Abstract
Long INterspersed Element-1 (LINE-1) retrotransposons comprise 17% of the human genome, and move by a potentially mutagenic “copy and paste” mechanism via an RNA intermediate. Recently, the retrotransposition-mediated insertion of a new transcript was described as a novel cause of genetic disease, Duchenne muscular dystrophy, in a Japanese male. The inserted sequence was presumed to derive from a single-copy, non-coding RNA transcribed from chr. 11q22.3 that retrotransposed into the dystrophin gene. Here we demonstrate that a non-reference full-length LINE-1 is situated in the proband and maternal genome at chr. 11q22.3, directly upstream of the sequence, whose copy was inserted into the dystrophin gene. This LINE-1 is highly active in a cell culture assay. LINE-1 insertions are often associated with 3’ transduction of adjacent genomic sequences. Thus, the likely explanation for the mutagenic insertion is a LINE-1-mediated 3’ transduction with severe 5’ truncation. This is the first example of LINE-1-induced human disease caused by an “orphan” 3’ transduction.
Keywords: LINE-1, retrotransposon, 3’ transduction, dystrophin, Duchenne muscular dystrophy
Retrotransposons (“jumping genes”) are highly abundant mobile genomic elements. In particular, the long interspersed element-1 (LINE-1 or L1) class comprises 17% of the human genome. A full-length human LINE-1 is about 6 kilobases and contains a 5’ untranslated region (UTR) encoding promoter activity (Swergold, 1990), two open reading frames (ORFs) separated by a spacer (Dombroski et al., 1991), a 3’ UTR, and a poly(A) tail. ORF1 encodes a protein with RNA binding (Martin, 1991; Hohjoh and Singer, 1996) and nucleic acid chaperone activity (Martin and Bushman, 2001), while ORF2 is a protein with endonuclease (Feng et al., 1996) and reverse transcriptase activities (Mathias et al., 1991). LINE-1s are autonomous non-long terminal repeat retrotransposons that move by a potentially mutagenic “copy and paste” mechanism via an RNA intermediate that is reverse transcribed and inserted into the genome (reviewed in Goodier and Kazazian, 2008). LINE-1s can also cause disease indirectly, through mobilization of the non-autonomous Alu and SVA retrotransposons (Dewannieux et al., 2003; Ostertag et al., 2003; Hancks et al., 2011).
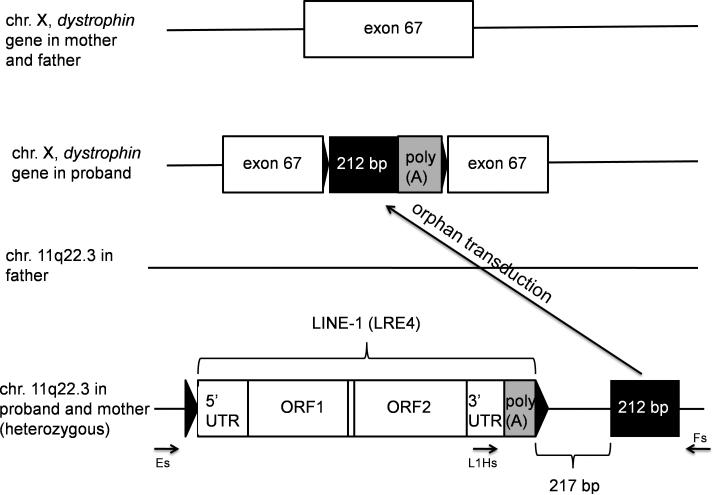
Recently, the retrotransposition-mediated insertion of a new cDNA was described as a novel cause of genetic disease, Duchenne muscular dystrophy (MIM# 310200), in a Japanese boy (Awano et al., 2010). In this work, it was presumed that the inserted sequence was derived from a non-repetitive non-coding RNA transcribed from chr. 11q22.3 that was reverse transcribed and integrated in the antisense orientation into exon 67 of the dystrophin gene on chr. X, causing exon 67 skipping. The whole insertion was 327 bp long, of which 212 bp was identical to a sequence on chr. 11q22.3 (chr11:105,479,198-105,479,409 of hg19/NCBI Build 37.1 Feb 2009), while the remaining 115 bp was a poly(T) stretch. The inserted sequence had hallmarks of LINE-1 retrotransposition, namely a poly(A) tail complementary to the poly(T) stretch, target site duplication flanking the insertion in dystrophin exon 67, and insertion at a near-consensus LINE-1 endonuclease site (TTTT/CA instead of TTTT/AA) (Awano et al., 2010). Another LINE-1-related phenomenon is 3’ transduction, the co-mobilization of DNA sequences downstream of LINE-1s as a consequence of transcriptional read-through due to the weak LINE-1 poly(A) signal (Holmes et al., 1994; Moran et al., 1996). Because LINE-1 insertions are often associated with 3’ transductions (Moran et al., 1999; Goodier et al., 2000; Pickeral et al., 2000), we hypothesized that the insertion in the patient might result from such an event. However, no LINE-1 was present in the DNA upstream of the single copy sequence from chr. 11q22.3 in the human reference genome (hg19).
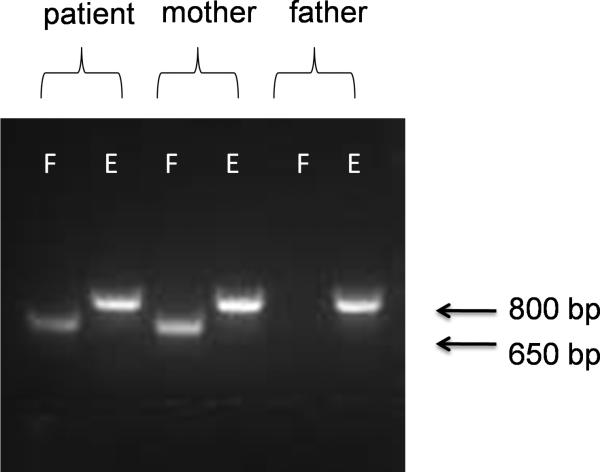
On the other hand, a LINE-1 directly upstream of the transduced sequence on chr. 11q22.3 was present in one Japanese individual out of 15 unrelated individuals in a LINE-1 targeted resequencing dataset generated in our laboratory (Ewing and Kazazian, 2010). Based on bioinformatic analysis, this LINE-1 was absent from the 185 HapMap phase I individuals, including 30 individuals with self-reported Japanese ancestry, whose genomes were sequenced by the 1000 Genomes Consortium (1000 Genomes Project Consortium, 2010; Stewart et al., 2011). Additionally, it was not detected in our independent whole-genome analysis of individuals included in the 1000 Genomes project (Ewing and Kazazian, 2011). The lack of evidence for this insertion in multiple analyses indicates that it may represent a population-restricted variant present at low allele frequency in the general population. To evaluate further its gene frequency in the Japanese population, we analyzed 80 Japanese chr. 11q22.3 sites by PCR and found 5 that contained the LINE-1, corresponding to a gene frequency of 6% (data not shown). Here, we demonstrate through PCR-based analysis that this non-reference LINE-1 is full-length and is situated in the maternal and proband genomes at chr. 11q22.3, 217 bp 5’ of the retrotransposed 212 bp sequence that was inserted into the dystrophin gene (Fig. 1A and 1B). Thus, the most likely explanation for the mutagenic insertion is a LINE-1-induced 3’ transduction event from chr. 11q22.3 with severe 5’ truncation upon insertion, such that only part of the 3’ transduced sequence was inserted (Fig. 1A). What is less clear is in which cell lineage and at what time-point the LINE-1 RNA was reverse transcribed and inserted into the genome. Integration into the dystrophin gene most likely occurred in one or more of the mother's germ cells or early during the proband's development, because the mother's blood was negative for the presence of the 3’ transduced sequence.
Fig. 1A.
Orphan 3’ transduction event in the patient. The transduced sequence is in black. Black triangles indicate target site duplications (3 bp in exon 67, 21 bp at chr. 11q22.3).
Fig. 1B.
PCR analysis of the 3’ junctions of the presumed progenitor (LRE4) of the DNA sequence inserted into the dystrophin gene with the primers indicated on Fig. 1A. Genotyping primer pairs L1Hs (5’-GGGAGATATACCTAATGCTAGATGACAC-3’) and Fs (5’-CGTTACATTTCACCACAGATTG-3’) amplify the filled (F) site (around 686 bp), while primers Es (5’-AGCACAATACCTTGCACATTAG-3’) and Fs amplify only the empty (E) site (828 bp) due to short primer extension time. 40 Japanese individuals (80 chr. 11s) were also genotyped for the LRE4 allele and five were positive (data not shown). Thus, this allele is an uncommon variant in the Japanese population. PCR conditions are available upon request.
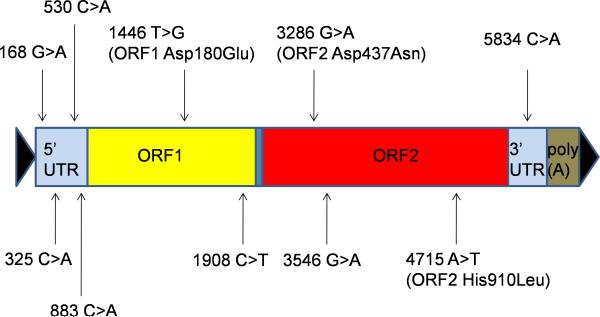
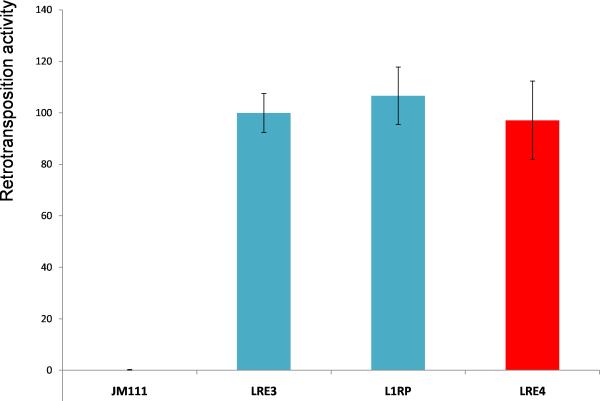
To characterize the LINE-1 progenitor element on chr. 11q22.3, hereafter referred to as LRE4 [LINE-1 Retrotransposable Element 4; BankIt1482137 LRE4 JN698885 (GenBank, http://www.ncbi.nlm.nih.gov/Genbank/)], we PCR-amplified it from the patient's blood DNA and sequenced it from multiple independent PCR products, all of which had the identical sequence. LRE4 is a full length Ta-1d element and contains 10 nucleotide alterations relative to LRE3, the most active LINE-1 isolated to date, and the only LINE-1 that is an exact match to the active LINE-1 amino acid consensus sequence (Brouha et al., 2002; Brouha et al., 2003). Three of the nucleotide substitutions in LRE4 resulted in amino acid changes relative to LRE3 (Fig. 2A). All LRE4 nucleotide changes were identified in other LINE-1s present in the hg19 assembly. However, no reference LINE-1 contained all 10 nucleotide alterations, nor was it present in a recent dataset of active LINE-1s (Beck et al., 2010). To determine the retrotransposition activity of LRE4, we cloned it into a plasmid with an EGFP retrotransposition indicator cassette, creating 99-LRE4-EGFP-Puro. This cassette was designed so that translation of the EGFP reporter gene occurs only after LINE-1 reverse transcription and integration of its cDNA copy into the genome – that is, after a retrotransposition event (Moran et al., 1996; Ostertag et al., 2000). Upon transfection of HEK293T cells with 99-LRE4-EGFP-Puro, and selection for transfected cells with puromycin, retrotransposition events were evaluated by FACS analysis of EGFP expressing cells (Ostertag et al., 2000). 99-LRE4-EGFP-Puro demonstrated a retrotransposition activity comparable to plasmids containing LRE3 or L1RP (Fig. 2B), indicating that it is a highly active or “hot” retrotransposon (Brouha et al., 2003).
Fig. 2A.
Schematic diagram of LRE4. Nucleotide changes differing from LRE3 (Brouha et al., 2002) are indicated. LRE4 was PCR-amplified from the patient's blood DNA with iProof High Fidelity DNA Polymerase (Bio-Rad) and sequenced. LRE4 belongs to the Ta-1d class of human-specific LINE-1s (Boissinot et al., 2000), and the Ta class elements cause most de novo pathogenic human insertions (reviewed in Chen et al., 2005).
Fig. 2B.
Retrotransposition activity of LRE4 (99-LRE4-EGFP-Puro) in HEK293T cells. LRE4 was PCR amplified with iProof High-Fidelity DNA Polymerase (Bio-Rad) using the primers 5’-CGTTACATTTCACCACAGATTG-3’ and 5’-AAGTAAAATAGAGGTTTTGGGGG-3’. The PCR product was cloned with the TOPO XL PCR Cloning Kit (Invitrogen) and was subsequently cloned to replace L1RP in the 99-RPS-EGFP-Puro reporter construct with BstZ17I and NotI-HF double digestion and ligation. HEK293T cells were transfected with 1 μg of each plasmid in a 6-well plate using Fugene 6 (Roche). Transfected cells were selected with puromycin, and retrotransposition events were evaluated four days later by FACS analysis of EGFP expressing cells. 99-RPS is the highly active L1RP cloned into a modified pCEP4 plasmid lacking the CMV promoter. Retrotransposition activity of 99-LRE3-EGFP-Puro is set to 100%. JM111 has the same sequence as 99-RPS, except that it contains two point mutations in ORF1 that abolish retrotransposition in cis (Moran et al., 1996) and serves as a negative control. Retrotransposition activity of L1RP and LRE3 was similar in HEK293T cells. Standard deviation of two independent experiments done in triplicate is shown.
This is the fifth case of LINE-1-driven insertional mutagenesis of the dystrophin gene (Narita et al., 1993; Holmes et al., 1994; Yoshida et al., 1998; Musova et al., 2006; and Awano et al., 2010 together with the current study). Therefore, mutation analyses of this gene should take into account large insertions mediated by LINE-1s. Although, LINE-1s are often truncated at their 5’ end, this is the first example of LINE-1-induced human disease caused by an orphan 3’ transduction, that is, a LINE-1-mediated insertion lacking LINE-1 sequence. Two non-disease causing retrotransposition events of gene fragments have also been described that may have arisen by LINE-1-mediated 3’ transduction, with the transducing LINE-1 being lost (Rozmahel et al., 1997; Ejima and Yang, 2003). In a previous report we showed that a mutagenic insertion into the α-spectrin gene was the result of an SVA-mediated orphan 3’ transduction (Ostertag et al., 2003). Therefore, any insertion of a non-repetitive sequence bearing the hallmarks of retrotransposition should be further investigated for a LINE-1- or SVA-mediated transduction event, as previously postulated by Moran et al. (1999). Our results indicate that LRE4 is a highly active, polymorphic retrotransposon with a pathogenic history.
Acknowledgments
We thank John L. Goodier and Prabhat K. Mandal for their great insights into the project and for their comments on the manuscript. Ling Cheung and David Sigmon are acknowledged for excellent technical assistance. We thank Dr. Christine Beck and Dr. John Moran for sharing their data concerning hot LINE-1s. Research in the Kazazian laboratory is funded by grants from the National Institutes of Health awarded to H.H.K.
Current grant: NIH RC4MH092880
Footnotes
Conflict of interest: None.
References
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano H, Malueka RG, Yagi M, Okizuka Y, Takeshima Y, Matsuo M. Contemporary retrotransposition of a novel non-coding gene induces exon-skipping in dystrophin mRNA. J Hum Genet. 2010;55:785–790. doi: 10.1038/jhg.2010.111. [DOI] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH., Jr. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr. Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Ejima Y, Yang L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet. 2003;12:1321–1328. doi: 10.1093/hmg/ddg138. [DOI] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH., Jr. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH., Jr. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr. Transduction of 3'-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr. Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Musova Z, Hedvicakova P, Mohrmann M, Tesarova M, Krepelova A, Zeman J, Sedlacek Z. A novel insertion of a rearranged L1 element in exon 44 of the dystrophin gene: Further evidence for possible bias in retroposon integration. Biochem Biophys Res Commun. 2006;347:145–149. doi: 10.1016/j.bbrc.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Narita N, Nishio H, Kitoh Y, Ishikawa Y, Ishikawa Y, Minami R, Nakamura H, Matsuo M. Insertion of a 5' truncated L1 element into the 3' end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest. 1993;91:1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozmahel R, Heng HH, Duncan AM, Shi XM, Rommens JM, Tsui LC. Amplification of CFTR exon 9 sequences to multiple locations in the human genome. Genomics. 1997;45:554–561. doi: 10.1006/geno.1997.4968. [DOI] [PubMed] [Google Scholar]
- Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, Stutz AM, Urban AE, Grubert F, Lam HY, Lee WP, Busby M, Indap AR, Garrison E, Huff C, Xing J, Snyder MP, Jorde LB, Batzer MA, Korbel JO, Marth GT, 1000 Genomes Project A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Nakamura A, Yazaki M, Ikeda S, Takeda S. Insertional mutation by transposable element, L1, in the DMD gene results in X-linked dilated cardiomyopathy. Hum Mol Genet. 1998;7:1129–1132. doi: 10.1093/hmg/7.7.1129. [DOI] [PubMed] [Google Scholar]