Abstract
The visualization and quantification of mitochondria-associated proteins with high power microscopy methods is of particular interest to investigate protein architecture in this organelle. We report the usage of a custom-made STimulated Emission Depletion (STED) fluorescence nanoscope with ~20 nm lateral resolution for protein mapping of Percoll-purified viable mitochondria from murine heart. Using this approach, we were able to quantify and resolve distinct protein clusters within mitochondria; specifically, cytochrome c oxidase subunit 2 is distributed in clusters of ~20 and ~28 nm; whereas the voltage dependent anion channel 1 displays four size distributions of ~22, ~33, ~55 and ~83 nm.
Keywords: STED, mitochondria, subdiffraction-resolution, imaging, VDAC1, Cox2
1. INTRODUCTION
Mitochondria participate in key metabolic reactions, regulate Ca2+ signaling, and play a central role in apoptosis (Dimmer and Scorrano, 2006; Liu et al., 1996; Yang et al., 1997). In the heart, mitochondria can determine myocardium survival and protection, whilst mitochondria dysfunction contributes to heart disease (Baines, 2010). To understand the details of these mechanisms it is important to map mitochondrial-associated proteins at nanometer resolution as localization is intimately related to functional performance.
STimulated Emission Depletion (STED) fluorescence nanoscopy with ~40-80 nm lateral resolution in biological samples offers the opportunity to scrutinize the distribution of proteins in subcellular compartments or organelles using common immunocytochemistry techniques and tagged proteins (Kellner et al., 2007; Neumann et al., 2010; Watanabe et al., 2011; Willig et al., 2006). STED has the advantage over electron microscopy that it reaches nanometer resolution while at the same time maintains the microscopic scale.
Only recently, has STED been utilized to image a few mitochondrial proteins. Native hexokinase I was observed in mitochondria of an osteosarcoma cell line (U2OS) (Neumann et al., 2010); while ectopically expressed TOM20 was imaged delineating mitochondria periphery in a ring-like arrangement in Caenorhabditis elegans (Watanabe et al., 2011), and expressed voltage dependent anion channels (VDAC) 1-3 were visualized in U2OS cells with VDAC3 forming clusters of ~40 to 90 nm (Neumann et al., 2010).
The objective of this work was to map with STED nanoscopy two classical mitochondria proteins in cardiac mitochondria: VDAC1, an outer mitochondrial membrane protein that serves as an interface between cellular and mitochondrial metabolism (Shoshan-Barmatz and Ben-Hail, 2011); and cytochrome c oxidase (complex IV of the respiratory chain) located in the mitochondrial inner membrane and assembled by 13 subunits in humans. Specifically, we imaged cytochrome c oxidase's subunit 2 (Cox2) that forms part of the catalytic core of the enzyme (Brzezinski and Johansson, 2010; Mick et al., 2011).
2. METHODS
2.1. Antibodies
Primary antibodies were used against VDAC1 (Ab14734, Abcam), Cox2 (A6407, Invitrogen), Cadherin (C1821, anti-Pan Cadherin antibody, Sigma), GM130 (ab1299, Abcam), Lamin b1 (ab16048, Abcam), GRP78 BiP (ab21685, Abcam), L-type Ca2+ channels (α1C) (ACC003, Alomone) and Src (sc-18, Santa Cruz). Secondary antibodies for Western blots were: Alexa Fluor® 680 goat anti-rabbit (A-21109, Invitrogen) and IRDye 800CW conjugated goat anti-mouse (926-32210, LI-COR); and for immunocytochemistry were: Atto 647N goat anti-mouse (50185, Sigma), and Atto 647N goat anti-rabbit (15048, Active Motif).
2.2. Animals
Protocols received institutional approval. Male 3 mo old C57BL/6NCrL mice were injected (i.p.) with heparin (200 IU/Kg). After 20 min, animals were anesthetized with sodium pentobarbital (70 mg/kg, i.p.). The heart was surgically removed and rapidly arrested in ice-cold phosphate buffer saline (PBS) (in mM): 2.7 KCl, 1.5 KH2PO4, 138 NaCl, 8.1 Na2HPO4.
2.3. Isolation and purification of mitochondria
The entire procedure was at 4°C and lasted approximately 75 min. One mouse heart was finely minced and homogenized in isolation buffer A (in mM): 70 sucrose, 210 mannitol, 1 EDTA-Na2, 50 Tris-HCl, pH 7.4 using a Potter-Elvejem homogenizer (10 rapid strokes). The homogenate was transferred into a 2.0 ml Eppendorf tube and centrifuged at 1,300 ×g for 3 min. The supernatant was carefully transferred into a clean 1.5 ml Eppendorf tube and centrifuged at 10,000 ×g for 10 min. The pellet containing crude mitochondria was resuspended in 55 μl of isolation buffer A.
The crude mitochondria preparation was carefully overlaid on 3 ml of 30% (v/v) Percoll (Graham, 2001) in buffer B (in mM): 250 sucrose, 10 HEPES-Na, 1 EDTA-Na2, pH 7.4. The sample was centrifuged at 50,000 ×g for 45 min. After ultracentrifugation, three clear layers were observed which were labeled as M1, M2 and M3 (Fig. 1A). The M1, M2 and M3 fractions were carefully isolated and resuspended in 1 ml of isolation buffer A. Samples were centrifuged at 12,000 ×g for 5 min, mitochondrial pellets were resuspended, and washed two times with 1 ml isolation buffer A. All the fractions were resuspended in buffer A for organelle immunochemistry and in buffer B for functional assays. Purified mitochondria were used within 2 hrs after isolation.
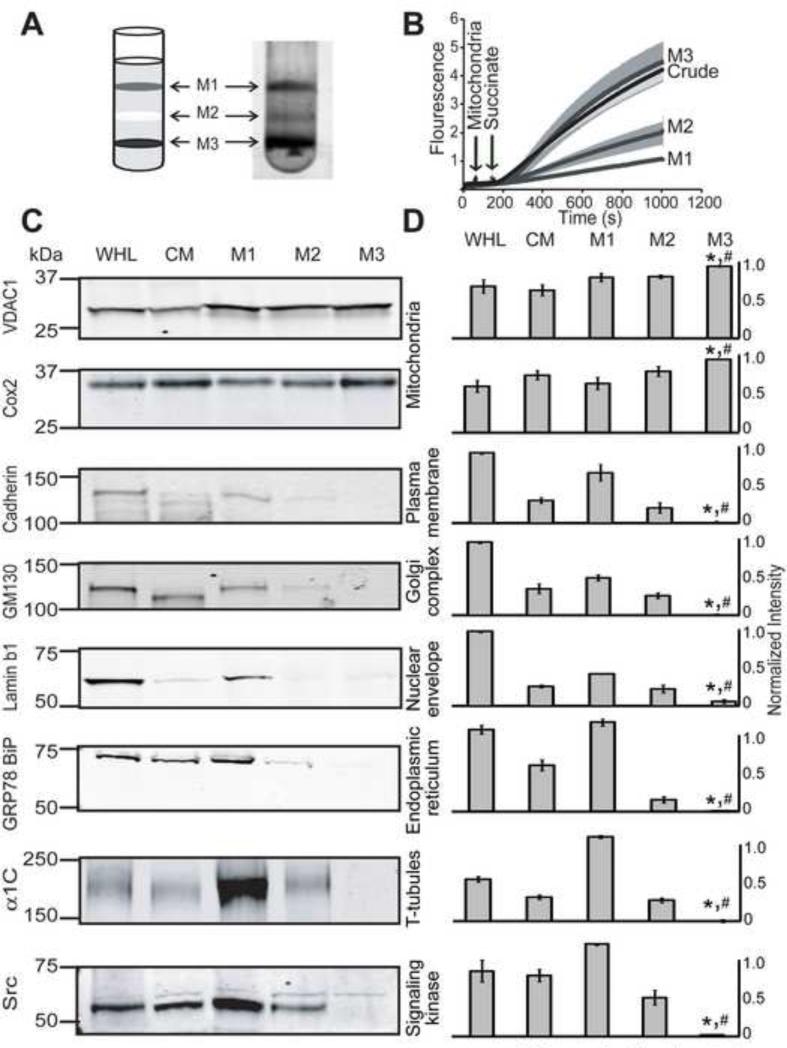
Figure 1. Purification and characterization of mitochondria fractions.
A. Mitochondrial fractions M1, M2 and M3 are shown in the 30% Percoll gradient. B. ROS was measured with the H2O2-sensitive dye amplex red (10 μM). Arrows mark the addition of cardiac mitochondria, crude, M1, M2 and M3 fractions (250 μg), and succinate (3 mM) to the reaction media. M3 but not M2 or M1 mitochondrial fractions produced normal amount of ROS as the crude mitochondria. C. Western blot analysis of whole heart lysate (WHL), crude mitochondria (CM), M1, M2 and M3 fractions. Antibodies and type of protein markers are given in each blot. Equal amounts (50 μg) of proteins were loaded on each lane. D. Quantification of Western blots normalized to the Ponceau S signal in each experiment (n=3 except for VDAC1, n=5 and Cox2, n=7). M3 mitochondrial fraction was enriched with VDAC1 and Cox2, and was practically devoid of other organelle markers including plasma membrane, Golgi, endoplasmic reticulum, T-tubules and nucleus. *, M3 significantly different from WHL; # M3 significantly different from CM (p<0.05).
2.4. Western blot analysis
Isolated mitochondrial pellets from the different fractions were resuspended in lysis buffer (in mM): Tris-HCl 50, NaCl 150, EDTA-Na2 1, EGTA-Na4 1, 1% (v/v) NP-40, 0.5% (w/v) Na-deoxycholate and 0.1% (w/v) SDS, pH 7.4 containing protease inhibitors (1 tablet/50 ml, Roche), and incubated for 1 hour with shaking. Samples were centrifuged at 10,000 xg for 5 min, the supernatants were collected, and the protein concentration was measured. Equal amounts of protein (50 μg) were loaded for 10% SDS-PAGE, and transferred to nitrocellulose membranes. Proper loading was corroborated with Ponceau S staining. Membranes were blocked with 5% (w/v) milk in TBS (150 mM NaCl, 20 mM Tris-Cl pH 7.4,) for 60 min at room temperature. Membranes were washed three times with TBS (5 min at room temperature) and incubated overnight with primary antibodies at 4°C. Primary antibodies were used at the following concentrations: for plasma membrane (Cadherin, 19.4 μg/ml), endoplasmic reticulum (GRP78 BiP, 0.2 μg/ml), nuclear envelop (Lamin b1, 0.2 μg/ml), Golgi (GM130, 0.5 μg/ml), mitochondria (VDAC1, 0.2 μg/ml and Cox 2, 0.1 μg/ml), T-tubules (α1C, 0.45 μg/ml) and for the signaling kinase (Src, 0.04 μg/ml). After washing three times with TBS (5 min, room temperature) membranes were incubated with 0.01 μg/ml secondary antibodies (anti-mouse/rabbit) for 1 hour at room temperature, and washed again three times with TBS for 5 min. Signals were visualized using an infrared fluorescence system (Odyssey™ Imaging System, Li-Cor). The intensity of the bands was measured with Metamorph (Molecular Devices) and normalized to loaded protein using Ponceau S signals.
2.5. Mitochondrial H2O2 measurement
Mitochondrial reactive oxygen species (ROS) generation was measured spectrophotometrically (560 nm excitation and 590 nm emission) by incubating 250 μg of mitochondrial protein in a solution containing (in mM): 20 Tris-HCl, 250 sucrose, 1 EGTA-Na4, 1 EDTA-Na2, and 0.15% (w/v) bovine serum albumin, pH 7.4 at 25°C with continuous stirring. Sodium succinate (3 mM) was used to activate complex II. ROS was measured with the H2O2-sensitive dye amplex red (10 μM) according to the manufacturer's instructions (Invitrogen).
2.6. Labeling of mitochondrial proteins
Purified mitochondria were incubated with 500 nM Mitotracker® Red CMXRos (Invitrogen) for 60 min with shaking, plated drop-wise onto coverslips and incubated at 4°C for 1 hr. Coverslips (0.17 mm thickness; Warner Instruments, Cat no. 64-0712) were coated with 0.1% Poly-L-Lysine (Sigma-Aldrich) for 2 hr at room temperature and washed with phosphate buffered saline (PBS) (in mM, 138 NaCl, 2.7 KCl, pH 7.4) before use. Attached mitochondria were then washed with PBS once and fixed with 4% (w/v) paraformaldehyde in PBS for 10 min at room temperature. After three washes with PBS, mitochondria were permeabilized with 0.5% (v/v) Triton-X 100 in PBS for 10 min at room temperature. Mitochondria were further colabeled with specific antibodies (in 1% (w/v) normal goat serum (NGS), 0.1% (v/v) Triton X-100 in PBS) for mitochondrial (0.2 μg/ml anti-VDAC1 and 0.2 μg/ml anti-Cox2 monoclonal antibodies) and nuclear envelope (0.2 μg/ml anti-Lamin b1 polyclonal antibody) markers. ATTO 647N-labelled secondary antibodies (1 μg/ml anti-mouse and anti-rabbit) in 1% (w/v) NGS, 0.2% (v/v) Triton X-100 in PBS were used. Some mitochondria were only incubated with secondary antibodies as controls. Preparations were sequentially treated with 10, 25, 50 and 97% (v/v) 2,2′-Thiodiethanol (TDE, Sigma-Aldrich) solutions in PBS (pH 7.5) for 5 min each at room temperature (Staudt et al., 2007). TDE solutions were added and aspirated gently to prevent uplifting of mitochondria from the coverslips. Coverslips were mounted onto slides and sealed with regular nail polish. Samples used only for regular confocal microscopy were mounted using ProLong®Gold (P36934, Invitrogen) instead of TDE. Confocal images were acquired with an Olympus confocal microscope using a 60X oil immersion objective with 1.42 NA (PlanAppoN) at a scanning resolution of 0.0575 μm per pixel. STED images were collected as described below.
2.7. STED Microscopy
STED images were acquired with a custom-made STED system using an oil immersion objective (HCX PL APO 100x/1.40-0.70 OIL CS, Leica Germany). A 635 nm pulsed diode laser (LDH-D-C-635, PicoQuant GmbH) was used for excitation. The pulses for STED depletion were delivered by a tunable Ti:sapphire laser (Mai Tai HP, Spectra Physics) set at 780 nm. Fluorescence emission from ATTO 647N–labeled secondary antibodies was collected through a Semrock BrightLine FF01-692/40-25 nm band pass filter in front of a photomultiplier (H7422PA-40, Hamamatsu Photonics K.K.). Images (955 × 960 pixels) were acquired with a 16 kHz line frequency (resonant mirror of 8 kHz) and summed 256 times. Pixel size was ~9.575 nm × 9.575 nm. For a fair comparison between conventional confocal images and STED images, all imaging parameters were kept identical except for the number of summations which was 64 when recording confocal images. For comparison, confocal images were acquired for the same field prior STED imaging.
2.8. Image analysis
Confocal double-labeled images were median filtered and analyzed with custom-made software to determine their protein proximity index as previously described (Li et al., 2010; Wu et al., 2010). For display, STED images were deconvolved by 20 iterations of the Richardson-Lucy deconvolution algorithm and a median filter with a radius of 2 pixels using the plug-in DeconvolutionLab developed for ImageJ by Cédric Vonesch, Raquel Terrés Cristofani and Guillaume Schmit at the Biomedical Image Group (BIG), EPFL, Switzerland. Analysis of cluster size was done using the raw images without any modifications by manually performing line scans across the clusters to measure pixel intensity as a function of distance and calculating the full width at half maximum (FWHM) of the light intensity peaks. Cluster sizes were measured from 500 clusters each from three independent experiments. Histograms were manually fitted using IGOR Pro (WaveMetrics Inc.).
2.9 Statistics
Error bars were plotted as standard errors of the mean (+SEM) for a minimum of three independent experiments. Two-tailed Student's t-test was performed and p<0.05 was considered significantly different.
3. RESULTS
To image VDAC1 and Cox2 in purified murine cardiac mitochondria at the nanoscopic level, our strategy was to first evaluate the purity and viability of our mitochondrial preparation using biochemical and immunochemical approaches followed by STED fluorescence nanoscopy analysis.
3.1. Mitochondrial fractions: purity and functional evaluation
Purified mitochondria were obtained using a 30% self-generated Percoll density gradient containing 250 mM sucrose (Graham, 2001; Zhang et al., 2008). Figure 1A shows three distinct fractions obtained after Percoll separation, named M1, M2 and M3 (Fig. 1A). The purity and viability of the mitochondrial fractions was assessed using three complementary parameters: a) production of reactive oxygen species (ROS), b) usage of organelle markers for Western blot analysis; and c) colocalization with mitotracker which labels mitochondria with intact membrane potential. The viability of M1, M2 and M3 fractions was first tested by measuring ROS production using amplex red, a H2O2-sensitive dye, and succinate (3 mM), a substrate of complex II. As shown in Fig. 1B, mitochondria from M3 fraction maintained the same efficiency to produce ROS as crude mitochondria; however, M1 and M2 had only 25% and 50% of crude mitochondria capacity to produce ROS, respectively. These results indicated to us that M3 fraction was the most viable of the three purified fractions.
Next, M1, M2 and M3 fractions along with crude mitochondria (CM) and whole heart lysate (WHL) were examined for the presence of classical protein markers of different organelles. We used antibodies against VDAC1 and Cox2 for mitochondria, Cadherin for plasma membrane, GM130 for Golgi, Lamin b1 for nuclear envelope, and against GRP78 BiP for endoplasmic reticulum. In addition, we used antibodies against the L-type Ca2+ channel α1C subunit, a T-tubule marker, and for Src, a family of signaling protein tyrosine kinases (SFKs). SFKs, known to be tethered to the plasma-membrane or with cytoplasmic localization have been observed in brain mitochondria by immunogold electron microscopy (Salvi et al., 2002; Tibaldi et al., 2008) and by immunocytochemistry in primary mouse pre-leptotene spermatocytes GC2 cells (Livigni et al., 2006); while in the heart, the presence of SFKs in mitochondria is supported by pharmacology and site- and phosphorylation state-directed antibodies showing increased Src-dependent phosphorylation of the mitochondrial protein, adenine nucleotide translocator 1 in isoflurane-preconditioned heart (Feng et al., 2010).
Western blots in Fig. 1C show that M3 fraction produced robust signals for VDAC1 and Cox2 but no significant signals for Cadherin, GM130, Lamin b1, GRP78 BiP, α1C, and Src. Quantification of normalized signals (to Ponceau S signals) demonstrates that M3 fraction is significantly enriched in VDAC and Cox2 with respect to whole heart lysate (WHL) and crude mitochondria (CM), and that it is practically free of non-mitochondrial protein markers with the largest contaminant being from the nuclear envelope (Lamin b1, 0.06 ± 0.03 in a scale 0-1; n=3), and the least being from the T-tubules (α1C, 0.001 ± 0.0003; n=3). Interestingly, SFKs signals were also low in M3 (0.02 ± 0.004; n=3).
Although M1 and M2 fractions contain similar amounts of VDAC1 and Cox2 than M3, indicating the presence of mitochondria in these fractions, the signals from other organelle markers in M1 and M2 were still significant. Electron microscopy of M1 and M2 fractions showed very few intact mitochondria when compared to M3 (n=3) suggesting that the strong signals of VDAC1 and Cox2 in M1 and M2 originate from damaged mitochondria. In summary, the degree of purity as reflected by the absence of extramitochondrial markers was optimal in M3 fraction. These results are consistent with the functional data in Fig. 1B where M3 had the best performance.
3.2. Confocal immunofluorescence of purified mitochondria
To further validate the suitability of M3 fraction for nanoscopy studies, we colabeled this mitochondria fraction with antibodies against Cox2 (Fig. 2A,A'), VDAC1 (Fig. 2D,D'), and Lamin b1 (Fig. 2G,G') together with mitotracker, which was efficiently uptaken by purified mitochondria (Fig. 2B,B', E,E', H,H'). Both Cox2 and VDAC1 immunofluorescence highly coincided with mitotracker signals (Fig. 2C,C' and Fig. 2F,F') supporting the mitochondrial nature of both types of signals. As expected from the Western blot analysis in Fig. 1C,D, the immunofluorescence signals of the nuclear envelope marker Lamin b1 were scarce and showed no colocalization with mitotracker (Fig. 2I,I'). Thus, organelle labeling by specific antibodies represents a further purification step as it can readily discern between scarce contaminant proteins and mitochondria-associated proteins. Controls with secondary antibodies in the absence of primary antibodies showed no labeling (Fig. 2J vs. Fig. 2K) and VDAC1 antibody signal could be blocked with recombinant VDAC1 protein (Fig. S1).
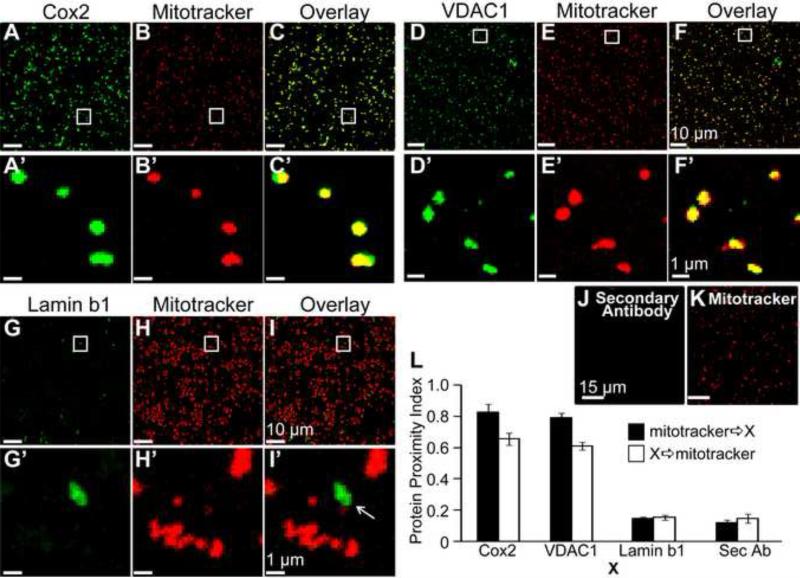
Figure 2. Visualization of mitochondrial proteins by confocal microscopy.
Purified M3 fraction mitochondria were loaded with mitotracker and counter-stained for Cox2, VDAC1 and Lamin b1 proteins. A-B. Anti-Cox2 monoclonal antibody and mitotracker signals. C. Overlay shows high correlation between mitotracker and Cox2 signals. D-E. Anti-VDAC1 monoclonal antibody and mitotracker signals. F. Overlay of D and E shows high signal correlation of mitotracker with VDAC1. A'-F'. Squares in A-F are shown at higher magnification for better examination. G-H. Lamin b1 (nuclear envelope marker, negative control) and mitotracker signals. I. Anti-Lamin b1 polyclonal antibody and mitotracker signals do not coincide; thus, contaminant nuclear envelope fragments can be clearly distinguished from mitotracker-loaded mitochondria. G'-I'. Squares in G-I are shown at higher magnification for better examination. J-K. Mitochondria were loaded with mitotracker and labeled with secondary antibody alone. Secondary antibody (anti-mouse) showed no specific signals (J), while mitotracker labeling was robust (K) confirming the presence of mitochondria. L. Protein Proximity Index (Wu et al., 2010) of mitotracker towards Cox2, VDAC1, Lamin b1, or secondary antibody alone (Sec Ab) (black bars) and vice versa (open bars) (n=5 mitochondria preparations). Values are given in the text. Confocal images were acquired at 0.0575 μm/pixel.
We next quantified the colocalization index of Cox2, VDAC1, Lamin b1 and secondary antibodies with mitotracker using the protein proximity index (PPI) method (Wu et al., 2010) (Fig 2L). This method isolates specific colocalization signals from those produced by image heterogeneity and non-specific colocalization as those arising from the antibody background signal. The PPI values were: mitotracker 21E8 Cox2 = 0.83 ± 0.05; Cox2 21E8 mitotracker = 0.66 + 0.04; mitotracker 21E8 VDAC1 = 0.79 ± 0.03; and VDAC1 21E8 mitotracker = 0.61 + 0.03 (n=5). These data indicate that about 80% of mitochondria labeled by mitotracker are also labeled by Cox2 and VDAC1; whereas, only about 65% of mitochondria labeled with Cox2 and VDAC1 were also labeled by mitotracker. The latter suggests that ~ 35% of mitochondria could not uptake or maintain mitotracker although they could be labeled for Cox2 and VDAC1. In contrast to Cox2 and VDAC1, PPI of Lamin b1 and secondary antibody alone (Sec Ab) with mitotracker were very low and similar in both directions. Values were: mitotracker 21E8 Lamin b1 = 0.14 ± 0.01; Lamin b1 21E8 mitotracker = 0.15 + 0.01; mitotracker 21E8 Sec Ab = 0.12 ± 0.02; Sec Ab 21E8 mitotracker = 0.14 + 0.03 (n=5). These results demonstrate as a reference that Cox2 and VDAC1 but not Lamin b1 are located in mitochondria from M3 fraction. Because mitotracker is concentrated by functional mitochondria with an intact membrane potential, the results also confirm that M3 fraction contains viable mitochondria.
We have also tested the presence of α1C and SFKs proteins in purified mitochondria from M3 fraction by colabeling with polyclonal antibodies against these proteins and with mitotracker (Fig. S2). As expected for M3 fraction, mitochondria were readily labeled with mitotracker; however, α1C produced scarce signals that did not coincide with mitotracker, while Src signals were undetectable and similar to secondary antibody alone. To validate the labeling properties of both antibodies, we labeled isolated murine cardiomyocytes under identical conditions and obtained clear labeling with the same antibodies. These results are consistent with the Western blot analysis showing that α1C and Src proteins are negligible in the purified M3 mitochondrial fraction isolated from murine heart.
3.3. Localization of mitochondrial proteins with STED
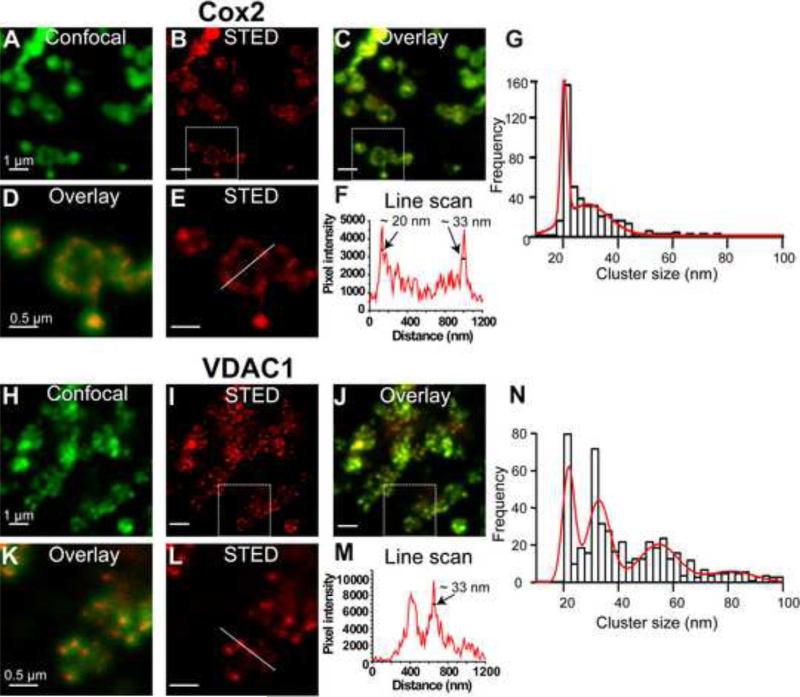
The diameter of mitochondria in M3 fraction is around 200-1000 nm, while the maximum resolution of conventional confocal microscopy (limited by the diffraction limit of the light) is ~200 nm in the focal plane and ~500 nm along the optic axis. Therefore, using traditional confocal microscopy, it is near impossible to differentiate protein clusters from the blurred mitochondrial image. However, with STED nanoscopy protein clusters can be readily identified. For comparison, we have acquired both confocal (Fig. 3A,H) and STED (Fig. 3B,E,I,L) images of the same preparation. The overlays (Fig. 3C,D,J,K) contrast the dramatic increase in resolution with STED nanoscopy, allowing the visualization of clusters of VDAC1 and Cox2 proteins. Cox2 showed a diffused cluster distribution (Fig. 3B, E), whereas VDAC1 had discreet clusters (Fig 3I, L). Figure 3F,M shows examples of resolution measurements using a line scan (corresponding lines in Fig. 3E,L) through clusters and measuring the Full Width Height Maximum (FWHM) of individual peaks (arrows). Similar measurements of 500 clusters from 50 isolated mitochondria (10 per mitochondrion from three independent experiments) were used to construct the cluster size frequency histograms. Histograms were fitted to multiple Gaussian functions (red lines) revealing that in purified cardiac mitochondria Cox2 clusters are distributed mostly in clusters of ~20-33 nm (Fig. 3N), while VDAC1 clusters had in addition larger size populations of ~55 and 83 nm (Fig. 3G).
Figure 3. STED nanoscopy of isolated mitochondria reveals Cox2 and VDAC1 clusters.
Purified M3 mitochondria were labeled with monoclonal antibodies directed against Cox2 and VDAC1. A. Confocal image of Cox2 labeled mitochondria. B. Corresponding STED image with clear clusters. C. Overlay of A and B highlights increased resolution with STED. D. Square in C was magnified for better visualization. E. Magnification of square in B. F. Intensity profile of the line in E. Arrows mark an intensity peaks with FWHM of ~20 nm and ~33 nm. G. Cluster size frequency histogram for Cox2 was manually fitted to the sum of two Gaussian distributions; cluster size peaks were at 20 and 29 nm. H-I. Confocal and STED images of mitochondria labeled for VDAC1. J. Overlay. K. Square in J was magnified highlighting increased resolution of STED image and VDAC1 clusters. L. Square in I at higher magnification. M. Intensity profile of line scan in L. Arrow marks a peak with FWHM of ~33 nm. N. Cluster size frequency histogram for VDAC1 was manually fitted to the sum of four Gaussian distributions; cluster size peaks were at 22, 33, 55, and 83 nm. n=3 independent mitochondria isolations.
4. DISCUSSION AND CONCLUSIONS
We have been able to visualize protein clusters of VDAC1 and Cox2 in cardiac mitochondria using a custom-made STED fluorescence microscope and distinguish specific cluster populations of the proteins examined.
The visual power of this method (hundreds of mitochondria can be visualized and analyzed in a single experiment) together with the usage of specific antibodies should allow unambiguous demonstration about the presence of proteins in mitochondria. In our M3 purified mitochondria fraction, VDAC1 and Cox2 were readily detected by Western blot and superresolution fluorescence microscopy. In contrast, neither SFKs nor the α1C subunit of the L-type Ca2+ channel gave significant signals (Fig. 1 and Fig. S2). These results are consistent with the LC/MS/MS analysis of Percoll (30%:60% gradient) purified murine cardiac mitochondria, where neither α1C nor SFKs peptides were detected (Zhang et al., 2008). However, the existence of SFKs in rat heart mitochondria has been supported by pharmacological and biochemical studies where Western blots of Percoll (25%)-separated mitochondria showed robust signals for Src (Feng et al., 2008; Feng et al., 2010). Src signals were also robust in our M1 and M2 mitochondrial fractions but were basically absent in the purest and functional M3 fraction using both Western blot and STED microscopy. It is possible that the concentration of Percoll influences the purification process or that species variation (rat vs. mouse) or perhaps mitochondria subpopulations may account for the differences.
Previous qualitative evaluation of Flag-tagged VDAC3 expressed in U2OS cells, gave similar large clusters (40 to 90 nm) (Neumann et al., 2010) in comparison to VDAC1 (~55 and 83 nm) in our studies. However, we identified additional smaller size clusters for VDAC1 (~22 and 30 nm). The usage of isolated mitochondria instead of cells may avoid out of focus fluorescence and contribute to increased resolution in our measurements. In our studies with 20 nm resolution limit, cluster sizes of 20 nm may be actually smaller.
Conclusions
Using STED microscopy and purified murine cardiac mitochondria, we have shown that native VDAC1 and Cox2 are distinctly organized in cardiac mitochondria forming clusters of different sizes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Jeff Abramson (UCLA) for VDAC1 recombinant protein. This work was supported by NIH grants, HL54970 (LT) and HL088640 (ES) and the American Heart Association Fellowships 09POST2190008 (JCB) and 10POST4230081 (YW).
Non-standard Abbreviations and Acronyms
- STED
STimulated Emission Depletion
- VDAC1
Voltage Dependent Anion Channel 1
- Cox2
Cytochrome c Oxidase subunit 2
- SFKs
Src family of tyrosine kinases
- PPI
protein proximity index
- FWHM
Full Width Half Maximum
- TDE
2,2'-Thiodiethanol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baines CP. The cardiac mitochondrion: nexus of stress. Annu. Rev. Physiol. 2010;72:61–80. doi: 10.1146/annurev-physiol-021909-135929. [DOI] [PubMed] [Google Scholar]
- Brzezinski P, Johansson AL. Variable proton-pumping stoichiometry in structural variants of cytochrome c oxidase. Biochim. Biophys. Acta. 2010;1797:710–723. doi: 10.1016/j.bbabio.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Scorrano L. (De)constructing mitochondria: what for? Physiology. (Bethesda. ) 2006;21:233–241. doi: 10.1152/physiol.00010.2006. [DOI] [PubMed] [Google Scholar]
- Feng J, Lucchinetti E, Enkavi G, Wang Y, Gehrig P, Roschitzki B, Schaub MC, Tajkhorshid E, Zaugg K, Zaugg M. Tyrosine phosphorylation by Src within the cavity of the adenine nucleotide translocase 1 regulates ADP/ATP exchange in mitochondria. Am. J. Physiol Cell Physiol. 2010;298:C740–C748. doi: 10.1152/ajpcell.00310.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhu M, Schaub MC, Gehrig P, Roschitzki B, Lucchinetti E, Zaugg M. Phosphoproteome analysis of isoflurane-protected heart mitochondria: phosphorylation of adenine nucleotide translocator-1 on Tyr194 regulates mitochondrial function. Cardiovasc. Res. 2008;80:20–29. doi: 10.1093/cvr/cvn161. [DOI] [PubMed] [Google Scholar]
- Graham JM. Purification of a crude mitochondrial fraction by density-gradient centrifugation. Curr. Protoc. Cell Biol. Chapter. 2001;3:3.4.1–3.4.22. doi: 10.1002/0471143030.cb0304s04. [DOI] [PubMed] [Google Scholar]
- Kellner RR, Baier CJ, Willig KI, Hell SW, Barrantes FJ. Nanoscale organization of nicotinic acetylcholine receptors revealed by stimulated emission depletion microscopy. Neuroscience. 2007;144:135–143. doi: 10.1016/j.neuroscience.2006.08.071. [DOI] [PubMed] [Google Scholar]
- Li M, Tanaka Y, Alioua A, Wu Y, Lu R, Kundu P, Sanchez-Pastor E, Marijic J, Stefani E, Toro L. Thromboxane A2 receptor and MaxiK-channel intimate interaction supports channel trans-inhibition independent of G-protein activation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19096–19101. doi: 10.1073/pnas.1002685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Livigni A, Scorziello A, Agnese S, Adornetto A, Carlucci A, Garbi C, Castaldo I, Annunziato L, Avvedimento EV, Feliciello A. Mitochondrial AKAP121 links cAMP and src signaling to oxidative metabolism. Mol. Biol. Cell. 2006;17:263–271. doi: 10.1091/mbc.E05-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann D, Buckers J, Kastrup L, Hell SW, Jakobs S. Two-color STED microscopy reveals different degrees of colocalization between hexokinase-I and the three human VDAC isoforms. PMC Biophys. 2010;3:4. doi: 10.1186/1757-5036-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi M, Brunati AM, Bordin L, La RN, Clari G, Toninello A. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim. Biophys. Acta. 2002;1589:181–195. doi: 10.1016/s0167-4889(02)00174-x. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, Ben-Hail D. VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion. 2011 doi: 10.1016/j.mito.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Staudt T, Lang MC, Medda R, Engelhardt J, Hell SW. 2,2'-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech. 2007;70:1–9. doi: 10.1002/jemt.20396. [DOI] [PubMed] [Google Scholar]
- Tibaldi E, Brunati AM, Massimino ML, Stringaro A, Colone M, Agostinelli E, Arancia G, Toninello A. Src-Tyrosine kinases are major agents in mitochondrial tyrosine phosphorylation. J. Cell Biochem. 2008;104:840–849. doi: 10.1002/jcb.21670. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davis MW, Hell SW, Jorgensen EM. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- Wu Y, Eghbali M, Ou J, Lu R, Toro L, Stefani E. Quantitative determination of spatial protein-protein correlations in fluorescence confocal microscopy. Biophys. J. 2010;98:493–504. doi: 10.1016/j.bpj.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li X, Mueller M, Wang Y, Zong C, Deng N, Vondriska TM, Liem DA, Yang JI, Korge P, Honda H, Weiss JN, Apweiler R, Ping P. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8:1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.