2 | 0.05 | 0.60 | 25 | 0.92 | 24.00 | 0.020 | Ochsner et al. 1995 [27] |
| E. coli W3110 | M9 | Glucose | 5 | not given | 0.04 | 0.12 | 48 | 0.10 | 2.51 | not calculable | Cabrera-Valladares et al. 2006 [29] |
| E. coli TnERAB | MS | Glucose | 4 | not given | 0.03 | 0.08 | 24 | 0.12 | 3.15 | not calculable | Wang et al. 2007 [28] |
| P. fluorescens ATCC 15453 | GS | Glycerol | 20 | not given | 0.02 | 0.25 | 168 | 0.06 | 1.49 | not calculable | Ochsner et al. 1995 [27] |
| E. coli DH5α | M9 | Glucose | 5 | not given | < 0.01 | < 0.02 | 168 | 0.00 | 0.06 | not calculable | Ochsner et al. 1995 [27] |
| P. oleovorans GPo1 | GS | Glycerol | 20 | not given | 0.00 | < 0.02 | 168 | 0.00 | 0.06 | not calculable | Ochsner et al. 1995 [27] |
A comprehensive overview of production performance of recombinant strains is presented. For comparison, wild type P. aeruginosa PAO1 is listed in line 1. The next three lines show data from experiments with hydrophobic substrates in contrast to the remaining entries. This data is based on hydrophilic substrate experiments.
1) For the calculation of yields during production on complex media, rhamnolipids were assumed to be synthesized from the used carbon source, while media compounds were utilized for cell growth. 2) The specific rhamnolipid production rate was calculated as average over the whole fermentation time. LB, Lysogeny broth medium; MS, mineral salt medium; GS, nitrogen-limited minimal medium.
An important improvement in comparison to the fermentation of P. aeruginosa is the utilization of glucose as substrate instead of hydrophobic substances such as plant oils (despite the theoretical yield benefits) (Figure 3). Notably, the carbon yield of 0.23 Cmolrhamnolipid/Cmolglucose on glucose here reported is 33% of the theoretical yield achievable on this substrate and in the same order as the highest yield on oily carbon substrate reported for P. aeruginosa (0.33 Cmolrhamnolipid/Cmolsoybean oil) [53]. The main advantage nevertheless is that purification of rhamnolipids from the fermentation broth can be significantly simplified, as the surfactant emulsifies the hydrophobic substrate in the P. aeruginosa fermentation in the aqueous phase. Avoidance of stable substrate/product emulsions using glucose as carbon source results in modest demands on sample preparation for analytical procedures and more importantly in reduced complexity of downstream processing.
Another benefit is that most bacteria grow faster with glucose as carbon source than with fatty acids. Particularly P. putida features a high growth rate when growing on glucose [34]. Furthermore, glucose as carbon source is a common substrate in biotechnological production processes, which makes it notably cheaper than for example soybean oil [54]. These advantages of glucose as carbon source for rhamnolipid production outweigh the downside that the theoretical yield is higher with fatty acids as substrate.
Comparison to recombinant strains
The majority of experiments using recombinant bacteria described so far utilize glucose as substrate for heterologous rhamnolipid production, while some reports with hydrophobic substances exist (Table 1). Disregarding the downsides accompanying the introduction of oily substances in the production process, these strains feature a major advantage. Like the wild type strain P. aeruginosa PAO1 they show higher titers than recombinant strains using glucose as carbon source. However, probably due to complex quorum sensing based regulation, these strains need 1 to 3 days to reach the maximal titer. In contrast, and partly also because sugars as carbon source can be taken up and metabolized faster than fatty acids most of the recombinant strains, which constitute a serious alternative to P. aeruginosa using hydrophilic substances as substrate reach the maximal concentration of rhamnolipids in the medium after 1 to 2 days. This reduction in process time results in an enhancement of the space-time yield. Notably, except P. putida KCTC 1067 [30], the strain engineered in this study features the highest reported space-time yield, making it appropriate for further industrial development of a biotechnological rhamnolipid production.
Comparing strain P. putida KT42C1 pVLT31_rhlAB further to other attempts of recombinant rhamnolipid production based on hydrophobic substrates, we achieved a two to ten times higher titer (1.5 g/L), which contributes to simplifying downstream processing and entails reduction of investment costs as a smaller reactor is required. The highest carbon yield reported with recombinant strains by now was 0.17 Cmolrhamnolipid/Cmolsubstrate [27,30]. The newly engineered strain P. putida KT42C1 pVLT31_rhlAB had a comparatively high yield of 0.23 Cmolrhamnolipid/Cmolsubstrate, which is in the range of yields achieved with P. aeruginosa (0.33 Cmolrhamnolipid/Cmolsubstrate) [53].
Logistic growth
Logistic growth, as it can be observed in our experiments (Figure 8) is based on population dynamics. This model is applied in growth situations where the increase in population size is limited by upper boundaries. The general shape of a growth curve is sigmoidal, resulting from zero growth at the beginning, increasing growth to a maximal growth rate and subsequent decreasing growth rates until an asymptote is reached. The logistic model can be utilized to describe such courses. The parameters incorporated in the equation on which logistic growth is based upon resemble the biological parameters of the population. Parameters used are the initial biomass concentration, the final biomass concentration, the time after which half of the biomass is formed, and furthermore a curve form coefficient, which has no direct biological counterpart.
Growth of P. putida while producing rhamnolipids can accurately be characterized via a logistic model, as was shown for P. aeruginosa [55]. Having in mind the phenomenon of multiauxic growth described earlier, it could be that under the chosen cultivation conditions P. putida is experiencing multiple, shifting limitations. One can speculate that these growth-limits are determined by the availability of the currently metabolized carbon source, which changes frequently, when P. putida grows on LB-medium supplemented with glucose. Again, the significant reduction in growth is independent of rhamnolipid production.
Conclusions
In this study, we successfully carried out mono-rhamnolipid synthesis by engineered P. putida. Featuring high resistance against rhamnolipids, simple and controllable production kinetics, and the metabolic ability to produce rhamnolipids with high yield and rate, P. putida proved to be an appropriate host for heterologous rhamnolipid production.
The fermentation of non-pathogenic P. putida was possible using glucose as substrate for the synthesis of mono-rhamnolipids, while constituents of LB-medium were the carbon sources for cell growth. By using quantitative fermentation kinetics monitoring including advanced off-gas analysis we could show that rhamnolipid production is uncoupled from biomass formation, which offers possibilities for substrate exploitation and process management.
In summary, an alternative, non-pathogenic host for rhamnolipid production utilizing glucose as carbon source was successfully developed. The insights into rhamnolipid production with P. putida will contribute to the further advance of this host; with the aim to develop an industrially viable process. This study thus adds to an increasing amount of literature regarding industrial importance [56] and the applicability for heterologous expression of complex secondary metabolites [57] of this bacterium.
Methods
Bacterial strains, culture conditions, and plasmids
The used bacteria strains Pseudomonas aeruginosa PAO1 and Pseudomonas putida KT2440 [31,58] were routinely cultivated in LB-medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl), while Escherichia coli DH5α [59], Bacillus subtilis TEB1030 [32], and Corynebacterium glutamicum ATCC 13032 [60] were cultivated in TB-medium (12 g/L tryptone, 24 g/L yeast extract, 5 g/L glycerol, 12,54 g/L dipotassium phosphate, 2,31 g/L monopotassium phosphate). All bacteria were cultivated at 37°C except P. putida and C. glutamicum, which were grown at 30°C. P. putida and E. coli containing the vector pVLT33 [61] and derivatives thereof were selected by adding 50 μg/mL kanamycin to LB-agar and liquid cultures. For selecting pVLT31 and derivates tetracycline with concentrations of 10 μg/mL for recombinant E. coli and 20 μg/mL for recombinant P. putida were added. Rhamnolipid production with P. aeruginosa and recombinant P. putida was carried out using LB-medium complemented with 10 g/L glucose.
Rhamnolipid toxicity determination
The experiments were carried out with a micro bioreactor system (BioLector, m2p-labs GmbH, Baesweiler, Germany), in 48 well plates (Flowerplates m2p-labs GmbH, Baesweiler, Germany). The biomass concentration was quantified by online light scattering. All bacteria apart from P. putida KT2440 were cultivated in 800 to 1000 μL TB-medium. P. putida KT2440 was grown in 500 μL LB-Medium supplemented with 10 g/L glucose and 90 mM potassium phosphate buffer (pH 7.4). The growth medium contained rhamnolipid concentrations between 0 g/L and 90 g/L. The cultures were shaken at 1,100 to 1,200 rpm (throw: 3 mm) and incubated at 37°C or 30°C, for E. coli DH5α, B. subtilis TEB1030, C. glutamicum ATCC13032, and P. putida KT2440, respectively.
Construction of the rhamnolipid production module
The rhlAB operon was amplified from the genomic DNA of P. aeruginosa PAO1 that was isolated with a DNA isolation Kit (DNeasy Blood and Tissue Kit, QIAGEN, Hilden, Germany), using DNA polymerase (PfuTurbo, Stratagene, Waldbronn, Germany) as described by the supplier. The used primer had the following sequences: sense 5'TTGAATTCCATCGGCTACGCGTGAACACGG'3, antisense 5'TTTTTCTAGATCAGGACGCAGCCTTCAGCC'3. The oligonucleotides were obtained from Eurofins MWG Operon (Ebersberg, Germany). The rhlAB PCR product was digested with EcoRI/Acc65I and subsequently ligated into pVLT33, which was digested with the same enzymes, creating the plasmid pVLT33_rhlAB. Restriction enzymes and T4 DNA ligase were obtained from Fermentas (St. Leon-Rot, Germany) and used as recommended. DNA manipulation was carried out as described in Sambrook and Russell [62]. Ligations were transformed into competent E. coli DH5α using a standard protocol [59]. Transformed cells were selected on LB-agar plates containing 50 μg/mL kanamycin. Experiments with the kanamycin resistant single gene deletion strain P. putida KT42C1, lacking the poly(3-hydroxyalkanoic acid) synthase 1 encoded by phaC1, required subcloning of rhlAB into pVLT31, which contains a gene for tetracycline resistance.
P. putida KT2440 was transformed using electroporation as described by Choi et al. [63]. Cells containing plasmid pVLT33 or the derivate pVLT33_rhlAB were selected on LB-Agar plates or liquid cultures containing 50 μg/mL kanamycin. A resulting strain was denominated P. putida KT2440 pVLT33_rhlAB. This strain was utilized for all further works.
Characterization of rhamnolipid production by P. putida KT2440 pVLT33_rhlAB
For the production of rhamnolipids a main culture of 50 mL LB-medium supplemented with 10 g/L glucose and 50 μg/mL kanamycin in a 500 mL Erlenmeyer flask was inoculated with 1 ml from a starter culture and incubated at 30°C and 200 rpm (throw: 25 mm). The expression of rhl-genes was induced by adding IPTG (isopropyl β-D-1-thiogalactopyranoside) to a final concentration of 0.4 mM (guaranteeing full induction) from the beginning of the fermentation. Rhamnolipids were extracted 24 h after induction.
P. aeruginosa was cultivated in 10 mL phosphate-limited protease peptone-glucose-ammonium salt medium (PPGAS) at pH 7.2, which promotes the production of rhamnolipids [64], containing 5 g/L glucose, 10 g/L peptone 0.02 M NH4Cl, 0.02 M KCl, 0.12 M Tris-HCl, and 0.0016 M MgSO4. After 24 h at 37°C, with an agitation of 150 rpm, rhamnolipids were harvested.
Cultivations of P. putida KT2440 pVLT33_rhlAB carried out in order to supply rhamnolipid-characterization via thin layer chromatography (TLC) and HPLC-ESI-MS featured slightly different process parameters. Only 10 ml of LB-Medium, supplemented with 10 g/L glucose and 50 μg/mL kanamycin in a 100 mL Erlenmeyer flask, were inoculated with an OD580 of 0.05 from a starter culture and incubated at 30°C and 150 rpm (throw: 25 mm). IPTG was added to a final concentration of 0.4 mM at an OD580 of 0.5.
To gain further insight into the fermentation kinetics of rhamnolipid producing P. putida experiments in 300 mL bioreactors (RALF, Bioengineering AG, Wald, Switzerland) were carried out. During 22 hours, the off-gas was analyzed with an off-gas sensor (BlueSens GmbH, Herten, Germany) applying dual wavelength infrared light. This sensor facilitated the simultaneous quantification of 12CO2 and 13CO2 concentrations in the off-gas. The reactor was filled with 200 mL LB-medium complemented with 10 g/L 100% labeled 13C6-glucose, IPTG to a final concentration of 0.4 mM, and 20 μg/mL tetracycline. The temperature was adjusted to 30°C and the aeration rate to 12.1 NL/h. The stirrer speed was set to 500 rpm and after seven hours of fermentation increased to 750 rpm.
The scale-up of rhamnolipid production was tested in a 3.2-liter fermenter vessel (KLF 2000, Bioengineering AG, Wald, Switzerland) with a working volume of 2 liters. The fermenter contained two 6-blade turbine stirrers, a temperature control, a pH control, and a gas inlet. The operating conditions were set to pH 6.8 and a temperature of 30°C, a constant gassing rate of 0.5 vvm and a stirrer speed in the range from 300 to 900 rpm depending on the online-determined pO2 signal. Additional glucose was fed using a peristaltic pump.
Quantification of rhamnolipids
For analysis, rhamnolipids were extracted using 100 μL for orcinol-assay and 500 μL for TLC respectively of cell-free culture broth and 500 μL of ethyl acetate. Samples were mixed by vortexing, with a subsequent phase separation by centrifugation in a tabletop centrifuge at maximum speed (30 sec). The upper, rhamnolipid-containing phase was transferred to a new reaction tube. This procedure was repeated three times. Finally, the organic solvent was removed by evaporation in a vacuum centrifuge.
Thin layer chromatography of rhamnolipids
For detection of rhamnolipids using TLC, the dried rhamnolipids were dissolved in 10 μL ethanol. 5 μL of this solution were spotted on a silica 60 TLC-plate (Macherey-Nagel, Düren, Germany). In addition, 5 μL of a 0.1% commercial rhamnolipid extract (JBR425, Jeneil Biosurfactant Co., LCC, Saukville, USA) containing mono- and di-rhamnolipids was spotted. The running buffer was a mixture of chloroform, methanol, and acetic acid in a ratio of 65:15:2. To visualize the rhamnolipids on the TLC-plates, the plates were covered with a detection agent consisting of 0.15 g orcinol, 8.2 mL sulfuric acid (60%), and 42 mL deionized water. For preservation, dried plates were incubated at 110°C for 10 min.
Rhamnolipid quantification using orcinol assay
The total amount of rhamnolipids was determined using the orcinol assay [65,66]. The evaporated rhamnolipids were dissolved in 100 μL deionized water. Subsequently 100 μL orcinol solution (1.6% orcinol in deionized water) and 800 μL sulphuric acid (60%) were added. The samples were incubated at 80°C for 30 min and 1000 rpm orbital shaking in a thermomixer (Eppendorf AG, Hamburg, Germany). After cooling to room temperature, the samples were measured at 421 nm in comparison to different concentrations of the commercial rhamnolipid extract using a Genesys 10 UV spectrophotometer (Thermo Fisher Scientific, Waltham, USA).
Rhamnolipid quantification using RP-HPLC-CAD
Reversed phase high performance liquid chromatography corona charged aerosol detection (RP-HPLC-CAD) was used for rhamnolipid quantification. Culture samples were centrifuged at 17,700 × g for 30 minutes. 100 μL supernatant were added to 900 μL deionized water, mixed on a vortex shaker, and analyzed on a gradient quaternary reversed phase HPLC system (LaChrom, VWR- Hitachi, Darmstadt, Germany). The system was equipped with an integrated C8(2) silica based column (Luna C8(2), 4.6 × 150 mm, 5 μ, 100 Å, Phenomenex, Inc. Torrance, CA, USA) and a corona CAD (ESA Biosciences Inc., MA, USA). The sample volume was set to 20 μL. The sample was eluted at a flow rate of 800 μL per minute and the temperature of the column oven was set to 40°C. The mobile phase contained filtered water with 0.4% trifluoracetic acid (TFA) (solvent A), acetonitrile (solvent B), and methanol with 0.2% TFA (solvent C) in different ratios. The method started with 20:0:80 (Vol.-% of solvent A:Vol.-% of solvent B:Vol.-% of solvent C) and switched at 7.5 minutes to 2:18:80 during 2 minutes. After 20.6 minutes, the starting concentration was reestablished, again during 2 minutes. The method ended after 24.6 minutes.
Rhamnolipid composition characterization by HPLC-ESI-MS
High performance liquid chromatography electrospray ionization mass spectrometry (HPLC-ESI-MS) was used for rhamnolipid characterization (Central Division of Analytical Chemistry/BioSpec, Forschungszentrum Jülich, Jülich, Germany). Rhamnolipids were extracted from 1 L culture broth (5 L Erlenmeyer flask) as described by Déziel et al. [8] with small modifications. Cells were removed by centrifugation for 30 min at 9,000 × g and 10°C. The supernatant was acidified with 37% HCL to a pH of 3 and incubated overnight at 4°C. The precipitated rhamnolipids were recovered by centrifugation (9,000 × g, 45 min, 4°C) and resuspended in 15 mL acidified water (pH 3). This suspension was extracted three times with 15 mL ethyl acetate. The combined organic phases were evaporated in a vacuum centrifuge. The residue was dissolved in 15 mL of 0.05 M NaHCO3, acidified to pH 2 with 37% HCl, and incubated overnight at 4°C. The precipitate was finally recovered by centrifugation for 60 min at 13,000 × g and 4°C.
For characterization, a binary HPLC system (Agilent 1100 series, Agilent Technologies, Waldbronn, Germany), assembled with a diode array detector (DAD) (190-400 nm), coupled with a triple quadrupole mass spectrometer (4000QTRAP™, Applied Biosystem/MDS SCIEX, Foster City, CA, USA) assembled with a turbo ion spray source was used.
For rhamnolipid separation, normal phase chromatography was used with column dimensions of 150 × 2 mm i.d., 3 μm particle size (ProntoSIL 120-C8-SH, Bischoff Chromatography, Leonberg, Germany) at 20°C. The gradient elution was done with deionized water with 0.1% formic acid (solvent A) followed by different concentrations of acetonitrile with 0.1% formic acid (solvent B). The elution started with 60% B isocratic for 4 min, from 4 to 24 min a linear increase from 60% B to 90% B was applied, subsequently followed by a second isocratic step (90% B for 10 min), and ended by a return to 60% B in one min. The re-equilibration was done with 60% B isocratic for 10 min. All steps were performed at a constant flow rate of 300 μL/min. The injection volume was 20 μL.
The MS was used in negative enhanced mass spectrum mode scanning from 200 - 1000 Da. A flow injection analysis with a standard was used at first to optimize the following parameters: IS -4500 V, declustering potential -100 V, curtain gas (N2) 10 arbitrary units (au), source temperature 500°C, nebulizer gas (N2) 50 au, and heater gas (N2) 20 au. Collision energy (CE) and third quadrupole-entry barrier were set to -5 V and 8 V, respectively. The negative enhanced product ion scan mode was used for structural elucidation MS/MS experiments, in which product ions are generated in the second quadrupole by collision-activated dissociation of selected precursor ions of the first quadrupole and mass analyzed in a linear ion trap. The CE ranged from 30 to 70 V.
The di-rhamnolipid standard (Rha-Rha-C10-C10) for HPLC analysis was a gift from Sanofi-Aventis Deutschland GmbH, former Hoechst AG (Frankfurt, Germany). Mono-rhamnolipid standard (Rha-C10-C10) was prepared as described before [67].
Rhamnolipid purification by adsorption
The medium was centrifuged in 200 mL cups for 60 min at 4,000 rpm (5810R Eppendorf AG, Hamburg, Germany) to remove cells and cell debris. The cell-free medium was loaded with five times the bed volume per hour by a peristaltic pump (MP-3 Micro Tube Pump, Eyela Inc., Tokyo, Japan) as specified by the manufacturer to a column packed with 90 g of conditioned hydrophobic polymeric adsorbent (Amberlite XAD-2, Sigma-Aldrich, St. Louis, MO, USA). After washing with bidistilled water, rhamnolipids were eluted with 99% isopropanol using a continuous flow (HPLC pump 114 M, Beckman Coulter, Inc., Brea, CA, USA). The organic solvent was evaporated in a freeze dryer (Alpha I-5, Martin Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz, Germany).
Theoretical capacity estimation
The flux balance analysis was carried out using the software Insilico Discovery (version 3.2.0, Insilico Biotechnology AG, Stuttgart, Germany). The provided metabolic network used for simulations was modified to represent the reaction network of P. putida (Additional file 1).
The following reactions were added to the P. putida model:
| (A) |
| (B) |
| (C) |
A linear optimization for rhamnolipid-production with simultaneous minimization of total fluxes was carried out. The rhamnolipid production rate was simulated with different carbon substrates (glucose, glycerol, sucrose, and octanoate). To ensure comparability of the results, the unit Cmol, which normalizes the rhamnolipid production rate to the amount of carbon atoms present in the carbon substrate was chosen. The substrate uptake was varied between 0 and 120 mCmol/(gCDW h). The maintenance metabolism, characterized through the simple reaction of ATP to ADP, was varied in the range of 0 to 50 mmol/(gCDW h). Blank et al. [34] described a value for the non-growth associated maintenance of 10.2 mmol ATP/(gCDW h) for P. putida DOT-T1E. The considerably higher upper limit of 50 mmol ATP/(gCDW h) accounts for scenarios of extra stress, e.g., for metabolic cost of handling high rhamnolipid concentrations. The chosen values for the growth rates were 0 1/h, 0.4 1/h and 0.8 1/h, reflecting ideal production condition, growth observed during rhamnolipid production, and maximal growth of P. putida on glucose [68]. Additionally all occurring fluxes were limited to a maximal value of 120 mCmol/(gCDW h). Furthermore, variation of the fluxes through the pathways ED pathway, TCA cycle, and PP pathway were examined. In addition, an alternative glucose uptake system, the phosphotransferase system, and a complemented EMP pathway (insertion of a phosphofructokinase reaction for example encoded on a fructose utilization operon by fruK (PP0794), catalyzing the conversion of glucose-6P to glucose-1,6P) were simulated.
Determination of fermentation kinetics
The growth kinetic was described mathematically using a logistic growth model. Logistic growth of pseudomonads had been previously reported for rhamnolipid producing wild type P. aeruginosa growing on sunflower oil [55].
The biomass concentration X was described using equation 1, where X0 is the initial biomass concentration, Xadd the additional biomass concentration, t0 the time after which half of Xadd is formed, and b is a curve form coefficient.
| (1) |
The experimental data for the rhamnolipid and glucose concentrations could be described with equations 2 and 3, where rRL is the specific rhamnolipid production rate [grhamnolipid/(gCDW h)] and rGlucose is the specific glucose uptake rate [gglucose/(gCDW h)].
| (2) |
| (3) |
A multivariable least squares fit was used to illustrate the development of all three fermentation parameters depending on each other.
Prior to utilizing the described procedure, to fit glucose and rhamnolipid concentrations, two more attempts applying different models were carried out (Additional file 2).
List of abbreviations used
ACP: Acyl carrier protein; HAA: 3-(3-hydroxyalkanoyloxy)alkanoate; au: Arbitrary units; CDW: Cell dry weight; CE: Collision energy; CoA: Coenzyme A; DAD: Diode array detector; ED: Entner-Doudoroff; EMP: Embden-Meyerhof-Parnas; HPLC-ESI-MS: High performance liquid chromatography electrospray ionization mass spectrometry; IPTG: Isopropyl β-D-1-thiogalactopyranoside; LB: Lysogeny broth; PHA: Polyhydroxyalkanoate; PP: Pentose phosphate; PPGAS: Protease peptone-glucose-ammonium salt medium; RP-HPLC-CAD: Reversed phase high performance liquid chromatography corona charged aerosol detection; TCA: Tricarboxylic acid; TFA: Trifluoracetic acid; TLC: Thin layer chromatography.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AW carried out the molecular biology and the early strain characterization and drafted parts of the manuscript, TT performed the computational flux balance analysis and drafted most of the manuscript, TTA carried out the growth experiments and fermentation kinetic analysis and drafted parts of the manuscript, PW and JH executed the toxicity experiments and also participated in drafting the manuscript, CM coordinated the toxicity experiments and critically read the manuscript, BK carried out the fermentations and some downstream processing experiments, RW coordinated the fermentations and the downstream processing and critically read the manuscript, MZ executed the preparation of the mono- and di-rhamnolipid standards including fermentation procedures and downstream processing, SW took part in initiating the research done for this manuscript and also in coordination of molecular biologic cloning experiments and critically read the manuscript, RH coordinated the preparation of the standards and took part in drafting the manuscript, CS read the manuscript, FR initiated the research and coordinated the molecular biologic cloning experiments and critically read the manuscript, and LMB coordinated the growth experiments and the flux balance analysis, critically read the manuscript, and designed and coordinated the study. All authors read and approved the final manuscript.
Supplementary Material
Tabular presentation of the in silico reaction network of Pseudomonas putida. A list of all reactions implemented in the in silico model of rhamnolipid producing P. putida.
Calculations and assumptions carried out in order to justify the supposition that rhamnolipid production is uncoupled from growth. Detailed presentation of the calculations carried out and the assumptions made prior to align the equations described in the present work leading to the conclusion that rhamnolipid production is independent of growth.
Contributor Information
Andreas Wittgens, Email: a.wittgens@fz-juelich.de.
Till Tiso, Email: till.tiso@rwth-aachen.de.
Torsten T Arndt, Email: torsten.arndt@bci.tu-dortmund.de.
Pamela Wenk, Email: wenk@m2p-labs.com.
Johannes Hemmerich, Email: hemmerich@m2p-labs.com.
Carsten Müller, Email: mueller@m2p-labs.com.
Rolf Wichmann, Email: rolf.wichmann@bci.tu-dortmund.de.
Benjamin Küpper, Email: Benjamin.Kuepper@bci.tu-dortmund.de.
Michaela Zwick, Email: michaela.zwick@kit.edu.
Susanne Wilhelm, Email: s.wilhelm@fz-juelich.de.
Rudolf Hausmann, Email: rudolf.hausmann@kit.edu.
Christoph Syldatk, Email: christoph.syldatk@kit.edu.
Frank Rosenau, Email: frank.rosenau@uni-ulm.de.
Lars M Blank, Email: lars.blank@rwth-aachen.de.
Acknowledgements
The authors are grateful to Isabel Escapa and Auxiliadora Prieto for providing strain P. putida KT42C1. We also express our gratitude to Holger Müller (BlueSens) for providing the 12C/13C CO2 off-gas sensor and for his invaluable insights into off-gas analysis. The Deutsche Bundesstiftung Umwelt (DBU) is gratefully acknowledged for providing financial support.
References
- Lang S, Wullbrandt D. Rhamnose lipids - biosynthesis, microbial production and application potential. Appl Microbiol Biotechnol. 1999;51:22–32. doi: 10.1007/s002530051358. [DOI] [PubMed] [Google Scholar]
- Maier RM, Soberon-Chavez G. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl Microbiol Biotechnol. 2000;54:625–633. doi: 10.1007/s002530000443. [DOI] [PubMed] [Google Scholar]
- Gautam KK, Tyagi VK. Microbial surfactants: A review. J Oleo Sci. 2006;55:155–166. doi: 10.5650/jos.55.155. [DOI] [Google Scholar]
- Rahman PKSM, Gakpe E. Production, characterisation and applications of biosurfactants - Review. Biotechnology. 2008;7:360–370. doi: 10.3923/biotech.2008.360.370. [DOI] [Google Scholar]
- Abalos A, Pinazo A, Infante MR, Casals M, García F, Manresa A. Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir. 2001;17:1367–1371. doi: 10.1021/la0011735. [DOI] [Google Scholar]
- Benincasa M, Contiero J, Manresa MA, Moraes IO. Rhamnolipid production by Pseudomonas aeruginosa LBI growing on soapstock as the sole carbon source. J Food Eng. 2002;54:283–288. doi: 10.1016/S0260-8774(01)00214-X. [DOI] [Google Scholar]
- Haba E, Pinazo A, Jauregui O, Espuny MJ, Infante MR, Manresa A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol Bioeng. 2003;81:316–322. doi: 10.1002/bit.10474. [DOI] [PubMed] [Google Scholar]
- Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim Biophys Acta. 1999;1440:244–252. doi: 10.1016/s1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- Jarvis FG, Johnson MJ. A glyco-lipide produced by Pseudomonas aeruginosa. J Am Chem Soc. 1949;71:4124–4126. doi: 10.1021/ja01180a073. [DOI] [Google Scholar]
- Abdel-Mawgoud AM, Lépine F, Déziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol. 2010;86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology. 2000;146:2803–2814. doi: 10.1099/00221287-146-11-2803. [DOI] [PubMed] [Google Scholar]
- Zhu K, Rock CO. RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J Bacteriol. 2008;190:3147–3154. doi: 10.1128/JB.00080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AJ. Differences between lipopolysaccharide compositions of plant pathogenic and saprophytic Pseudomonas species. Appl Environ Microbiol. 1984;48:31–35. doi: 10.1128/aem.48.1.31-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal O, Lüderitz O. Chemische Erforschung von Lipopolysacchariden gramnegativer Bakterien. Angew Chem. 1954;66:407–417. doi: 10.1002/ange.19540661312. [DOI] [Google Scholar]
- Déziel E, Lépine F, Milot S, Villemur R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology. 2003;149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Fiechter A, Reiser J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberón-Chávez G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol. 2001;40:708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- Rehm BHA, Mitsky TA, Steinbüchel A. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by Pseudomonads: Establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl Environ Microbiol. 2001;67:3102–3109. doi: 10.1128/AEM.67.7.3102-3109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitermann FP. PhD thesis. Universität Karlsruhe, Fakultät für Chemieingenieurwesen und Verfahrenstechnik; 2008. Entwicklung und Optimierung eines biotechnologischen Prozesses zur Herstellung mikrobieller Rhamnolipide auf Basis nachwachsender Rohstoffe. [Google Scholar]
- Müller MM, Hörmann B, Syldatk C, Hausmann R. Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl Microbiol Biotechnol. 2010;87:167–174. doi: 10.1007/s00253-010-2513-7. [DOI] [PubMed] [Google Scholar]
- Soberón-Chávez G, Lépine F, Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- Medina G, Juárez K, Díaz R, Soberón-Chávez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR encoding a quorum-sensing regulatory protein. Microbiology. 2003;149:3073–3081. doi: 10.1099/mic.0.26282-0. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim L, Ward OP, Li ZY. Production and characterisation of a biosurfactant isolated from Pseudomonas aeruginosa UW-1. J Ind Microbiol Biotechnol. 1997;19:232–238. doi: 10.1038/sj.jim.2900450. [DOI] [PubMed] [Google Scholar]
- Reiling HE, Thanei-Wyss U, Guerra-Santos LH, Hirt R, Kappeli O, Fiechter A. Pilot plant production of rhamnolipid biosurfactant by Pseudomonas aeruginosa. Appl Environ Microbiol. 1986;51:985–989. doi: 10.1128/aem.51.5.985-989.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toribio J, Escalante AE, Soberón-Chávez G. Rhamnolipids: Production in bacteria other than Pseudomonas aeruginosa. Eur J Lipid Sci Technol. 2010;112:1082–1087. doi: 10.1002/ejlt.200900256. [DOI] [Google Scholar]
- Ochsner UA, Reiser J, Fiechter A, Witholt B. Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl Environ Microbiol. 1995;61:3503–3506. doi: 10.1128/aem.61.9.3503-3506.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Fang X, Bai B, Liang X, Shuler PJ, Goddard WA III, Tang Y. Engineering bacteria for production of rhamnolipid as an agent for enhanced oil recovery. Biotechnol Bioeng. 2007;98:842–853. doi: 10.1002/bit.21462. [DOI] [PubMed] [Google Scholar]
- Cabrera-Valladares N, Richardson A-P, Olvera C, Treviño LG, Déziel E, Lépine F, Soberón-Chávez G. Monorhamnolipids and 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Appl Microbiol Biotechnol. 2006;73:187–194. doi: 10.1007/s00253-006-0468-5. [DOI] [PubMed] [Google Scholar]
- Cha M, Lee N, Kim M, Kim M, Lee S. Heterologous production of Pseudomonas aeruginosa EMS1 biosurfactant in Pseudomonas putida. Bioresour Technol. 2008;99:2192–2199. doi: 10.1016/j.biortech.2007.05.035. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP, Fouts DE, Gill SR, Pop M, Holmes M. et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Eggert T, van Pouderoyen G, Dijkstra BW, Jaeger KE. Lipolytic enzymes LipA and LipB from Bacillus subtilis differ in regulation of gene expression, biochemical properties, and three-dimensional structure. FEBS L. 2001;502:89–92. doi: 10.1016/S0014-5793(01)02665-5. [DOI] [PubMed] [Google Scholar]
- Timmis KN. Pseudomonas putida: A cosmopolitan opportunist par excellence. Environ Microbiol. 2002;4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- Blank LM, Ionidis G, Ebert BE, Bühler B, Schmid A. Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS J. 2008;275:5173–5190. doi: 10.1111/j.1742-4658.2008.06648.x. [DOI] [PubMed] [Google Scholar]
- Hervas AB, Canosa I, Santero E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol. 2008;190:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder P, Dutzler R, Dumas F, Koebnik R, Schirmer T. Sucrose transport through maltoporin mutants of Escherichia coli. Protein Eng. 2001;14:943–948. doi: 10.1093/protein/14.11.943. [DOI] [PubMed] [Google Scholar]
- Joardar V, Lindeberg M, Jackson RW, Selengut J, Dodson R, Brinkac LM, Daugherty SC, Deboy R, Durkin AS, Giglio MG. et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol. 2005;187:6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF. et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Zhou XR, Liu Q, Chen GQ. Biosynthesis of polyhydroxyalkanoate homopolymers by Pseudomonas putida. Appl Microbiol Biotechnol. 2011;89:1497–1507. doi: 10.1007/s00253-010-2964-x. [DOI] [PubMed] [Google Scholar]
- Choi MH, Xu J, Gutierrez M, Yoo T, Cho YH, Yoon SC. Metabolic relationship between polyhydroxyalkanoic acid and rhamnolipid synthesis in Pseudomonas aeruginosa: Comparative 13C NMR analysis of the products in wild-type and mutants. J Biotechnol. 2011;151:30–42. doi: 10.1016/j.jbiotec.2010.10.072. [DOI] [PubMed] [Google Scholar]
- de Eugenio LI, Escapa IF, Morales V, Dinjaski N, Galán B, García JL, Prieto MA. The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ Microbiol. 2010;12:207–221. doi: 10.1111/j.1462-2920.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Rehm BHA, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- Soberón-Chávez G, Aguirre-Ramírez M, Sánchez R. The Pseudomonas aeruginosa RhlA enzyme is involved in rhamnolipid and polyhydroxyalkanoate production. J Ind Microbiol Biotechnol. 2005;32:675–677. doi: 10.1007/s10295-005-0243-0. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Zhang MJ, Zhang G, Chen GQ. Production of 3-hydroxydecanoic acid by recombinant Escherichia coli HB101 harboring phaG gene. Antonie van Leeuwenhoek. 2004;85:93–101. doi: 10.1023/B:ANTO.0000020275.23140.ca. [DOI] [PubMed] [Google Scholar]
- Timm A, Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxyalkanoic acid) gene locus of Pseudomonas aeruginosa PAO1. Eur J Biochem. 1992;209:15–30. doi: 10.1111/j.1432-1033.1992.tb17256.x. [DOI] [PubMed] [Google Scholar]
- Franklin FC, Bagdasarian M, Bagdasarian MM, Timmis KN. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baev MV, Baev D, Radek AJ, Campbell JW. Growth of Escherichia coli MG1655 on LB medium: determining metabolic strategy with transcriptional microarrays. Appl Microbiol Biotechnol. 2006;71:323–328. doi: 10.1007/s00253-006-0392-8. [DOI] [PubMed] [Google Scholar]
- Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends V, Ebbels TM, Williams HD, Bundy JG. Time-resolved metabolic footprinting for nonlinear modeling of bacterial substrate utilization. Appl Environ Microbiol. 2009;75:2453–2463. doi: 10.1128/AEM.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GJ, Lee IY, Choi DK, Yoon SC, Park YH. High cell density cultivation of Pseudomonas putida BM01 using glucose. J Microbiol Biotechnol. 1996;6:221–224. [Google Scholar]
- Lee SY, Wong HH, Choi J, Lee SH, Lee SC, Han CS. Production of medium-chain-length polyhydroxyalkanoates by high-cell-density cultivation of Pseudomonas putida under phosphorus limitation. Biotechnol Bioeng. 2000;68:466–470. doi: 10.1002/(SICI)1097-0290(20000520)68:4<466::AID-BIT12>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Sun Z, Ramsay JA, Guay M, Ramsay BA. Automated feeding strategies for high-cell-density fed-batch cultivation of Pseudomonas putida KT2440. Appl Microbiol Biotechnol. 2006;71:423–431. doi: 10.1007/s00253-005-0191-7. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Bakhit R, Daniels L, Mayerl F, Pickenhagen W. Microbially produced rhamnolipid as a source of rhamnose. Biotechnol Bioeng. 1989;33:365–368. doi: 10.1002/bit.260330316. [DOI] [PubMed] [Google Scholar]
- List of resources: soft commodities. http://www.onvista.de/rohstoffe/rohstoffliste/soft-commodities/
- Müller MM, Hörmann B, Kugel M, Syldatk C, Hausmann R. Evaluation of rhamnolipid production capacity of Pseudomonas aeruginosa PAO1 in comparison to the rhamnolipid over-producer strains DSM 7108 and DSM 2874. Appl Microbiol Biotechnol. 2010;89:585–592. doi: 10.1007/s00253-010-2901-z. [DOI] [PubMed] [Google Scholar]
- Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Wenzel SC, Gross F, Zhang Y, Fu J, Stewart AF, Müller R. Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via red/ET recombineering. Chem Biol. 2005;12:349–356. doi: 10.1016/j.chembiol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Ramos-Diáz MA, Ramos JL. Combined physical and genetic map of the Pseudomonas putida KT2440 chromosome. J Bacteriol. 1998;180:6352–6363. doi: 10.1128/jb.180.23.6352-6363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L. et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Eltis L, Kessler B, Timmis KN. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Wild M, Caro AD, Hernández AL, Miller RM, Soberón-Chávez G. Selection and partial characterization of a Pseudomonas aeruginosa mono-rhamnolipid deficient mutant. FEMS Microbiol Lett. 1997;153:279–285. doi: 10.1111/j.1574-6968.1997.tb12586.x. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran EV, BeMiller JN. Constituent analysis of glycosaminoglycans. New York: Academic Press; 1980. [Google Scholar]
- Ochsner UA. PhD thesis. ETH Zürich, Swiss Federal Institute of Technology Zurich; 1993. Genetics and biochemistry of Pseudomonas aeruginosa rhamnolipid biosurfactant synthesis. [Google Scholar]
- Trummler K, Effenberger F, Syldatk C. An integrated microbial/enzymatic process for production of rhamnolipids and L-(+)-rhamnose from rapeseed oil with Pseudomonas sp. DSM 2874. Eur J Lipid Sci Technol. 2003;105:536–571. doi: 10.1002/ejlt.200300782. [DOI] [Google Scholar]
- del Castillo T, Ramos JL. Simultaneous catabolite repression between glucose and toluene metabolism in Pseudomonas putida is channeled through different signaling pathways. J Bacteriol. 2007;189:6602–6610. doi: 10.1128/JB.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tabular presentation of the in silico reaction network of Pseudomonas putida. A list of all reactions implemented in the in silico model of rhamnolipid producing P. putida.
Calculations and assumptions carried out in order to justify the supposition that rhamnolipid production is uncoupled from growth. Detailed presentation of the calculations carried out and the assumptions made prior to align the equations described in the present work leading to the conclusion that rhamnolipid production is independent of growth.