Abstract
Detailed understanding of mesenchymal stromal cells (MSC) migration is imperative for future cellular therapies. To identify genes involved in the process of MSC migration, we generated gene expression profiles of migrating and nonmigrating fetal bone marrow MSC (FBMSC). Only 12 genes showed differential expression in migrating versus nonmigrating FBMSC. The nuclear receptors Nur77 and Nurr1 showed the highest expression in migratory MSC. Nur77 and Nurr1 are members of NR4A nuclear orphan receptor family, and we found that their expression is rapidly increased upon exposure of FBMSC to the migratory stimuli stromal-derived factor-1α (SDF-1α) and platelet-derived growth factor-BB. Lentiviral expression of Nur77 or Nurr1 resulted in enhanced migration of FBMSC toward SDF-1α compared with mock-transduced FBMSC. Analysis of the cell cycle, known to be involved in MSC migration, revealed that expression of Nur77 and Nurr1 decreases the proportion of cells in S-phase compared with control cells. Further, gain-of-function experiments showed increased hepatocyte growth factor expression and interleukin (IL)-6 and IL-8 production in MSC. Despite the altered cytokine profile, FBMSC expressing Nur77 or Nurr1 maintained the capacity to inhibit T-cell proliferation in a mixed lymphocyte reaction. Our results demonstrate that Nur77 and Nurr1 promote FBMSC migration. Modulation of Nur77 and Nurr1 activity may therefore offer perspectives to enhance the migratory potential of FBMSC which may specifically regulate the local immune response.
Introduction
Mesenchymal stromal cells (MSC) are a key component of new cellular therapies, due to their multilineage differentiation potential and immunosuppressive capacities [1]. MSC can be derived from various adult and fetal tissues [2–4]. Since no specific markers are available to exclusively isolate MSC from tissues, MSC are currently defined as plastic adherent cells that are capable of in vitro differentiation toward osteoblasts, chondrocytes, and adipocytes [5]. In addition, they express CD105, CD73, and CD90, but do not express hematopoietic markers [5].
MSC have been successfully used in clinical trials to treat osteogenesis imperfecta [6] and graft-versus-host disease [7] and many other clinical applications are currently explored. For current clinical trials, tissue culture-based expansion of MSC is required to obtain a sufficient number of cells to allow successful transplantation. This cell culture step decreases the homing capacity of MSC [8]. Correspondingly, in animal studies the majority of the culture-expanded MSC do not engraft and disappear within a few days after intravenous transplantation [9,10]. Therefore, it is suggested that the benefits of MSC transplantation are not only caused by locally engrafted cells but also by systemic effects of secreted factors.
It is often assumed that common mechanisms of cell migration also apply to MSC migration. Previous studies have identified chemotactic stimuli for MSC, such as stromal-derived factor-1α (SDF-1α) [11,12], platelet-derived growth factor (PDGF) [13], hepatocyte growth factor (HGF) [14], and basic fibroblast growth factor [15]. These stimuli induce migration of MSC derived from various adult and fetal tissues [16–18]. In addition, MSC have the capacity to migrate across endothelial monolayers [19], an important feature since MSC are administered intravenously in most clinical trials. However, we and others observed that only a small fraction of MSC shows efficient migration [9,16,20]. Thus, it is important to elucidate why only a small proportion of all culture-expanded MSC is able to migrate and whether these migratory cells can be discriminated from nonmigratory MSC in terms of function and phenotypic markers. In search for differences between migratory and nonmigratory MSC, we reported that the molecular machinery involved in migration was functional in both groups. In addition, migratory and nonmigratory cells could not be distinguished by cell surface markers such as integrins, adhesion molecules or chemokine and growth factor receptors [16]. Interestingly, we found that the cell cycle correlated with MSC migration. Similar to human hematopoietic stem cells, MSC in S- and G2/M-phase showed reduced migration compared with MSC in G1-phase [16,21].
In the current study, we report on a search for novel factors that regulate MSC migration. We performed a microarray-based gene expression analysis of migratory and nonmigratory fetal bone marrow MSC (FBMSC) and identified 2 genes involved in FBMSC biology, which are significantly increased in migratory FBMSC compared with nonmigratory cells: Nur77 and Nurr1. Overexpression of these nuclear receptors increased migration toward SDF-1α and enhanced cytokine production, whereas the immunosuppressive capacities of MSC were maintained.
Materials and Methods
Isolation and culture of MSC
Fetal bone derived from 4 individual donors was obtained after informed consent from legally terminated second trimester pregnancies. The protocol for collecting fetal tissues for research purposes was approved by the medical ethics review board of the AMC (MEC: 03/038). To obtain FBMSC, fetal bones were flushed with Iscove’s Modified Dulbecco’s Media (IMDM) (Lonza) containing 10% fetal calf serum (FCS; Bodinco), 50 U/mL penicillin, and 50 μg/mL streptomycin (Gibco). The remaining erythrocytes were lysed using NH4Cl for 10 min on ice. Subsequently, cells were rinsed in phosphate-buffered saline (PBS). About 1.6×106 cells were seeded per well in 6-well dishes in M199 (Gibco) supplemented with 10% FCS, penicillin (50 IU/mL), streptomycin (50 μg/mL), 20 μg/mL ECGF (Roche Diagnostics), and 8 IU/mL heparin (Leo Pharma), hereafter referred to as M199c. The obtained cells were considered to be FBMSC. Upon reaching 80% confluency after initial plating, FBMSC were replated and further cultured in T80 tissue culture flasks at an initial density of 2,500 cells/cm2. For all experiments, 80% confluent passage 3–5 FBMSC were used.
Flow cytometry
MSC were characterized for surface expression by flow cytometry. Cells were rinsed, trypsinized, washed, and resuspended in PBS containing 0.2% bovine serum albumin (BSA) before incubation (20 min at room temperature) with the following monoclonal antibodies. Antibodies purchased from BD: CD73 (clone AD2), CD90 (clone 5E10), and CD45 (clone HI30). Antibodies from other companies: CD105 (clone SN6; Ancell) and CD34 (clone 581; IQ-Products). As a negative control, cells were labeled with IgG1 isotype controls (monoclonal antibodies, Sanquin; BD). A minimum of 10,000 events was recorded, using a FACS LSR II flow cytometer (BD).
Differentiation experiments
To study the multilineage differentiation capacity, MSC were cultured under conditions that promote differentiation toward osteoblasts or adipocytes as previously described [22]. MSC were plated in a 24-well dish at a plating density of 2.5×104 cells/cm2 in α-MEM (Gibco). For osteogenic induction, α-MEM was supplemented with 10% FCS and penicillin (50 IU/mL)–streptomycin (50 μg/mL) to which ascorbic acid (50 μg/mL; Sigma) and dexamethasone (10−7 M; Sigma) were added. From day 7 onward, β-glycero-phosphate (5 mM; Sigma) was added. Cultures were incubated in a humidified atmosphere of 5% CO2 at 37°C. Medium was replaced every 4th and 7th day of the week. For induction of adipogenesis, indomethacine (50 μM; MP Biomedicals), IBMX (0.5 mM; Sigma), and insulin (1.6 μM; Sigma) were used.
At day 21, the cells induced toward osteogenic differentiation were stained for alkaline phosphatase and calcium deposition. Cells were incubated with a substrate solution [0.2 mg/mL α-naphthyl-1-phosphate (Sigma)], 3 mg/mL sodium borate, 0.3 mg/mL magnesium sulfate and 0.8 mg/mL fast blue RR acid (Sigma) for 15 min, resulting in the formation of an insoluble purple reaction product. To detect calcium deposition, cells were fixed with 3.7% formaldehyde (Merck) for 10 min, and stained with 2% Alizarin Red S (ICN Biomedicals) and 0.1 NH4OH [pH 5.4] for 1 min. Mineralization was indicated by the presence of red depositions. To demonstrate the presence of adipocytes, expanded cells were fixed as described above. Cytoplasmic inclusions of neutral lipids were stained with Oil-Red-O (3 mg Oil-Red-O/mL 60% isopropanol; Sigma) for 10 min.
In vitro migration experiments
Migration experiments for collecting migratory and nonmigratory MSC toward SDF-1α (600 ng/mL; Peprotech) were performed using 12 μm pore size Transwell plates (Corning Costar). Optimal SDF-1α concentration for migration was determined previously [16]. The apical compartment was coated overnight with fibronectin (20 μg/mL; Sigma) in PBS. One lakh cells were seeded into the apical compartment in 500 μL IMDM supplemented with 0.25% BSA (Sigma), and the stimulus was added to the basolateral compartment in 1.5 mL IMDM with 0.25% BSA. Cells were allowed to migrate for 4 h.
For collection of migratory cells for RNA extraction and subsequent microarray analysis, 21 Transwell inserts were used. To obtain a pure migratory fraction, nonmigratory MSC were removed from the apical side of the membrane using a cotton swap. After removing the nonmigratory MSC, the inserts were carefully rinsed in PBS twice before trypsinizing the migratory MSC from the membrane. The cells were collected in en eppendorf tube and lysed in RLT buffer (Qiagen) supplemented with β-mercaptoethanol according to the manufacturer's instructions. Nonmigratory MSC were collected from 3 Transwell inserts. To collect a pure nonmigratory FBMSC fraction, migratory MSC on the basolateral compartment of the Transwell membrane were reoved with a cotton swap. The filters were rinsed in PBS and nonmigratory cells were collected by Trypsin detachment, collected, and lysed as described.
To evaluate the percentage of migratory cells, 4 inserts (2 negative controls and 2 SDF-1α-stimulated wells) were fixed in 3.7% formaldehyde and further stained with Hoechst 33258 (1:500 dilution; Invitrogen). The Transwell filter membranes were mounted onto glass slides using Mowiol (Sigma). The total number of migrating cells per view field was qualified by counting nuclei using fluorescence microscopy. Twenty-three view fields per filter were counted. Unless stated otherwise, data were expressed as the percentage of migrating cells related to the total number of cells loaded into the upper compartment.
RNA extraction and microarray analysis
RNA was extracted using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions, and RNA integrity was analyzed on a Agilent 2100 bioanalyzer (Agilent Technologies, Inc.) using the Agilent RNA 6000 Non-Assay protocol. Three hundred nanograms RNA was hybridized to an Affymetrix Human Exon 1.0 ST array (Affymetrix, Inc.). Hybridized assays were scanned with a GeneChip scanner 3000 7G (Affymetrix). Affymetrix CEL-files were imported into Partek® (Partek® Genomic Suite software, version 6.4 Copyright © 2008 Partek, Inc.) where only core probe sets were extracted and normalized using the RMA algorithm with GC background correction. Core transcript summaries were calculated using the mean intensities of the corresponding probe sets, representing the quantitative expression levels of all genes. The correspondence of the replicate samples was confirmed using principle component analysis and Pearson correlation analysis. We performed an analysis of variance on the log 2 probe intensities, representing the gene expression intensities, of 6 samples (migratory and nonmigratory FBMSC derived from 3 independent donors) and did a post hoc analysis to compare migratory and nonmigratory samples. Data are deposited in Gene Expression Omnibus under the accession no. GSE24892 (www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE24892).
cDNA synthesis and real-time quantitative–polymerase chain reaction
A maximum of 1 μg total RNA was used for cDNA synthesis using random hexamers (25 μM; Gibco-BRL Life Technologies). real-time quantitative (RQ)–polymerase chain reactions (PCRs) were performed on a StepOne Plus (Applied Biosystems) using Sybrgreen dye for detection (Sybrgreen mastermix; Applied Biosystems). Primer sequences are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/scd). Abelson (ABL) was used for normalization [normalized threshold cycle (ΔCt)=Ctgene−CtABL]. Fold changes were normalized to the control sample in each experiment: 2−(ΔCt sample-ΔCt value control).
Production of lentiviral particles
Lentiviral vectors for knock down and overexpression of Nur77 and Nurr1 have been described previously [23]. Briefly, 293T cells were cultured in DMEM (25 mM HEPES; Gibco) containing 10% FCS, penicillin (50 IU/mL), streptomycin (50 μg/mL), and l-glutamine (2 mM; Gibco) and transfected with pMDL-pRRE, pVSV-G and pRSV-REV and Nur77 or Nurr1 using the Calcium Phosphate method. Viral particles containing supernatant was harvested at days 1 and 2 after transfection. After ultracentrifugation, viral titers were determined as described by Sastry et al. [24]. Briefly, serial dilutions of concentrated virus were used for transduction of 293T cells. Twenty-four hour after transduction, total genomic DNA was isolated using a DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's instructions. The number of inserted vector copies was determined by RQ-PCR, using the vector DNA for the standard curve: forward primer, 5′ CTGCAGCAGCAGAACAATTTG 3′; reverse primer, 5′ CCCCAGACTGTGAGTTGCAA 3′.
Transduction of MSC
One lakh MSC per well were plated in a fibronectin coated 6-well plate and allowed to adhere for 24 h. For transduction, medium was replaced by IMDM+0.25% BSA containing viral particles [multiplicity of infection (MOI) 100 and 600 for overexpression and knockdown, respectively]. After over-night exposure to the virus, the medium was replaced with M199c and the cells were cultured for another 24 h. Transduction efficiency was evaluated by RQ-PCR.
Cell cycle analysis
Three days after transduction, MSC were plated at 10,000 cells/cm2 and harvested for cell cycle analysis after 24 h. Cells were trypsinized and fixed in 4% paraformaldehyde (PFA) at room temperature or in 70% EtOH on ice and subsequently permeabilized in 0.1% Triton X-100. MSC were then incubated with Ki67 (Clone MIB-1; Dako) for 30 min at 4°C in PBS containing 0.1% Triton X-100. Finally, both overexpression and knockdown MSC were incubated for 15 mins at 37°C with labeling reagent containing Hoechst 33258 (1 μg/mL), RNase A (Sigma), and 0.1% Triton X-100. A minimum of 10,000 cells was analyzed by flow cytometry as described above and further analyzed using Modfit LT 3.0 software (Verity Software House). Ratios were calculated for each cell cycle phase from the percentage of cells in a certain phase in migrating MSC divided by the percentage of cells in the same phase in nonmigrating MSC.
Cytokine treatment of MSC and quantification of cytokines in culture supernatant
Three days after transduction, MSC were plated at a density of 15,000 cells/cm2 in 24-well plates (Nunc) in M199c. After 24 h, the medium was refreshed and cells were stimulated with tumor necrosis factor-α (TNF-α) (1,000 U/mL; Peprotech) or interferon-γ (IFN-γ) (1,000 U/mL; Peprotech) or left untreated. Conditioned medium and RNA samples were collected 6 and 24 h after stimulation. Expression levels of transforming growth factor-β1 (TGF-β1), HGF, and indoleamine-2,3-dioxygenase (IDO) were determined by RQ-PCR as described above. Protein levels of interleukin (IL)-6, IL-8, and IL-10 were measured by ELISA using the PeliKine compact ELISA (Sanquin Reagents).
Immunofluorescent microscopy
Three days after transduction, 10,000 transduced or control MSC were seeded on fibronectin-coated coverslips and were allowed to adhere for 24 h. The cells were fixed for 10 min on ice, then permeabilized using 0.2% Triton X-100 (Sigma) and stained with anti-Nur77 (M210, Rabbit polyclonal; Santa Cruz Biotechnologies) or anti-Nurr1 (N20, Rabbit polyclonal; Santa Cruz), followed by secondary goat-anti-rabbit Alexa 568 (20 μg/mL), Alexa 488 phalloidin, and DAPI (all form Invitrogen Molecular Probes). Coverslips were then mounted using Mowiol (Sigma). Immunofluorescent staining was detected using a LSM 510 META confocal microscope (Zeiss) using a 40×oil-objective. Images were captured by ZEN 2007 confocal software (Zeiss).
Cocultures MSC and peripheral blood mononuclear cells
Immunosuppressive capacity of MSC was tested in a coculture of MSC and peripheral blood mononuclear cells (PBMC). MSC from 3 individual donors were plated in graded doses in a 96-well flat-bottom plate in DMEM-Glutamax-I (Invitrogen) containing 50 U/mL penicillin, 50 U/mL streptomycin (Lonza), and 10% FBS (Greiner Bio-One). Cells were allowed to adhere overnight. PBMC (1.0×105/well) from 1 donor were added to the MSC and stimulated with human T-activator CD3/CD28 dynabeads (Invitrogen Dynal AS) in a bead:cell ratio 1:5. After 5 days of coculture, cells were pulsed with [3H]thymidine (0.5 μCi/well) and incubated for 16 h at 37°C. The cultures were harvested on a glass fiber filter, and thymidine incorporation was measured with a liquid scintillation counter (Wallac). Data were expressed as median counts per minute of triplicate cocultures.
Statistical analysis
Statistical significance was determined by Man–Whitney U test, using SPSS 15.0 (SPSS, Inc.), except for cell cycle data, which were analyzed by one-sample Kolmogorov–Smirnov Test. Results were considered significant when P≤0.05.
Results
Characterization of FBMSC
The FBMSC isolated from all 4 donors had a spindle-shaped morphology and differentiated toward osteoblasts and adipocytes upon induction of differentiation (Supplementary Fig. S1). In addition, these FBMSC expressed CD73, CD90, and CD105, and did not express the hematopoietic markers CD34, CD45 (Supplementary Table S2), CD11b, CD14, CD19, or HLA-DR (data not shown). Thus, based on these currently accepted criteria, these cells were designated as MSC.
Gene expression profiling of migratory MSC
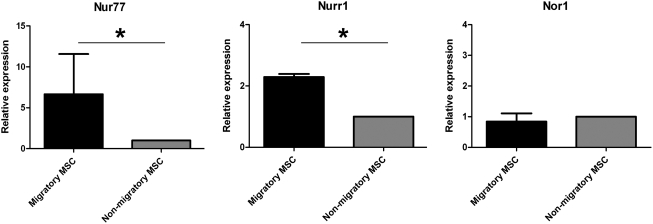
Transwell migration experiments were performed to separate migratory from nonmigratory FBMSC fractions before transcriptome analysis using Affymetrix Human Exon 1.0 ST arrays. Whereas 5.4%±0.6% (n=4) of the cells showed spontaneous migration, SDF-1α stimulated migration of 11.2%±0.2% (n=4) of the cells (Supplementary Fig. S2). These data are in agreement with our previous results [16]. RNA was isolated from FBMSC that did or did not show a chemotactic response toward SDF-1α and this RNA was hybridized to Affymetrix Human Exon 1.0 ST array. We performed an analysis of variance on the log 2 probe intensities (representing the gene expression intensities) of 6 samples from 3 FBMSC donors and did a post hoc analysis to compare migratory and nonmigratory samples. Only 12 genes were differentially expressed with statistical significance (P<0.05) with a minimum fold change of 1.6 between migratory and nonmigratory MSC (Table 1). The nuclear orphan receptors NR4A1 (Nur77/TR3) and NR4A2 (Nurr1/NOT), hereafter referred to as Nur77 and Nurr1, respectively, were the only 2 genes that were increased by more than 2-fold in migratory FBMSC as compared with nonmigratory FBMSC. The array data were successfully confirmed by RQ-PCR on the samples used for the microarray as well as on samples obtained from independent migration experiments (Fig. 1).
Table 1.
Differentially Expressed Genes Migrating Versus Nonmigrating Mesenchymal Stromal Cells
| Symbol//gene name | NCBI accession no. | Fold change (±SEM) |
|---|---|---|
| NR4A1//nuclear receptor subfamily 4, group A, member 1 | NM_002135 | 2.3±0.48 |
| NR4A2//nuclear receptor subfamily 4, group A, member 2 | NM_006186 | 2.3±0.67 |
| OBP2B//odorant binding protein 2B | NM_014581 | 2.0±0.14 |
| GADD45B//growth arrest and DNA-damage-inducible, beta | NM_015675 | 1.9±0.25 |
| ZNF808//zinc finger protein 808 | NM_001039886 | 1.7±0.23 |
| CYR61//cysteine-rich, angiogenic inducer, 61 | NM_001554 | 1.7±0.32 |
| KIAA1274//KIAA1274 | — | 1.7±0.28 |
| OR4D1//olfactory receptor, family 4, subfamily D, member 1 | NM_012374 | 1.6±0.26 |
| SMAD7//SMAD family member 7 | NM_005904 | 1.6±0.33 |
| HIST1H4B//histone cluster 1, H4b | — | −1.6±0.18 |
| BDH2//3-hydroxybutyrate dehydrogenase, type 2 | BC037277 | −1.6±0.26 |
| HIST1H2AK//histone cluster 1, H2ak | NM_003510 | −1.6±0.30 |
n=3.
SEM, standard error of the mean.
FIG. 1.
Microarray confirmation by real-time quantitative–polymerase chain reaction (RQ-PCR). The increased expression of Nur77 and Nurr1 in migratory FBMSC compared with nonmigratory MSC was confirmed by RQ-PCR for Nur77 (left) and Nurr1 (middle). Bars depict the fold increase (mean±SD, n=3) of migratory FBMSC normalized to nonmigratory FBMSC. The third family member Nor1 (right) was confirmed not differentially expressed. *P≤0.05. FBMSC, fetal bone marrow mesenchymal stromal cells; MSC, mesenchymal stromal cells.
Nur77 and Nurr1 are members of the NR4A subfamily of nuclear orphan receptors that comprises 3 members. They are believed to function as ligand-independent early response genes that are involved in proliferation, apoptosis, and inflammation (reviewed in Refs. [25,26]). The third member of the NR4A family, NOR-1 (NR4A3), was not differentially expressed between migratory and nonmigratory FBMSC (Fig. 1).
SDF-1α and PDGF-BB induce Nur77 and Nurr1 mRNA expression in FBMSC
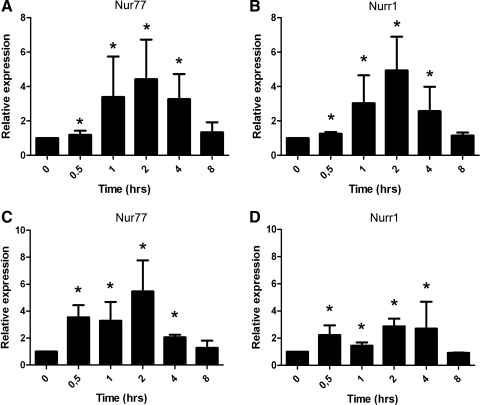
Gene expression of all 3 NR4A family members is enhanced in response to various growth factors, chemokines, and cytokines in, among others, vascular smooth muscle cells [27], endothelial cells [28], and macrophages [23,29]. To study whether exposure to SDF-1α increases Nur77 and Nurr1 expression in MSC, FBMSC were exposed to 600 ng/mL SDF-1α for different time-periods. A maximum increase in Nur77 (4.4-fold) and Nurr1 (4.9-fold) expression was observed already after 2 h (Fig. 2A, B). This effect was transient, as expression of both Nur77 and Nurr1 declined to control levels after 8 h. To test whether this induction was specific for SDF-1α, FBMSC were also exposed to platelet-derived growth factor-BB (PDGF-BB), which is a growth factor that is a potent chemoattractant for MSC [13,30]. Similar to SDF-1α, exposure to PDGF-BB induced mRNA expression of Nur77 with a maximum around 2 h (Fig. 2C), whereas Nurr1 was induced to a lesser extent (Fig. 2D).
FIG. 2.
SDF-1α and PDGF-BB exposure induces Nur77 and Nurr1 expression in FBMSC. (A, B) A rapid significant induction (P<0.05) of Nur77 (A) and Nurr1 (B) was observed after SDF-1α (600 ng/mL) stimulation for 0.5 h and persisted for 4 h. (C, D) PDGF-BB (5 ng/mL) significantly induced expression (P<0.05) of Nur77 (C) and to a lesser extent Nurr1 (D) from 0.5 to 4 h. Bars represent the fold increase (mean±SD) of stimulated FBMSC normalized to unstimulated cells at indicated points. Basal expression of Nur77 and Nurr1 is similar and it is approximately128-fold lower than that of ABL (data not shown). SDF-1α, stromal cell-derived factor 1α; PDGF-BB, platelet-derived growth factor–BB. *p<0.05.
Overexpression of Nur77 and Nurr1 enhances MSC migration
Subsequently, FBMSC were transduced with lentivirus to induce overexpression of Nur77 and Nurr1. Cells were analyzed 72 h after viral transduction. In Nur77-or Nurr1-transduced FBMSC, immunofluorescent staining was observed predominantly in the nucleus as expected (Supplementary Fig. S3A). Overexpression was also validated by RQ-PCR (Supplementary Fig. S3B). As Nur77 and Nurr1 expression under basal conditions is very low (7±0.9 Ct lower than ABL), transduction results in very high expression of these genes. Lentiviral transduction did not influence the classical MSC phenotype (Supplementary Table S2). Nur77 and Nurr1 expression was increased upon induction of differentiation toward osteoblasts or adipocytes (Nur77, 16-fold; Nurr1, 3-fold) in wild type and mock-transduced MSC. This elevated expression persisted during the complete differentiation period (data not shown). In agreement with these findings, the differentiation capacity of FBMSC toward osteoblasts or adipocyte upon induction of differentiation was not affected (Supplementary Fig. S1). Importantly, overexpression of Nur77 or Nurr1 did not induce spontaneous differentiation (Supplementary Fig. S1). Nur77 induces apoptosis in T-cells [31] and other cell types [32,33]; however, overexpression of Nur77 or Nurr1 did not induce apoptosis in FBMSC (data not shown).
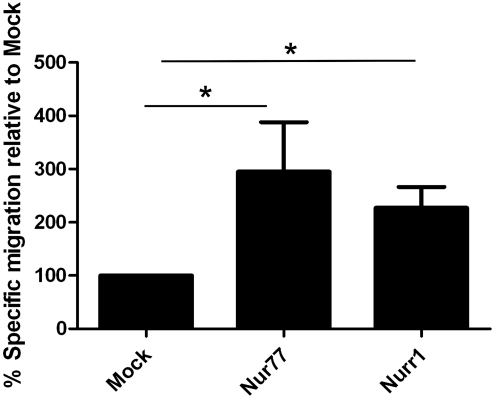
Because expression levels of Nur77 and Nurr1 were increased in migratory MSC, we tested for a causal relationship by determining whether overexpression of Nur77 or Nurr1 in FBMSC results in enhanced migration toward SDF-1α. Importantly, overexpression of Nur77 or Nurr1 did not influence expression of CXCR4, the receptor for SDF-1α. Three days after transduction, Nur77, Nurr1, or mock-transduced FBMSC were allowed to migrate toward SDF-1α for 4 h in a Transwell-based chemotaxis assay. As depicted in Fig. 3, expression of Nur77 and Nurr1 significantly increased chemotaxis as compared with mock-transduced FBMSC (Nur77 mean 297%±186% relative to mock, P≤0.014, n=4; Nurr1 mean 227%±79%, P≤0.014, n=4).
FIG. 3.
Overexpression of Nur77 and Nurr1 increases specific FBMSC migration. Mock-, Nur77-, and Nurr1-transduced FBMSC were allowed to migrate for 4 h toward 600 ng/mL SDF-1α or control medium. The percentage of migratory cells was evaluated by fluorescence microscopy. The bars represent the percentage (mean±SD) specific migration (% SDF-induced migration −% spontaneous migration) relative to mock-transduced FBMSC. *P≤0.05.
Nur77 and Nurr1 are involved in FBMSC cell cycle regulation
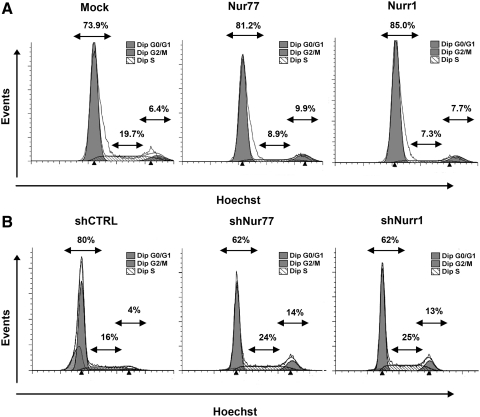
We previously showed that migration of MSC depends on cell cycle status. Cells in S- and G2/M-phase have a reduced migratory capacity [16]. Nur77 and Nurr1 are known to be involved in cell cycle regulation of endothelial cells [28,34] and smooth muscle cells [35]. We therefore studied the role of Nur77 and Nurr1 in cell cycle regulation of FBMSC. Three days after lentiviral transduction, mock-, Nur77-, or Nurr1-transduced FBMSC were seeded at identical cell densities in fibronectin-coated 6-well plates. The next day, cells were harvested for cell cycle analysis by flow cytometry. Overexpression of Nur77 or Nurr1 significantly reduced the proportion of cells in S-phase from 19.7% to 8.9% and 7.3%, respectively (a reduction of 38%±10% [P<0.04, n=4] and 33%±17% [P<0.039, n=4]) (Fig. 4A). The percentage of cells in G0/G1 was concomitantly increased (Fig. 4A). The altered cell cycle distribution in Nur77- and Nurr1-transduced cells suggests decreased proliferation in these groups. Although cell numbers were increased in all treatment groups 3 days after transduction, fewer cells were harvested from Nur77-transduced (fold increase in 3 days 1.5±0.6, n=4) and Nurr1-transduced (1.4±0.6, n=4) groups compared with mock (1.8±0.7, n=4) or nontransduced cells (1.9±0.8, n=4). Because overexpression of Nur77 or Nurr1 did not increase apoptosis (data not shown), the decreased cell numbers in Nur77- and Nurr1-transduced cells are most likely due to decreased proliferation. ShRNA-based reduction of Nur77 and Nurr1 expression was achieved by lentiviral transduction. Transduction efficiency determined by flow cytometry was 90%–100% (data not shown) and this reduced expression of Nurr77 with 2.2±0.3-fold and Nurr1 11±8-fold (Supplementary Fig. S3C). Unfortunately, we were not able with the currently available antibodies to detect endogenous Nur77/Nurr1 protein expression in these cells. The specificity of our shRNA sequences has, however, been tested extensively both at mRNA and at protein level [36]. In agreement with the decreased percentage of cells in S-phase observed in cells that overexpress Nur77 and Nurr1, loss of their expression by shRNA transduction induced an increase in the fraction of cells in S-phase from 16% in shCTRL-transduced cells to 24% (shNur77) or 25% (shNurr1), an increase of 37%±11% P<0.036, n=3 and 45%±12%, P<0.03, n=3) (Fig. 4B). Correspondingly, the percentage of cells in G2/M-phase was also increased [percentage increase G2/M 76%±3% P<0.021, n=3 (Nur77); 72%±3%, P<0.021, n=3 (Nurr1)] and the fraction of cells in G0/G1-phased was decreased (Fig. 4B).
FIG. 4.
Nur77 and Nurr1 regulate cell cycle progression in FBMSC. Three days after transduction, FBMSC were seeded at identical densities. The next day, cell cycle distribution was measured by flow cytometry. (A) Overexpression of Nur77 (middle) or Nurr1 (right) decreased the percentage in S-phase of the cell cycle compared with mock-transduced FBMSC (left). Histograms represent the mean fluorescence of indicated FBMSC groups (representative experiment shown). (B) Complementary to overexpression, reduction of Nur77 (middle) or Nurr1 (right) expression increased the percentages in S- and G2/M-phase compared with short-hairpin control-transduced FBMSC (left) (representative experiment shown).
These data show that Nur77 and Nurr1 are involved in cell cycle regulation in MSC. Moreover, our findings that high expression levels lead to decreased proliferation and increased chemotaxis are in full agreement with our earlier results [16] showing that decreased proliferation correlates positively with increased migration.
Nur77 and Nurr1 increase cytokine and growth factor production
MSC secrete a wide variety of cytokines, chemokines, and growth factors [37] that contribute to MSC-mediated immune suppression [38,39]. IL-6, IL-8, IL-10, HGF, TGF-β1, and IDO secreted by MSC are key factors in suppressing T-cell proliferation and monocyte differentiation [1,40]. Overexpression of NR4A family members in macrophages resulted in an anti-inflammatory secretory profile [23]. Thus, we studied whether Nur77 and Nurr1 affect cytokine and growth factor release in FBMSC under basal and TNF-α- or IFN-γ-stimulated conditions.
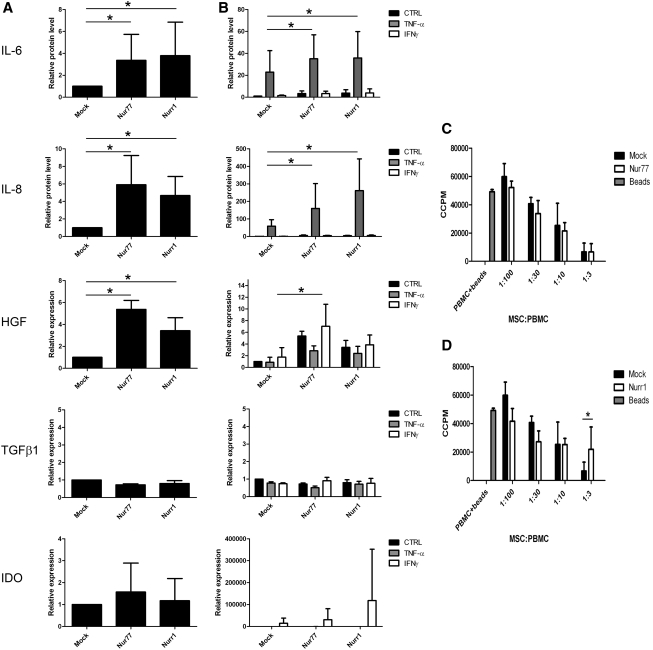
Expression of Nur77 and Nurr1 leads to significantly increased secretion of IL-6 and IL-8 protein and an increase in mRNA for HGF in otherwise unstimulated cells (Fig. 5A). IL-10 protein was not detected (data not shown). IL-6 and IL-8 protein levels were enhanced by TNF-α, whereas HGF expression was increased by IFN-γ. Under all conditions, Nur77 and Nurr1 expression resulted in a statistically significant, further increase in IL-6, IL-8 release, and HGF mRNA levels (Fig. 5B). TGF-β1 mRNA expression was not influenced by overexpression of Nur77 or Nurr1, nor by TNF-α or IFN-γ stimulation (Fig. 5A, B). Expression of IDO, important for inhibition of T-cell proliferation [41], was strongly induced by IFN-γ exposure as described previously [42]. IFN-γ induced IDO expression was not changed in case of Nur77 or Nurr1 gain of function (Fig. 5A, B). In contrast to overexpression, reduction of Nur77 or Nurr1 expression did not consistently reduce the production of IL-6 and IL-8 or expression of HGF, TGF-β1, or IDO (data not shown). This may be expected as expression of Nur77 and Nurr1 is low under homeostatic conditions and expression is rapidly induced upon stimulation of a wide range of cytokines. These data show that upon increase of Nur77 or Nurr1 expression, production of IL-6 and IL-8, and expression of HGF in FBMSC is enhanced; however, expression of Nur77 or Nurr1 is not required for production of these cytokines.
FIG. 5.
Increased cytokine production in Nur77- and Nurr1-transduced FBMSC does not influence T-cell proliferation. (A) Four days after transduction, IL-6 and IL-8 protein secreted in supernatant of unstimulated FBMSC cultures were measured by ELISA after 24 h incubation and mRNA levels of HGF, TGF-β1, and IDO were analyzed by RQ-PCR. Basal protein levels of IL-6 and IL-8 and HGF mRNA expression were significantly increased in Nur77- or Nurr1-transduced FBMSC compared with mock-transduced FBMSC. Data (mean±SD) normalized to unstimulated mock-transduced MSC. (B) Transduced FBMSC were stimulated with TNF-α and IFN-γ for 24 h or left untreated. Again the highest levels of IL-6 and IL-8 protein and HGF mRNA were observed in Nur77- and Nurr1-tranduced FBMSC. Data (mean±SD) normalized to unstimulated mock-transduced MSC. (C, D) Transduced FBMSC were cocultured with CD3/CD28 bead-activated PBMC. FBMSC with enhanced expression of Nur77 (C) or Nurr1 (D) have at least a similar capacity to inhibit T-cell proliferation compared with mock-transduced FBMSC. Beads were added to PBMC as negative control (CCPM count 49.157). Data represent pooled data (mean±SD) of 3 individual FBMSC donors cocultured with the same allogeneic PBMC. IL-6, interleukin-6; IL-8, interleukin-8; HGF, hepatocyte growth factor; TGF-β1, transforming growth factor-β1; IDO, indoleamine-2,3-dioxygenase; TNF-α, tumor necrosis factor-α; IFN-γ, interferon-γ; PBMC, peripheral blood mononuclear cells; CCPM, radioactivity counts per minute. *P≤0.05.
Overexpression of Nur77 or Nurr1 does not affect immune suppression by FBMSC
In a final series of experiments, we tested whether increased expression of Nur77 and Nurr1, which results in increased migration and cytokine production by MSC (Figs. 3 and 5), alters the capacity of FBMSC to inhibit T-cell proliferation. FBMSC from 3 individual donors were cocultured with CD3/CD28 bead-activated PBMC from 1 donor. Similar to mock-transduced FBMSC, Nur77- and Nurr1-transduced cells were able to inhibit T-cell proliferation as determined by 3H-thymidine incorporation (Fig. 5C, D). Only at a very high ratio (1:3) of PBMC versus FBMSC, Nurr1-transduced FBMSC showed a significantly decreased capacity to inhibit T-cell proliferation (n=3, P<0.030, Fig. 5D) compared with the mock-transduced cells.
Thus, the altered cytokine profile in FBMSC caused by overexpression of Nur77 or Nurr1 does not impair MSC-mediated suppression of T-cell proliferation.
Discussion
MSC are interesting candidates for cellular therapy due to their multilineage differentiation potential and immunosuppressive capacities. Culture expansion of MSC is required to obtain the required quantity for clinical application; however, culture expansion impairs the homing capacity of MSC to the bone marrow [8]. Thus, insight into the process of MSC migration and targeting is imperative for improving cellular therapies. In the present study, we identified the nuclear receptors Nur77 and Nurr1 as 2 genes regulating FBMSC migration and cytokine production.
In our transcriptome analysis of migratory and nonmigratory MSC, only 12 genes were differentially expressed. This set did not include cell surface markers that enable selection for a migratory subset, nor did it include previously reported genes associated with enhanced MSC migration such as CD49f and PODXL [43]. The transcription factors Nur77 and Nurr1 gave the most specific expression in migratory cells. Nur77 and Nurr1 are members of the NR4A subfamily of nuclear orphan receptors that function as ligand-independent early response genes involved in proliferation, apoptosis, and inflammation. NR4A members are thought to act as constitutively active transcription factors that bind the promoter of target genes on consensus NBRE sites. Only a few direct target genes have been identified to date (reviewed in Refs. [25,26]). Gene expression of NR4A members is induced by various growth factors and cytokines in a variety of cell types [23,27–29]. Transcriptional activation of NR4A nuclear receptors is most likely dependent on the expression level of the receptors themselves and on post-translational control (reviewed in Refs. [25,26]). Expression of Nur77 has not been described in MSC before, whereas increased expression of Nurr1 was observed in adult BMSC exposed to SDF-1α [44] or CXCL7 [45]. In these 2 studies, no functional experiments on Nurr1 were performed. The findings in adult BMSC are in good agreement with our data obtained with FBMSC, in which exposure to SDF-1α or PDGF-BB alone rapidly induced an increase in expression of Nur77 and Nurr1.
Since we found Nur77 and Nurr1 to be upregulated in migratory MSC, a lentiviral expression approach was used to study the contribution of these proteins to FBMSC migration. Indeed, expression of Nur77 or Nurr1 in FBMSC significantly increased migration compared with mock-transduced cells. These observations correspond well to impaired migration observed in dopaminergic neurons from Nurr1-null mice [46] and the reduced migration observed after reduction of Nurr1 expression in endothelial cells [28] and tumor cell lines [47]. Further, Nur77 was described to be among the 17 genes associated with primary solid tumor metastasis [48]. No previous study has reported that overexpression of Nur77 can enhance cell migration.
Although the mechanism by which Nur77 and Nurr1 enhance cell migration is unknown, these proteins could favor MSC migration by modulating the cell cycle. In endothelial cells, proliferation is decreased either when Nur77 is highly expressed [34] or Nurr1 expression is low [28]. In vascular smooth muscle cells, Nur77 decreases proliferation [35]. Recently, Sirin et al. demonstrated that Nurr1 overexpression drives hematopoietic stem cells to quiescence [49]. Thus, involvement of Nur77 and Nurr1 in cell cycle regulation is well established; promoting or inhibitory effects may be cell type dependent.
Our data show that in FBMSC, Nur77 and Nurr1 decrease the proportion of cells in S-phase, whereas a loss of expression promotes cell cycle progression. The observations that Nur77 and Nurr1 promote MSC migration and decrease proliferation are in agreement with our previous finding that MSC in S- and G2/M-phase of the cell cycle are less prone to migrate [16].
Several studies revealed that the NR4A can inhibit the NFkB pathway and as such regulate cytokine synthesis [23,50,51]. The interaction between NFkB and NR4A may be a common mechanism, as this was reported in macrophages [23,29], synoviocytes [50], microglia, and asterocytes [51].
As Nur77 and Nurr1 enhance FBMSC migration and regulate inflammatory cytokine production in macrophages [23,52], it was important to test if Nur77 and Nurr1 expression in FBMSC would promote the immune modulatory capacities of these cells. Therefore, we studied the effect of Nur77 and Nurr1 on cytokine and growth factor production in FBMSC under basal and inflammatory conditions. In both conditions, Nur77 and Nurr1 increased IL-6 and IL-8 production and HGF mRNA expression, showing that also in FBMSC, Nur77 and Nurr1 promote the production of certain cytokines. A strong increase of IL-8 production by Nurr1 with a synergistic effect of TNF-α was also found in synoviocytes [50].
Based on the increased levels of HGF, IL-6, and IL-8, it cannot be predicted which effect Nur77 and Nurr1 would exert on MSC-mediated immune suppression. Increased production of HGF may result in stronger immune suppression by FBMSC overexpressing Nur77 or Nurr1 [38]. Elevated levels of IL-6 are associated with both pro- and anti-inflammatory processes such as neutrophil and leukocyte infiltration [53] or inhibition of monocyte maturation and differentiation by MSC [54], whereas IL-8 enhances inflammation by inducing neutrophil chemotaxis [55]. Our coculture experiments of transduced MSC with PBMC showed that Nur77- or Nurr1-transduced FBMSC were able to efficiently inhibit T-cell proliferation similar to control cells at low ratios of MSC versus PBMC. Only at a nonphysiological ratio, inhibition of T-cell proliferation by Nurr1-transduced FMBSC was significantly reduced.
In summary, we have identified 2 genes, Nur77 and Nurr1, that promote migration of FBMSC. In addition, Nur77 and Nurr1 expression increases MSC cytokine production, whereas these MSC retained the capacity to inhibit T-cell proliferation. Because expression of Nur77 and Nurr1 can be induced by various growth factors and cytokines, studies on cytokine pretreatment for optimal transient upregulation of Nur77 and Nurr1 in MSC will contribute to enhancing MSC migration in a preclinical and clinical setting. In conclusion, modulation of Nur77 and Nurr1 expression in MSC therefore offers perspectives to endow migratory MSC with the capacity to specifically regulate the local immune response.
Supplementary Material
Acknowledgments
The authors would like to thank C. Mudde Jadra and S.van Reijmerdal for technical support, Ms. T. Jorritsma for TGF-β1 primer design, and the staff of the Bloemenhove Kliniek (Heemstede, The Netherlands) for providing fetal tissues. We also thank Dr. P.L Hordijk and Dr. M.M. von Lindern for critically reading the article. M.W.M. is financially supported by DPTE grant no. 06728, and S.M.M. and H.R. by DPTE grant no. 06725.
Disclosure Statement
No competing financial interests exist.
References
- 1.Nauta AJ. Fibbe WE. Immunomodulatory properties of mesenchymal stromal cells. Blood. 2007;110:3499–3506. doi: 10.1182/blood-2007-02-069716. [DOI] [PubMed] [Google Scholar]
- 2.da Silva ML. Chagastelles PC. Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 3.Fan CG. Tang FW. Zhang QJ. Lu SH. Liu HY. Zhao ZM. Liu B. Han ZB. Han ZC. Characterization and neural differentiation of fetal lung mesenchymal stem cells. Cell Transplant. 2005;14:311–321. doi: 10.3727/000000005783983070. [DOI] [PubMed] [Google Scholar]
- 4.In't Anker PS. Noort WA. Scherjon SA. Kleijburg-van der Keur C. Kruisselbrink AB. van Bezooijen RL. Beekhuizen W. Willemze R. Kanhai HH. Fibbe WE. Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica. 2003;88:845–852. [PubMed] [Google Scholar]
- 5.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 6.Horwitz EM. Prockop DJ. Fitzpatrick LA. Koo WW. Gordon PL. Neel M. Sussman M. Orchard P. Marx JC. Pyeritz RE. Brenner MK. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 7.Le Blanc K. Rasmusson I. Sundberg B. Gotherstrom C. Hassan M. Uzunel M. Ringden O. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet. 2004;363:1439–1441. doi: 10.1016/S0140-6736(04)16104-7. [DOI] [PubMed] [Google Scholar]
- 8.Rombouts WJ. Ploemacher RE. Primary murine MSC show highly efficient homing to the bone marrow but lose homing ability following culture. Leukemia. 2003;17:160–170. doi: 10.1038/sj.leu.2402763. [DOI] [PubMed] [Google Scholar]
- 9.Francois S. Bensidhoum M. Mouiseddine M. Mazurier C. Allenet B. Semont A. Frick J. Sache A. Bouchet S, et al. Local irradiation not only induces homing of human mesenchymal stem cells at exposed sites but promotes their widespread engraftment to multiple organs: a study of their quantitative distribution after irradiation damage. Stem Cells. 2006;24:1020–1029. doi: 10.1634/stemcells.2005-0260. [DOI] [PubMed] [Google Scholar]
- 10.Lee RH. Pulin AA. Seo MJ. Kota DJ. Ylostalo J. Larson BL. Semprun-Prieto L. Delafontaine P. Prockop DJ. Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell. 2009;5:54–63. doi: 10.1016/j.stem.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhakta S. Hong P. Koc O. The surface adhesion molecule CXCR4 stimulates mesenchymal stem cell migration to stromal cell-derived factor-1 in vitro but does not decrease apoptosis under serum deprivation. Cardiovasc Revasc Med. 2006;7:19–24. doi: 10.1016/j.carrev.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Son BR. Marquez-Curtis LA. Kucia M. Wysoczynski M. Turner AR. Ratajczak J. Ratajczak MZ. Janowska-Wieczorek A. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells. 2006;24:1254–1264. doi: 10.1634/stemcells.2005-0271. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler J. Etzel N. Brenner RE. To go or not to go: migration of human mesenchymal progenitor cells stimulated by isoforms of PDGF. J Cell Biochem. 2004;93:990–998. doi: 10.1002/jcb.20219. [DOI] [PubMed] [Google Scholar]
- 14.Forte G. Minieri M. Cossa P. Antenucci D. Sala M. Gnocchi V. Fiaccavento R. Carotenuto F. De Vito P, et al. Hepatocyte growth factor effects on mesenchymal stem cells: proliferation, migration, and differentiation. Stem Cells. 2006;24:23–33. doi: 10.1634/stemcells.2004-0176. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt A. Ladage D. Schinkothe T. Klausmann U. Ulrichs C. Klinz FJ. Brixius K. Arnhold S. Desai B, et al. Basic fibroblast growth factor controls migration in human mesenchymal stem cells. Stem Cells. 2006;24:1750–1758. doi: 10.1634/stemcells.2005-0191. [DOI] [PubMed] [Google Scholar]
- 16.Maijenburg MW. Noort WA. Kleijer M. Kompier CJ. Weijer K. van Buul JD. van der Schoot CE. Voermans C. Cell cycle and tissue of origin contribute to the migratory behaviour of human fetal and adult mesenchymal stromal cells. Br J Haematol. 2010;148:428–440. doi: 10.1111/j.1365-2141.2009.07960.x. [DOI] [PubMed] [Google Scholar]
- 17.Ozaki Y. Nishimura M. Sekiya K. Suehiro F. Kanawa M. Nikawa H. Hamada T. Kato Y. Comprehensive analysis of chemotactic factors for bone marrow mesenchymal stem cells. Stem Cells Dev. 2007;16:119–129. doi: 10.1089/scd.2006.0032. [DOI] [PubMed] [Google Scholar]
- 18.Sengenes C. Miranville A. Maumus M. De Barros S. Busse R. Bouloumie A. Chemotaxis and differentiation of human adipose tissue CD34+/CD31- progenitor cells: role of stromal derived factor-1 released by adipose tissue capillary endothelial cells. Stem Cells. 2007;25:2269–2276. doi: 10.1634/stemcells.2007-0180. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt A. Ladage D. Steingen C. Brixius K. Schinkothe T. Klinz FJ. Schwinger RH. Mehlhorn U. Bloch W. Mesenchymal stem cells transmigrate over the endothelial barrier. Eur J Cell Biol. 2006;85:1179–1188. doi: 10.1016/j.ejcb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Ponte AL. Marais E. Gallay N. Langonne A. Delorme B. Herault O. Charbord P. Domenech J. The in vitro migration capacity of human bone marrow mesenchymal stem cells: comparison of chemokine and growth factor chemotactic activities. Stem Cells. 2007;25:1737–1745. doi: 10.1634/stemcells.2007-0054. [DOI] [PubMed] [Google Scholar]
- 21.Glimm H. Oh IH. Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0) Blood. 2000;96:4185–4193. [PubMed] [Google Scholar]
- 22.Noort WA. Kruisselbrink AB. in't Anker PS. Kruger M. van Bezooijen RL. de Paus RA. Heemskerk MH. Lowik CW. Falkenburg JH. Willemze R. Fibbe WE. Mesenchymal stem cells promote engraftment of human umbilical cord blood-derived CD34(+) cells in NOD/SCID mice. Exp Hematol. 2002;30:870–878. doi: 10.1016/s0301-472x(02)00820-2. [DOI] [PubMed] [Google Scholar]
- 23.Bonta PI. van Tiel CM. Vos M. Pols TW. van Thienen JV. Ferreira V. Arkenbout EK. Seppen J. Spek CA, et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol. 2006;26:2288–2294. doi: 10.1161/01.ATV.0000238346.84458.5d. [DOI] [PubMed] [Google Scholar]
- 24.Sastry L. Johnson T. Hobson MJ. Smucker B. Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- 25.Pols TW. Bonta PI. de Vries CJ. NR4A nuclear orphan receptors: protective in vascular disease? Curr Opin Lipidol. 2007;18:515–520. doi: 10.1097/MOL.0b013e3282ef77d1. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y. Bruemmer D. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol. 2010;30:1535–1541. doi: 10.1161/ATVBAHA.109.191163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomiyama T. Nakamachi T. Gizard F. Heywood EB. Jones KL. Ohkura N. Kawamori R. Conneely OM. Bruemmer D. The NR4A orphan nuclear receptor NOR1 is induced by platelet-derived growth factor and mediates vascular smooth muscle cell proliferation. J Biol Chem. 2006;281:33467–33476. doi: 10.1074/jbc.M603436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao D. Desai S. Zeng H. VEGF stimulates orphan nuclear receptor nurr1 expression via PKC-dependent PKD-mediated CREB activation in endothelial cells. Int J Cancer. 2010;128:2602–2612. doi: 10.1002/ijc.25600. [DOI] [PubMed] [Google Scholar]
- 29.Pei L. Castrillo A. Chen M. Hoffmann A. Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280:29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- 30.Fiedler J. Roderer G. Gunther KP. Brenner RE. BMP-2, BMP-4, and PDGF-bb stimulate chemotactic migration of primary human mesenchymal progenitor cells. J Cell Biochem. 2002;87:305–312. doi: 10.1002/jcb.10309. [DOI] [PubMed] [Google Scholar]
- 31.Winoto A. Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl):S57–S66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- 32.Lin B. Kolluri SK. Lin F. Liu W. Han YH. Cao X. Dawson MI. Reed JC. Zhang XK. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 33.Kim SO. Ono K. Tobias PS. Han J. Orphan nuclear receptor Nur77 is involved in caspase-independent macrophage cell death. J Exp Med. 2003;197:1441–1452. doi: 10.1084/jem.20021842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arkenbout EK. van Bragt M. Eldering E. van BC. Grimbergen JM. Quax PH. Pannekoek H. de Vries CJ. TR3 orphan receptor is expressed in vascular endothelial cells and mediates cell cycle arrest. Arterioscler Thromb Vasc Biol. 2003;23:1535–1540. doi: 10.1161/01.ATV.0000084639.16462.7A. [DOI] [PubMed] [Google Scholar]
- 35.Arkenbout EK. de Waard V. van Bragt M. van Achterberg TA. Grimbergen JM. Pichon B. Pannekoek H. de Vries CJ. Protective function of transcription factor TR3 orphan receptor in atherogenesis: decreased lesion formation in carotid artery ligation model in TR3 transgenic mice. Circulation. 2002;106:1530–1535. doi: 10.1161/01.cir.0000028811.03056.bf. [DOI] [PubMed] [Google Scholar]
- 36.de Waard V. Arkenbout EK. Vos M. Mocking AI. Niessen HW. Stooker W. de Mol BA. Quax PH. Bakker EN, et al. TR3 nuclear orphan receptor prevents cyclic stretch-induced proliferation of venous smooth muscle cells. Am J Pathol. 2006;168:2027–2035. doi: 10.2353/ajpath.2006.050932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner W. Roderburg C. Wein F. Diehlmann A. Frankhauser M. Schubert R. Eckstein V. Ho AD. Molecular and secretory profiles of human mesenchymal stromal cells and their abilities to maintain primitive hematopoietic progenitors. Stem Cells. 2007;25:2638–2647. doi: 10.1634/stemcells.2007-0280. [DOI] [PubMed] [Google Scholar]
- 38.Di Nicola M. Carlo-Stella C. Magni M. Milanesi M. Longoni PD. Matteucci P. Grisanti S. Gianni AM. Human bone marrow stromal cells suppress T-lymphocyte proliferation induced by cellular or nonspecific mitogenic stimuli. Blood. 2002;99:3838–3843. doi: 10.1182/blood.v99.10.3838. [DOI] [PubMed] [Google Scholar]
- 39.Rasmusson I. Ringden O. Sundberg B. Le BK. Mesenchymal stem cells inhibit the formation of cytotoxic T lymphocytes, but not activated cytotoxic T lymphocytes or natural killer cells. Transplantation. 2003;76:1208–1213. doi: 10.1097/01.TP.0000082540.43730.80. [DOI] [PubMed] [Google Scholar]
- 40.Uccelli A. Moretta L. Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 41.Meisel R. Zibert A. Laryea M. Gobel U. Daubener W. Dilloo D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood. 2004;103:4619–4621. doi: 10.1182/blood-2003-11-3909. [DOI] [PubMed] [Google Scholar]
- 42.Krampera M. Cosmi L. Angeli R. Pasini A. Liotta F. Andreini A. Santarlasci V. Mazzinghi B. Pizzolo G, et al. Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells. 2006;24:386–398. doi: 10.1634/stemcells.2005-0008. [DOI] [PubMed] [Google Scholar]
- 43.Lee RH. Seo MJ. Pulin AA. Gregory CA. Ylostalo J. Prockop DJ. The CD34-like protein PODXL and alpha6-integrin (CD49f) identify early progenitor MSCs with increased clonogenicity and migration to infarcted heart in mice. Blood. 2009;113:816–826. doi: 10.1182/blood-2007-12-128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stich S. Haag M. Haupl T. Sezer O. Notter M. Kaps C. Sittinger M. Ringe J. Gene expression profiling of human mesenchymal stem cells chemotactically induced with CXCL12. Cell Tissue Res. 2009;336:225–236. doi: 10.1007/s00441-009-0768-z. [DOI] [PubMed] [Google Scholar]
- 45.Kalwitz G. Endres M. Neumann K. Skriner K. Ringe J. Sezer O. Sittinger M. Haupl T. Kaps C. Gene expression profile of adult human bone marrow-derived mesenchymal stem cells stimulated by the chemokine CXCL7. Int J Biochem Cell Biol. 2009;41:649–658. doi: 10.1016/j.biocel.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Wallen A. Zetterstrom RH. Solomin L. Arvidsson M. Olson L. Perlmann T. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp Cell Res. 1999;253:737–746. doi: 10.1006/excr.1999.4691. [DOI] [PubMed] [Google Scholar]
- 47.Inamoto T. Czerniak BA. Dinney CP. Kamat AM. Cytoplasmic mislocalization of the orphan nuclear receptor Nurr1 is a prognostic factor in bladder cancer. Cancer. 2010;116:340–346. doi: 10.1002/cncr.24737. [DOI] [PubMed] [Google Scholar]
- 48.Ramaswamy S. Ross KN. Lander ES. Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 49.Sirin O. Lukov GL. Mao R. Conneely OM. Goodell MA. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat Cell Biol. 2010;12:1213–1219. doi: 10.1038/ncb2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aherne CM. McMorrow J. Kane D. FitzGerald O. Mix KS. Murphy EP. Identification of NR4A2 as a transcriptional activator of IL-8 expression in human inflammatory arthritis. Mol Immunol. 2009;46:3345–3357. doi: 10.1016/j.molimm.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 51.Saijo K. Winner B. Carson CT. Collier JG. Boyer L. Rosenfeld MG. Gage FH. Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pei L. Castrillo A. Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol. 2006;20:786–794. doi: 10.1210/me.2005-0331. [DOI] [PubMed] [Google Scholar]
- 53.Jones SA. Directing transition from innate to acquired immunity: defining a role for IL-6. J Immunol. 2005;175:3463–3468. doi: 10.4049/jimmunol.175.6.3463. [DOI] [PubMed] [Google Scholar]
- 54.Nauta AJ. Kruisselbrink AB. Lurvink E. Willemze R. Fibbe WE. Mesenchymal stem cells inhibit generation and function of both CD34+-derived and monocyte-derived dendritic cells. J Immunol. 2006;177:2080–2087. doi: 10.4049/jimmunol.177.4.2080. [DOI] [PubMed] [Google Scholar]
- 55.Baggiolini M. Novel aspects of inflammation: interleukin-8 and related chemotactic cytokines. Clin Invest. 1993;71:812–814. doi: 10.1007/BF00190326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.