Abstract
There is increasing awareness of a role of mtDNA alterations in the development of cancer since mtDNA point mutations are found at high frequency in a variety of human tumors. To determine the biological effects of mtDNA mutations in UV-induced skin tumors, hairless mice were irradiated to produce tumors and the tumor mtDNAs were screened for single nucleotide changes using temperature gradient capillary electrophoresis (TGCE) followed by direct sequencing. A mutation hot spot (9821insA) in mt-Tr locus (tRNAArg) was discovered in approximately one third of premalignant and malignant skin tumors. To determine the functional relevance of this particular mutation in vitro, cybrid cell lines containing different mt-Tr (tRNAArg) alleles were generated. The resulting cybrid cell lines contain the same nuclear genotype and differ only in their mtDNA. The biochemical analysis of the cybrids revealed that the mutant haplotype is associated with diminished levels of complex I protein resulting in lower levels of baseline oxygen consumption and lower cellular ATP production. We hypothesize that this specific mtDNA mutation alters cellular biochemistry supporting the development of keratinocyte neoplasia.
Introduction
Theories of mitochondrial involvement on cancer date back to Warburg’s theories that impairment of OXPHOS (oxidative phosphorylation) and increased use of glycolysis in tumors was thought to be a critical step in the development of cancer (Warburg, 1956). There is growing evidence that some nuclear-encoded OXPHOS genes have dual functions in energy generation as well as tumor suppression (Eng et al., 2003). Mutations in the fumarate hydratase gene have been linked to uterine leiomyomas and renal cell carcinomas and mutations in subunits of the succinate dehydrogenase gene (SDHB, SHDC, SDHD) have been associated with paragangliomas, pheochromocytomas and renal cell carcinomas (Baysal et al., 2000; Niemann and Muller 2000; Astuti et al., 2001; Lehtonen et al., 2004; Vanharanta et al., 2004). The role for mtDNA point mutations in the development of cancer has been suggested (Petros et al., 2005; Polyak et al., 1998) since they have been found in high frequency in tumors. These mtDNA mutations were often homoplasmic (the exclusive mtDNA species), implying a genetic selection for these mtDNA changes in the development of cancers. mtDNA mutations have been identified in epithelial tumors, tumors of musculoskeletal, central nervous and endocrine system (Brandon et al., 2006; Verma and Kumar 2007; Chatterjee et al., 2006).
mtDNA mutations have been studied in human skin cancer (Eshaghian et al., 2006; Birch-Machin 2006, Berneburg et al., 2006). One of the limitations in analysis of mtDNA changes in humans is that humans have many mtDNA polymorphisms which represent diverse cultural origins. The hairless mouse model system is useful for analysis of mtDNA changes in non melanoma skin cancer (NMSC) since this model is a well established way to generate epidermal tumors that are similar to those seen in human squamous cell carcinoma and the mtDNA is similar in size and structure to that of humans. Studying mtDNA changes in the laboratory mouse offers a distinct advantage of mtDNA homogeneity which arises from a recent common female ancestor and allows for a facilitated analysis of what mtDNA changes may by pathogenic (Ferris et al., 1983). To determine if mtDNA changes play a role in NMSC formation, tumors were induced in hairless mice by UV radiation. The DNA of these tumors was analyzed for mtDNA changes. A specific mutation in mt-Tr locus occurred in high frequency and this mutation was studied in a cybrid system to identify the specific biochemical changes imparted by the mtDNA mutation that are supportive of a tumorigenic phenotype.
Results
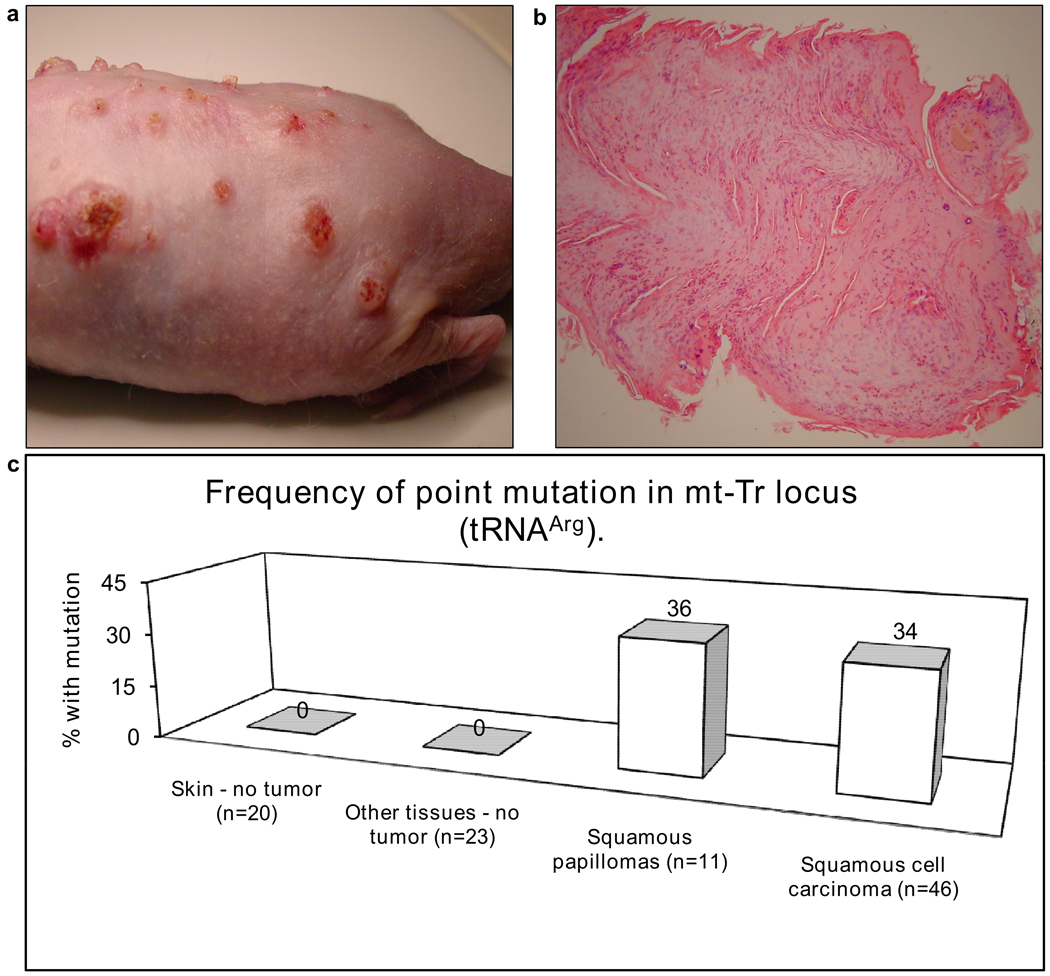
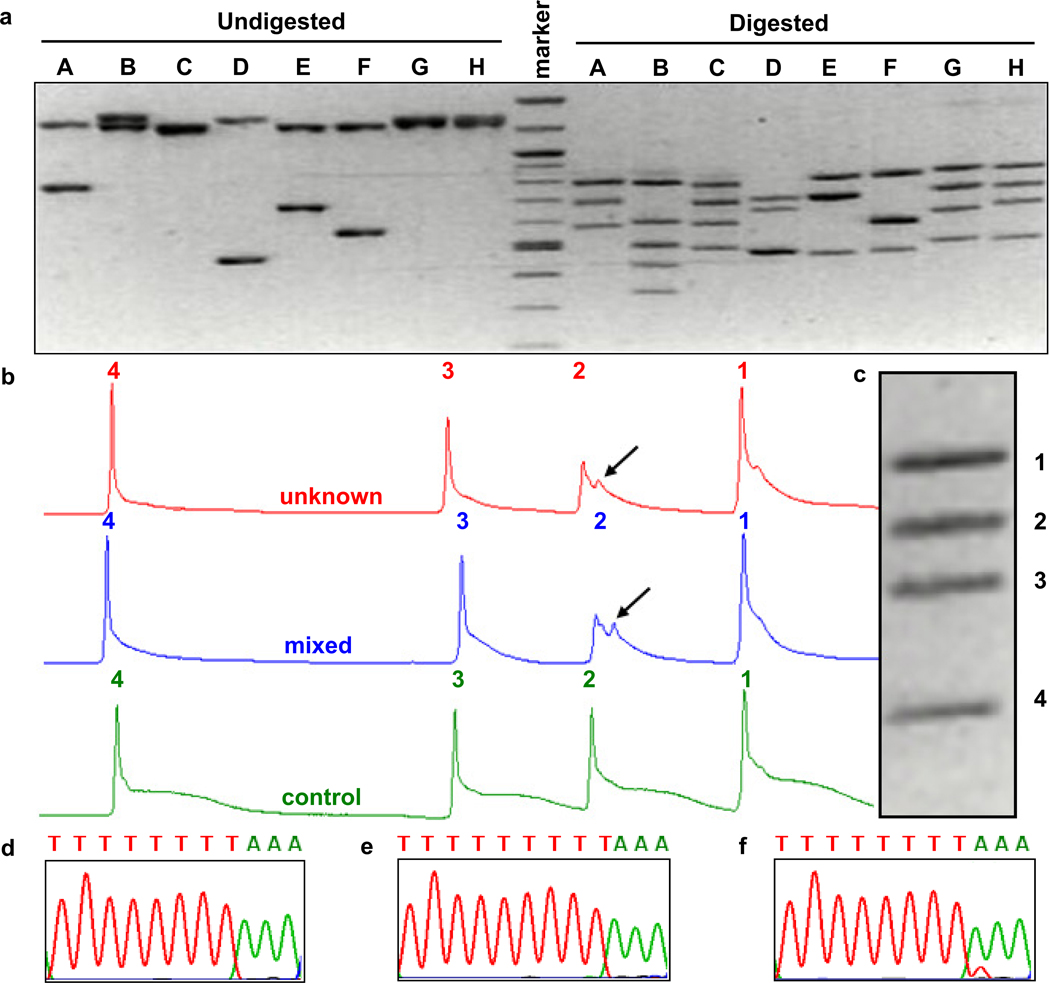
To determine if mtDNA changes occur in murine NMSCs, tumors were induced in hairless SKH1 mice by UV irradiation (Figure 1a, b). The entire mouse mtDNA was screened for mutations by genomic DNA isolation from tumors, multiplexed PCR of the entire mtDNA, restriction endonuclease digestion (Figure 2a) and heteroduplex analysis by multiplexed TGCE (Figure 2b, c). A mutation hotspot was identified in the gene encoding the mitochondrial tRNA for Arginine. The presence of an additional peak for fragment 2 (arrow in Figure 2b) is indicative of heteroduplex formation and the detection of a somatic mutation. Minimally irradiated skin from ventral surface of each mouse was used as a control and found to contain only homoduplexes, indicating the lack of any mutation. A 9821insA mutation in mt-Tr locus was defined by DNA sequencing (Figure 2d, e, f). This mutation predicts an insertion of an extra A in a homopolymeric tract in the dihydrouridine loop of the mitochondrial tRNAArg. The B6 mt-Tr gene contains a homopolymeric tract of 8 consecutive adenosine residues (Figure 2d). Tumors often contained an additional adenosine residue in this tract (Figure 2e). This change was confirmed by reamplification and sequencing the gene in the reverse orientation, as well by sequencing with Pfu DNA polymerase. This mutation was commonly seen in heteroplasmy (Figure 2f) as well as in homoplasmy (Figure 2e). mt-Tr 9821insA mutation was found in about one third of pre-malignant and malignant samples but was absent in normal skin and other non-tumor internal organs. The alleles present in the tumors include both heteroplasmic and homoplasmic 9821insA containing 9 consecutive A bases instead of the 8 normally found in this strain. Figure 1c shows the frequency of mutant mt-Tr 9821insA alleles in SKH1 mice that were irradiated with UV to induce cutaneous tumors. The “no tumor” skin is normal skin taken from the belly at the time of excision. “Other tissues” are healthy brain, liver, spleen, heart, kidney and lungs tissues taken from the same mice at the time of harvesting the skin tissues.
Figure 1. Induced cutaneous tumors and frequency of point mutation in mt-Tr locus (tRNAArg).
(a) Dorsal surface of a hairless mouse with UV-induced cutaneous tumors. (b) Hematoxylin and eosin histology stain of one of these tumors is consistent with a diagnosis of squamous papilloma. The height of the papilloma equals 2 mm. (c) The percentage of samples with various diagnoses harboring a mutant mt-Tr 9821insA allele is shown.
Figure 2. Restriction analysis, TGCE multiplex analysis and DNA sequencing detect mtDNA mutation hotspot.
(a) Resolution of undigested PCR products covering the entire mtDNA (left) and digested restriction fragments (right) are shown on an inverted image of a 1.5% agarose gel stained with ethidium bromide. (b) TGCE output from a single capillary. Unknown (from a tumor, top), control (bottom) and mixed (center) mouse DNA samples were analyzed. Fragments 1, 3, and 4 have only a single peak which is indicative of complete homoduplex formation and the lack of a mutation. The presence of an additional peak for fragment 2 in the unknown and mixed samples is indicative of heteroduplex formation and the detection of a somatic mutation. (c) The gel image of all four fragments. DNA sequencing of mt-Tr locus in control and tumor DNA samples in reverse orientation. (d) Partial DNA sequence of wild type allele and (e) homoplasmic or (f) heteroplasmic of 9821insA mutant allele.
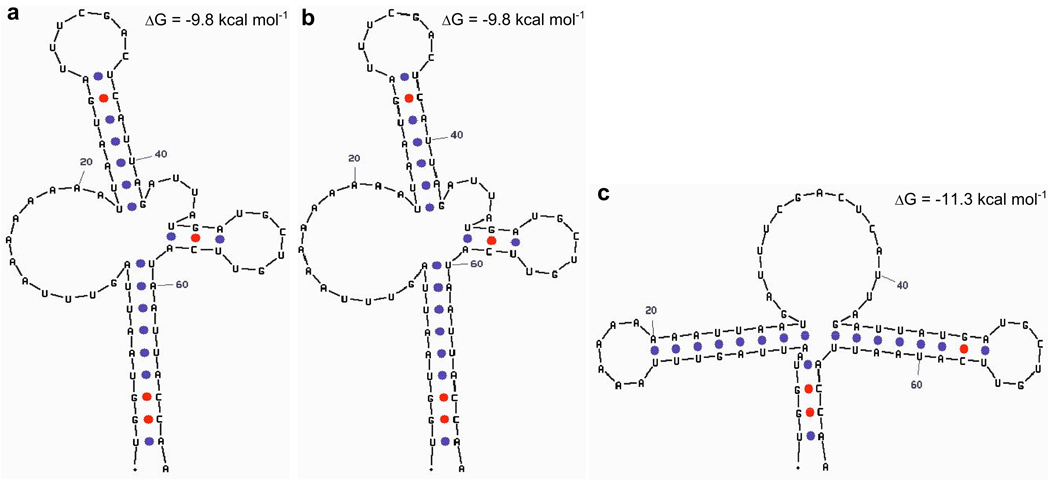
Structural analysis modeled with mfold software (Zuker 2003) predicts the murine mt-Tr 9821insA allele containing nine adenosine residues to have potential altered secondary structures (Figure 3b, c). One of these structures is predicted to be similar to the mt-Tr wild type allele containing eight consecutive adenosine residues (Figure 3a) whereas the other structure (Figure 3c) is dramatically different and energetically more stable (ΔG=−11.3 kcal/mol) than the former structure (ΔG=−9.8kcal/mol). Since tRNA structure is critical to its function, it is proposed that this altered structure would be more abundant and unlikely to be functional. At temperatures slightly lower than 37°C, as are encountered in the skin, non-functional isoforms may be even more energetically favored over the typical wild type allele structure. Thus it is expected that cells containing mt-Tr 9821insA mutation allele would have affected protein synthesis of all 13 mtDNA-encoded peptides.
Figure 3. Predicted tRNA structures based on sequence.
The predicted structures for the tRNA variants generated by mfold software. (a) wild type tRNA isoform with ΔG=−9.8kcal/mol (b) 9821insA tRNA isoform with ΔG=−9.8kcal/mol and (c) 9821insA tRNA isoform with ΔG=−11.3kcal/mol.
We utilized a naturally occurring allelic variant contained within Balb/c mouse strain as the source of 9821insA allele. To determine the functional relevance of this single nucleotide mutation, cybrid cells were generated. We produced two cybrid cell lines which have identical nuclear backround from murine L cells but differ only in their mtDNA. In this way, the two alleles could be compared in a neutral nuclear genetic backround. We analyzed the effects of this nucleotide insertion in the mtDNA by studying cybrids that harbor alterations at this locus.
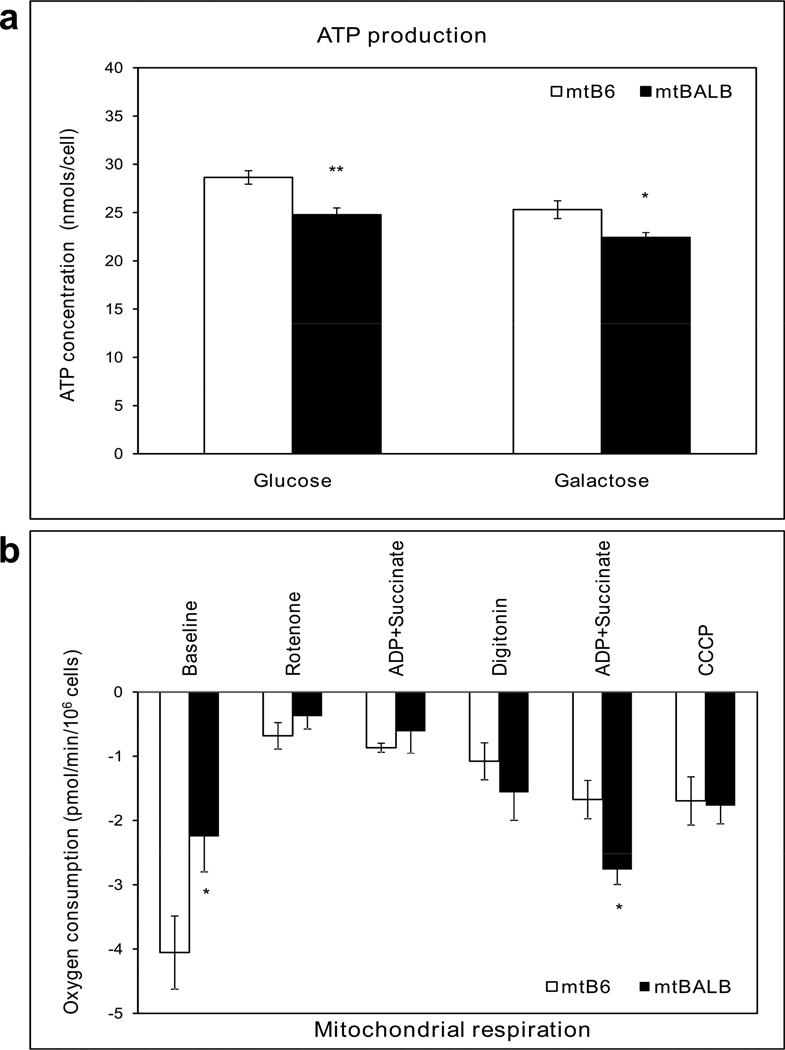
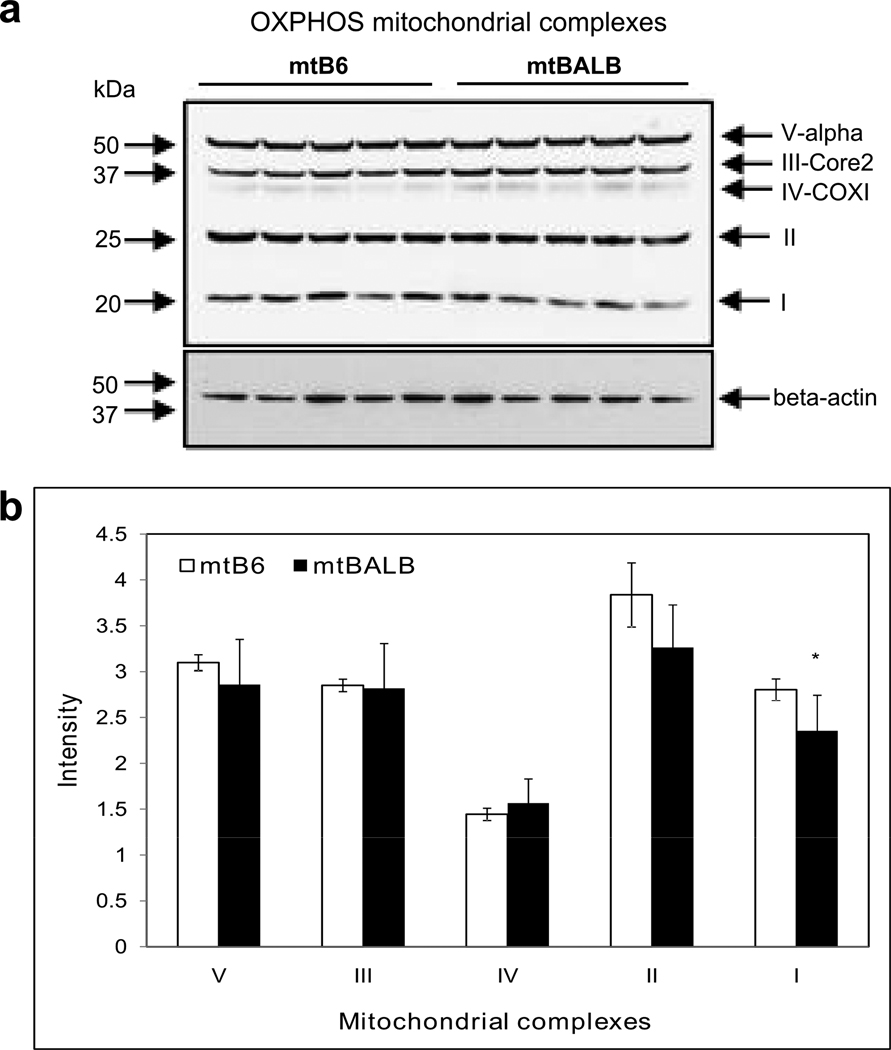
We characterized the biochemistry of the cybrid cells, first examining cellular ATP levels. Figure 4a shows that the cellular ATP levels were significantly lower in mtBALB cybrids compare to mtB6 cybrids in media containing glucose or galactose. Next, we examined respiratory activity of the cybrid lines by measuring cellular oxygen consumption. We observed that mtBALB cybrids consumed less oxygen than the mtB6 cybrid cells overall under the basal conditions. After rotenone was added to inhibit CI, digitonin was used to permeabilize the cells and the activity of CII was measured after adding the substrates ADP and succinate. Although the baseline levels of respiration were reduced in the mtBALB cybrids compared to mtB6, they displayed significantly enhanced oxygen consumption in the presence of CII substrates (Figure 4b). In order to better understand the observed changes in respiration, we sought to characterize the levels of OXPHOS proteins contained within the mitochondria. Using western blotting, we compared the protein levels of all mitochondrial OXPHOS complexes of mtB6 and mtBALB cybrid cells (Figure 5a). The levels of CI were significantly reduced in mtBALB cybrids compared to mtB6 measured by densitometry. The levels of complexes II, III, IV and V showed no significant differences between the cell lines (Figure 5b). Thus, the mtBALB cybrids have a diminished overall respiratory activity compared to the mtB6 cybrids, but they are more responsive to CII substrates.
Figure 4. mtBALB cybrid cells have lower ATP production and altered respiratory function compared to mtB6 cybrids.
(a) Cybrid cells were incubated in DMEM supplemented with either glucose or galactose. The levels of cellular ATP are shown. (b) Fiber optic investigation of mitochondrial respiratory chain function showing oxygen consumption was performed without addition of exogenous substrate, and with the addition of specific OXPHOS substrates or inhibitors. Values represent the mean ± SEM of three independent experiments of five clones. Asterisks indicate statistical difference between mtB6 and mtBALB cybrid cells (*P value< .05, **P value< .005).
Figure 5. Complex I levels are lower in mtBALB cybrid cells compared to mtB6.
(a) Five mouse monoclonal antibodies to mitochondrial OXPHOS subunits were used to detect protein levels by western blot analysis. (b) Densitometry of western blot analysis showing reduced levels of complex I protein. The level of complex I, II, III, IV and V subunits were expressed as the relative intensity of the immunoblots normalized to β-actin loading control. Data are mean ± SEM of three independent experiments of five clones. Asterisk indicates statistical difference between mtB6 and mtBALB cybrid cells (*P value< .05).
Discussion
The tumors generated in the mouse occurred primarily on the UV-exposed dorsal surfaces of the animals and were malignant SCC or pre-malignant squamous papillomas by microscopic analysis performed by a dermatopathologist. One particular point mutation in mt-Tr locus (tRNAArg) was very common, occurring in approximately one third of squamous papilloma samples and SCC samples (Figure 1c) but was not detected in normal skin from minimally irradiated ventral surface of animals or in normal appearing skin from the irradiated dorsal surface of the animals or in internal organs indicating that these induced changes occurred in the process of tumor development. The mouse mitochondrial mt-Tr (tRNAArg) gene appears to be a hotspot for UV-induced mutations having the sequence containing a homopolymeric tract of eight consecutive adenosine residues. The occurrence of an insertion mutation within a homopolymeric tract may highlight a potential weakness in the ability of mitochondrial DNA polymerase γ to repair UVA induced damage within the mtDNA (Longley et al., 2001). Such mutations may be important in tumor progression since mtDNA changes lead to increased oxidative stress which can be mitogenic to cells (Jandova at al., 2011, see footnote 4). We generated cybrid cell lines to identify biochemical and molecular changes due to this genetic alteration in mt-Tr gene. Since tRNAs are required for protein synthesis, the levels of all mtDNA-encoded polypeptides can be altered. The western blot analysis revealed that mutant haplotype is associated with diminished levels of complex I protein which is likely caused because a portion of the 9821insA tRNAs are non-functional. These decreased levels of complex I protein resulted in lower levels of baseline oxygen consumption and lower cellular ATP production. The mtBALB cybrids are more responsive to CII substrates. CII subunits are entirely encoded within the nuclear DNA and these proteins are synthesized on cytoplasmic ribosomes prior to import into the mitochondria. Enhanced CII substrate sensitivity may reflect a compensatory change by the nucleus to compensate for lower overall cellular ATP levels.
The biochemical differences between the cybrids suggest that energetic metabolism might be directly linked to neoplastic behavior. This notion of glycolytic metabolism as a key component of cellular transformation was proposed more than 50 years ago (Warburg 1956). Recent study of Ye et al. (Ye at al., 2011) reported that some mitochondrial and energy metabolism-related properties such as lower oxygen/glucose consumption, and lower intracellular concentrations of ATP can be used as indicators of lung cancer stem cells. In breast cancer cells were found much lower rates of ATP synthesis, lower cellular oxygen consumption and decreased activities of the respiratory chain complexes (Ma Y et al., 2010). Moreover, downregulation of the catalytic subunit of the mitochondrial ATP synthase is found to be a hallmark of many types of cancer such as liver, kidney, colon, breast, lung, squamous esophageal carcinomas and gastric adenocarcinomas (Formentini et al., 2010). Oncocytic tumors of the thyroid, kidney and parathyroids have been shown to accumulate mtDNA disruptive mutations, the large majority of which map in respiratory CI genes (Porcelli et al., 2010; Costa-Guda et al., 2007; Gasparre et al., 2007; Gasparre et al., 2008; Gasparre et al., 2009; Mayr et al., 2008; Zimmermann et al., 2009). In oncocytomas, a consistent loss of complex I was found along with compensatory up-regulation of the other respiratory chain complexes (Zimmermann et al., 2011). The involvement of Complex I mutations in cancer is demonstrated in a recent study of sporadic breast cancer (Czarnecka et al., 2010).
The oxidative damage generated by UV radiation may also increase the levels of ROS to the point where the cells start hyperproliferate and other carcinogenic phenotypes such migration and invasiveness can be triggered. The measurement of proliferative and migratory changes may be valuable to explain the higher incidence of this genetic change in tumors. Additionally, the possible differences in ROS production between the two cybrid lines should be determined since ROS levels can support a role of ROS as specific second messengers (Burdon 1995; Irani et al., 1997; Sauer et al., 2001; Liu et al., 2002; Nishikawa 2008; Mesquita et al., 2010) in signaling cascades involved in cell proliferation, differentiation and migration.
Inheritance of this mtDNA changes at mt-Tr locus in mice are associated with deafness (Johnson et al., 2001) as well as modulation of complex phenotypes such as cognitive functions (Roubertoux et al., 2003). Different phenotypes associated with mtDNA variation at the same locus is common among human mtDNA associated disease where tRNA mutations in particular often produce very pleomorphic phenotypes such as seen in human tRNALeu gene (Wittenhagen and Kelley 2003). The 9821insA allele may be expected to form different structurally distinct isoforms which may have normal or possibly non-existent function. The balance of functional to non-functional isoforms may be quite tissue specific depending on local factors such as temperature or ionic concentrations. Homeostasis imbalance in the organism may predispose to organ specific tumor development.
We demonstrate here that a hotspot for mutations was identified and that this mt-Tr 9821insA allele can alter the biochemical characteristics of murine cybrid cells. These profound differences in cellular physiology displayed by the murine cybrids may contribute to the overall tumorigenic phenotype of the tumors that harbor this mtDNA change and may also explain some of the varying predispositions to developing tissue specific malignancies in mouse strains (Krupke et al., 2008). Furthermore, these studies on mouse mtDNA haplotypes contribute to a better understanding of human phenotypes associated with mtDNA variation.
Materials & Methods
Mice
To study hairless mice with the mitochondrial background of C57BL/6J (B6) SKH1 hairless male mice (Charles River laboratories Inc., Wilmington, MA) were crossed with female B6 mice (The Jackson laboratory, Bar Harbor, ME) and the resulting female progeny were backcrossed ten times to SKH1. Female F2 mice that were hairless and albino were selected of each cross for continued backcrossing to SKH1 male mice. BALB/cJ mice (The Jackson laboratory, Bar Harbor, ME) have identical mtDNA to B6 mice. The only differences between their mtDNA are a T to C polymorphism at 9461 and a 9348G to A change resulting in the amino acid change V248I which is thought to be a neutral polymorphism (Moreno-Loshuertos et al., 2006).
Animal experiments have been approved by the Institutional Animal Care and Use Committees at the Vanderbilt University and the University of Arizona.
Tumor induction
Cutaneous tumors were induced in hairless SKH1 mice by exposure to 720mJ UVA and 60mJ UVB five days per week for ten weeks. Tumor formation began several weeks later. Tumors were removed from the dorsal surface of the animals along with minimally- irradiated skin from the ventral surface of the animal, and in some cases, tumor-free irradiated skin from the dorsal surface of the animal, as a control. After sacrifice, non UV irradiated tissues including brain, heart, skeletal muscle, gut, liver, spleen, gonads and lungs were isolated. mtDNA of these UV protected organs was also used as controls. Genomic DNA was isolated from each tumor and mtDNA was analyzed for single nucleotide changes using TGCE.
Temperature Gradient Capillary Electrophoresis (TGCE)
DNA extraction was performed using DNeasy kits (QIAGEN, Valencia, CA) or by alkaline lysis and protein precipitation after proteinase K tissue digestion.
The entire mouse mtDNA was PCR amplified in overlapping fragments in 8 tubes using 16 mtDNA-specific primer pairs (Table 1). Each tube contained two primers pair and two PCR products were produced per tube. Thirty cycles of multiplexed PCR was performed in Ampli-Taq Gold buffer (Applied Biosystems, Carlsbad, CA) with 15mM MgCl2 according to manufacturer’s instructions, supplemented with an additional 25mM MgCl2, 2mM dNTP (New England BioLabs, Ipswich, MA), 5µM primers, 10µg/µl BSA (Promega Corporation, Madison, WI), and 9:1 Ampli-Taq Gold: Pfu DNA polymerase (Promega Corporation, Madison, WI). PCR parameters: an initial denaturation at 95°C for 10 min followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 57°C for 1 min and elongation at 72°C for 3 min with a final elongation at 72°C for 10 min.
Table 1. Overview of multiplex system for mouse mtDNA genome sequencing.
Eight tubes (A–H) per sample were used to amplify the entire murine mitochondrial genome. Two PCR products (1, 2) were generated in each tube. These pairs of PCR products were subsequently digested with various restriction enzymes (RE) to generate restriction fragments (RF) and single nucleotide changes were detected using TGCE. The Start, End and Size of each PCR product is listed along with the primers used to generate the PCR products, the restriction enzymes used and the sizes of the fragments generated via the restriction enzymes.
| Tube ID |
Start | End | Size | Forward Primer | Reverse Primer | Restriction Enzyme |
Restriction Fragment |
|---|---|---|---|---|---|---|---|
| A1 | 1050 | 2340 | 1291 | TCTGGCCTACACCCAGAAGATTTCA | CTGGGTCAATAAGATATGTTGATTTTACTTTGACT | EcoRI | 759,700,591 |
| A2 | 8465 | 9223 | 759 | TATTAATAAATATTAGCCCACCAACAGCTACCATT | AGTCCATGGAATCCAGTAGCCATGA | ||
| B1 | 7168 | 8461 | 1294 | TACAAGCACAATAGATGCACAAGAAGTTGA | GAGTAGCTCCTCCGATTAGGTGTATTAATAAGTGT | TaqI | 794,597,500,431,338 |
| B2 | 14915 | 16280 | 1365 | TAATCCACTAAACACCCCACCCCATATT | GCGTAATAGAGTATGATTAGAGTTTTGGTTCACG | ||
| C1 | 11399 | 12666 | 1267 | CACCCAACGCGGCAAACTAAC | CTAGTTGGCTTGATGTAGAGAAGGCAATG | TaqI | 770,681,587,487 |
| C2 | 16159 | 1121 | 1257 | TCAAACCCTATGTCCTGATCAATTCTAGTAGTTC | TTTGTGTAGGGCTAGGGCTAGGATTAGTT | ||
| D1 | 5828 | 6299 | 472 | TATTATCAACATGAAACCCCCAGCCATA | AAGTCAGCTAAATACTTTGACACCGGTAGG | DraI | 694,595,472 |
| D2 | 9220 | 10508 | 1289 | GACTCCATGTAATTATTGGATCAACATTCCT | AATTAGTTCAGTTGCTGAAAAGGTTATGATTAGG | ||
| E1 | 2248 | 3507 | 1260 | GGTTGGGGTGACCTCGGAGAAT | CTGGTAAATTGATATAGTATAGGGGTCCTAGGAAGA | DraI | 793,701,467 |
| E2 | 6550 | 7250 | 701 | CATGAGCAAAAGCCCACTTCG | AGGGGAGAGCAATTATGATAAGGATTACAGC | ||
| F1 | 6123 | 6700 | 578 | GAACCTTTCGGCTATATAGGAATAGTATGAGCAAT | GATCCTATAGAAGAGACAGTGTTTCATGTGGTG | DraI | 801,578,468 |
| F2 | 13701 | 14969 | 1269 | CTCCCAAACCATCAAGATTAATTACTCCAA | TGCAAATAGGAAATATCATTCGGGTTTAAT | ||
| G1 | 4661 | 5988 | 1328 | TCCCTAGGAGGCCTTCCACCAC | CAAAGAAAGTTGTGTTTAGGTTGCGGTCT | DraI | 809,715,613,501 |
| G2 | 10194 | 11503 | 1310 | CTACCACTAACCTGACTATCAAGCCCTAAAA | GTTAGAAGAATAAGTGGAATTATGTGAAGGGCTAT | ||
| H1 | 3407 | 4700 | 1294 | ATTCGCGTTATTCTTTATAGCAGAGTACACTAACA | TTGGTAAGAATCCTGTTAGTGGTGGAAG | DraI | 807,703,633,487 |
| H2 | 12487 | 13822 | 1336 | CAATAGTAGTTGCAGGAATTTTCCTACTGGTC | TTGGGGATCTAACTGATTAATTTTGGGTTT | ||
Restriction endonuclease digestions of crude PCR products were performed in the augmented Ampli-Taq Gold buffer without further supplementation at 37°C except for TaqI, which is digested at 65°C. Two µl of the appropriate restriction enzyme (Table 1) was added to every PCR product. Samples were incubated at 37°C for 2 hrs, then at 65°C for 60 min. To determine the resolution of all restriction fragments, 5 µl of restriction fragments from each tube were run on 1.5% agarose gel electrophoresis at 10V/cm for 90 min.
Heteroduplex species are formed by mixing the digested unknown DNA fragments with the control DNA fragments at a 1:1 ratio in a thermal cycler. The mixed samples are denatured at 95° and annealed using this profile: a 3 minutes denaturation step at 95°C followed by a stepwise reduction in temperature as follows: decrease from 95°C to 80°C at 3°C/min, decrease from 80°C to 55°C at 1°C/min, hold 20 min at 55°C, decrease from 55°C to 45°C at 1°C/min, decrease from 45°C to 25°C at 2°C/min prior to storage at 4°C.
TGCE analysis was performed on a Reveal Discovery System (SpectruMedix, State College, PA). This system performs temperature gradient electrophoresis for DNA variant detection on an automated capillary array instrument. DNA samples consisting of homoduplexes and heteroduplexes are separated by capillary electrophoresis, during which a thermal ramp from 50C° to 60C° for 25 minutes is applied. The injection parameters are 6 kV for 90 seconds and the capillary length is 70 cm. The gel in the capillary is 2% w/v 7-million mol wt polyethylene oxide dissolved in 1X TBE buffer with ethidium bromide added at a final concentration 0.5 µg/ml. SpectruMedix Check Mate Software was used for instrument control and data acquisition (acquisition time- 65 minutes.) Data were analyzed using the SpectruMedix Reveal Software to generate mutation scoring results for all samples in a plate.
DNA Sequencing
Forward (5’- TTCTAGTCACAATTCTATCTCTAGGC) and reverse (5’-GCATTGTAGTAGGTTGAGATTTTGG) primers were designed for PCR to amplify the mt-Tr gene from total genomic DNA. PCR samples were prepared in a total volume of 50 uL. Each tube contained 1xTaq master Mix (New England BioLabs, Ipswich, MA) containing 10mM Tris-HCl, 50mM KCl, 1.5mM MgCl2, 25 units/ml Taq DNA polymerase, 0.2mM dNTPs each, 5% glycerol, 0.08%NP-40 and 0.05% Tween-20; 250ng of DNA template and 5uM of both primers. PCR parameters: an initial denaturation at 95°C for 5 minutes followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 60 seconds and elongation at 68°C for 1 minute; with a final elongation at 68°C for 5 minutes. PCR products were then purified using QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced on an Applied Biosystems 3730XL DNA analyzer (Life Technologies Corporation, Carlsbad, CA).
Cybrid cell lines generation
LMEB3(mtBALB) and LMEB3(mtB6) cells were generated by harvesting the mitochondria from brain synaptosomes of B6 and BALB mice and electrofusing them to a mouse fibroblast LMEB3ρ0 cell line that lacked its own mtDNA (Trounce and Wallace 1996; Trounce et al., 2000). All cell lines were grown in high glucose (4.5g/L) or in galactose (4.5g/L) DMEM (Invitrogen Corporation, Carlsbad, CA) supplemented with 10% FBS (Invitrogen Corporation, Carlsbad, CA) as indicated.
ATP Assay
The amount of cellular ATP was determined by using ATPlite 1step kit (PerkinElmer, Waltham, MA) based on firefly luciferin/luciferase (Cree & Andreotti 1997). Cells were seeded in 96-well plates (5 × 104 /well) and allowed to attach for 4 h, then were washed with DPBS and incubated in DMEM (Invitrogen Corporation, Carlsbad, CA) with glucose or galactose for 16h. The plates were removed from the incubator and allowed to equilibrate at room temperature for 30 min. Next 100 µL of ATPlite 1step reagent was added to each well and the plates were shaken for 10 min at 700 rpm using a rotary plate shaker. The measurements of mitochondrial ATP levels were carried out with a Veritas luminometric plate reader (Turner Biosystems, Medison, WI). The ATP levels were expressed in nmoles/cell.
Respiration Assay
Oxygen consumption rates were measured in whole cells by using a Fiber optic oxygen monitor (Instech laboratories, Plymouth Meeting, PA). Briefly, cells were suspended in 1 ml of respiratory buffer (10 mM Hepes, pH 7.4, 225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM KH2PO4, 5 mM MgCl2, 1 mg/ml BSA at a concentration 2 × 106 cell/ml at 37°C in a magnetically stirred 1.2 ml chamber. The decrease of O2 concentration in the sealed chamber was measured as cellular oxygen consumption by an O2 fiber optic sensor. Baseline respiration was recorded for 2 min before 2 µl aliquots of 0.1 mM rotenone (Sigma Aldrich, St. Louis, MO) were added until total inhibition of complex I was obtained. Cell membrane integrity was controlled by measuring succinate-driven respiration (10 µl of 0.1 M ADP plus 20 µl of 1 M succinate, pH 7.4) before and after digitonin permeabilization. Cells were permeabilized by using 2 µl of 5 mg/ml digitonin (Sigma Aldrich, St. Louis, MO) per 1 × 106 cells. Maximum uncoupled respiration rate was obtained by the addition of 2 µl of 2.5 mM carbonyl cyanide m-chlorophenylhydrazone (CCCP) (data not shown). Finally, inhibited respiration was measured after addition of 2 µl of 200 mM potassium cyanide (data not shown).
Western Blotting analysis of OXPHOS complexes
Cybrid cells were collected and resuspended in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1mM PMSF, 1 mM EDTA, 5 µg/ml Aprotinin, 5 µg/ml Leupeptin, 1% Triton x-100, 1% Sodium deoxycholate, 0.1% SDS (Sigma Aldrich, St. Louis, MO). Protein content was assessed by Coomassie staining. Twenty micrograms of protein were separated by 12% SDS-PAGE electrophoresis and transferred onto nitrocellulose membrane (Bio-Rad, Hercules, CA). MitoProfile® Total OXPHOS Rodent WB Antibody Cocktail (MitoSciences, Eugene, OR) containing five monoclonal antibodies against the 20 kDa subunit of complex I- ND6, CII-30kDa, CIII-Core protein 2, CIV subunit I and CV α subunit respectively was used to detect complex I–V subunits. The membrane was stripped and re-blotted with rabbit β-actin mAb (Cell Signaling Technology, Danvers, MA) to normalize protein loading levels. Dilutions of different antibodies for immunoblotting and subsequent detection by the enhanced chemiluminescence ECL plus system (GE Healthcare Biosciences, Pittsburgh, PA) were done according to manufacturer’s protocol.
Statistical analysis
Data were represented as mean ± SEM of five clones. All experiments were performed at least three times. Statistical significance between any two groups was determined by the two-tailed Student’s t-test, P values less than 0.05 were considered to be significant.
Acknowledgements
This work was supported by an NCI Cancer Center Support Grant P30 CA023074 (CCSG) and R01 AR 0501552 to JS, by the VA Advanced Career Development Award and VA Merit Award to JS and by the Vanderbilt Skin Diseases Research Center Core Grant (NIH P30AR41943).
Abbreviations
- mtDNA
mitochondrial DNA
- OXPHOS
oxidative phosphorylation
- UV
Ultraviolet
- TGCE
Temperature Gradient Capillary Electrophoresis
- NMSC
Non-melanoma Skin Cancer
- SCC
Squamous Cell Carcinoma
- CI, II, III, IV, V
mitochondrial complexes I, II, III, IV, V
- DPBS
Dulbecco's Phosphate Buffered Saline
- DMEM
Dulbecco's Modified Eagle’s Medium
- FBS
Fetal Bovine Serum
- BSA
Bovine Serum Albumin
Footnotes
Jandova et al., 2011: (manuscript in preparation) Somatic alterations in mitochondrial DNA produce changes in cell growth and metabolism supporting a tumorigenic phenotype (Approved by all authors).
Conflict of Interest
The authors have no conflict of interest.
References
- Astuti D, Latif F, Dallol A, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69(1):49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287(5454):848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Berneburg M, Kamenisch Y, Krutmann J, et al. 'To repair or not to repair - no longer a question': repair of mitochondrial DNA shielding against age and cancer. Exp Dermatol. 2006;15(12):1005–1015. doi: 10.1111/j.1600-0625.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- Birch-Machin MA. The role of mitochondria in ageing and carcinogenesis. Clin Exp Dermatol. 2006;31(4):548–552. doi: 10.1111/j.1365-2230.2006.02161.x. [DOI] [PubMed] [Google Scholar]
- Birch-Machin MA, Tindall M, Turner R, et al. Mitochondrial DNA deletions in human skin reflect photo- rather than chronologic aging. J Invest Dermatol. 1998;110:149–152. doi: 10.1046/j.1523-1747.1998.00099.x. [DOI] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. (abstr.) [DOI] [PubMed] [Google Scholar]
- Burdon RH. Superoxide and Hydrogen-Peroxide in Relation to Mammalian-Cell Proliferation. Free Radical Bio Med. 1995;18(4):775–794. doi: 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25(34):4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- Costa-Guda J, Tokura T, Roth SI, et al. Mitochondrial DNA mutations in oxyphilic and chief cell parathyroid adenomas. BMC Endocr. Disord. 2007;7:8. doi: 10.1186/1472-6823-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree IA, Andreotti PE. Measurement of cytotoxicity by ATP-based luminescence assay in primary cell cultures and cell lines. Toxicol in Vitro. 1997;11(5):553–556. doi: 10.1016/s0887-2333(97)00060-x. [DOI] [PubMed] [Google Scholar]
- Czarnecka AM, Krawczyk T, Zdrozny M, et al. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res. Treat. 2010;121:511–518. doi: 10.1007/s10549-009-0358-5. [DOI] [PubMed] [Google Scholar]
- Eshaghian A, Vleugels RA, Canter JA, et al. Mitochondrial DNA deletions serve as biomarkers of aging in the skin, but are typically absent in nonmelanoma skin cancers. J Invest Dermatol. 2006;126(2):336–344. doi: 10.1038/sj.jid.5700088. [DOI] [PubMed] [Google Scholar]
- Eng C, Kiuru M, Fernandez MJ, et al. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat Rev Cancer. 2003;3:193–202. doi: 10.1038/nrc1013. [DOI] [PubMed] [Google Scholar]
- Ferris SD, Sage RD, Prager EM, et al. Mitochondrial-DNA Evolution in Mice. Genetics. 1983;105(3):681–721. doi: 10.1093/genetics/105.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formentini L, Martínez-Reyes I, Cuezva JM. The mitochondrial bioenergetic capacity of carcinomas. IUBMB Life. 2010;62(7):554–560. doi: 10.1002/iub.352. [DOI] [PubMed] [Google Scholar]
- Gasparre G, Hervouet E, de Laplanche E, et al. Clonal expansion of mutated mitochondrial DNA isassociated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum. Mol. Genet. 2008;17:986–995. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- Gasparre G, Iommarini L, Porcelli AM, et al. An inherited mitochondrial DNA disruptive mutation shifts to homoplasmy in oncocytic tumor cells. Hum. Mutat. 2009;30:391–396. doi: 10.1002/humu.20870. [DOI] [PubMed] [Google Scholar]
- Gasparre G, Porcelli AM, Bonora E, et al. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc. Natl Acad. Sci. USA. 2007;104:9001–9006. doi: 10.1073/pnas.0703056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani K, Xia Y, Zweier JL, et al. Mitogenic signaling mediated by oxidants in ras-transformed fibroblasts. Science. 1997;275(5306):1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Bykhovskaya Y, et al. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat Genet. 2001;27(2):191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupke DM, Begley DA, Sundberg JP, et al. The mouse tumor biology database. Nat Rev Cancer. 2008;8(6):459–465. doi: 10.1038/nrc2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Lin X, Shi DY, et al. Reactive oxygen species stimulated human hepatoma cell proliferation via cross-talk between PI3-K/PKB and JNK signaling pathways. Arch Biochem Biophys. 2002;406(2):173–182. doi: 10.1016/s0003-9861(02)00430-7. [DOI] [PubMed] [Google Scholar]
- Lehtonen R, Kiuru M, Vanharanta S, et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol. 2004;164(1):17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, et al. The fidelity of human DNA polymerase γ with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276(42):38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19(7):802–815. doi: 10.1038/cr.2009.69. (abstr.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bai RK, Trieu R, et al. Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta. 2010;1797(1):29–37. doi: 10.1016/j.bbabio.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr JA, Meierhofer D, Zimmermann F, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin. Cancer Res. 2008;14:2270–2275. doi: 10.1158/1078-0432.CCR-07-4131. [DOI] [PubMed] [Google Scholar]
- Mesquita FS, Dyer SN, Heinrich DA, et al. Reactive Oxygen Species Mediate Mitogenic Growth Factor Signaling Pathways in Human Leiomyoma Smooth Muscle Cells. Biol Reprod. 2010;82(2):341–351. doi: 10.1095/biolreprod.108.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38(11):1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26(3):268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- Nishikawa M. Reactive oxygen species in tumor metastasis. Cancer Lett. 2008;266(1):53–59. doi: 10.1016/j.canlet.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, et al. mtDNA mutations increase tumorigenicity in prostate cancer. PNAS. 2005;102(3):719–724. doi: 10.1073/pnas.0408894102. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Li Y, Zhu H, Lengauer C, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20(3):291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- Porcelli AM, Ghelli A, Ceccarelli C, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet. 2010;19(6):1019–1032. doi: 10.1093/hmg/ddp566. [DOI] [PubMed] [Google Scholar]
- Roubertoux PL, Sluyter F, Carlier M, et al. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nat Genet. 2003;35(1):65–69. doi: 10.1038/ng1230. [DOI] [PubMed] [Google Scholar]
- Sauer H, Wartenberg M, Hescheler J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell Physiol Biochem. 2001;11(4):173–186. doi: 10.1159/000047804. [DOI] [PubMed] [Google Scholar]
- Trounce I, Wallace DC. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somat Cell Molec Gen. 1996;22(1):81–85. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- Trounce I, Schmiedel J, Yen HC, et al. Cloning of neuronal mtDNA variants in cultured cells by synaptosome fusion with mtDNA-less cells. Nucleic Acids Res. 2000;28(10):2164–2170. doi: 10.1093/nar/28.10.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Buchta M, McWhinney SR, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet. 2004;74(1):153–159. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M, Kumar D. Application of mitochondrial genome information in cancer epidemiology. Clin Chim Acta. 2007;383(1–2):41–50. doi: 10.1016/j.cca.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Warburg O. Origin of Cancer Cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wittenhagen LM, Kelley SO. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci. 2003;28(11):605–611. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Ye XQ, Li Q, Wang GH, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer. 2011;129(4):820–831. doi: 10.1002/ijc.25944. [DOI] [PubMed] [Google Scholar]
- Zimmermann FA, Mayr JA, Neureiter D, et al. Lack of complex I is associated with oncocytic thyroid tumours. Br. J. Cancer. 2009;100:1434–1437. doi: 10.1038/sj.bjc.6605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann FA, Mayr JA, Feichtinger R, et al. Respiratory chain complex I is a mitochondrial tumor suppressor of oncocytic tumors. Front Biosci. 2011;3:315–325. doi: 10.2741/e247. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]