Abstract
Lineage reporters of human embryonic stem cell (hESC) lines are useful for differentiation studies and drug screening. Previously, we created reporter lines driven by an elongation factor 1 alpha (EF1α) promoter at a chromosome 13q32.3 locus in the hESC line WA09 and an abnormal hESC line BG01V in a site-specific manner. Expression of reporters in these lines was maintained in long-term culture at undifferentiated state. However, when these cells were differentiated into specific lineages, reduction in reporter expression was observed, indicating transgene silencing. To develop an efficient and reliable genetic engineering strategy in hESCs, we used chromatin insulator elements to flank single-copy transgenes and integrated the combined expression constructs via PhiC31/R4 integrase-mediated recombination technology to the chromosome 13 locus precisely. Two copies of cHS4 double-insulator sequences were placed adjacent to both 5′ and 3′ of the promoter reporter constructs. The green fluorescent protein (GFP) gene was driven by EF1α or CMV early enhancer/chicken β actin (CAG) promoter. In the engineered hESC lines, for both insulated CAG-GFP and EF1α-GFP, constitutive expression at the chromosome 13 locus was maintained during prolonged culture and in directed differentiation assays toward diverse types of neurons, pancreatic endoderm, and mesodermal progeny. In particular, described here is the first normal hESC fluorescent reporter line that robustly expresses GFP in both the undifferentiated state and throughout dopaminergic lineage differentiation. The dual strategy of utilizing insulator sequences and integration at the constitutive chromosome 13 locus ensures appropriate transgene expression. This is a valuable tool for lineage development study, gain- and loss-of-function experiments, and human disease modeling using hESCs.
Introduction
Efficient and reliable genetic manipulation protocols are key to successfully study gene functions using human embryonic stem cells (hESCs) [1] and induced pluripotent stem cells (hiPSCs) [2,3] [collectively called human pluripotent stem cells (hPSCs)]. Random integration by viral- or non-viral-mediated delivery methods is easy to perform but lacks control over copy number, site of integration, and gene expression level, which often results in undesired transgene silencing and poor reproducibility. Recently, the human ROSA26 [4], ENVY [5], and 2 other loci [6] have been reported to mimic the mouse ROSA26 site, which is constitutive in almost any lineages tested in mice. However, a thorough demonstration of appropriate regulation of theses loci in hESCs during different biological activities toward mature cell types is lacking.
We have previously identified a constitutive locus at chromosome 13q32.3 and designed a retargeting strategy. We inserted transgenes [eg, an emerald green fluorescent protein (EmGFP) cassette] precisely and efficiently to this site via recombination mediated by integrases of the PhiC31 family [7,8]. We have shown that expression at the chromosome 13q32.3 locus remained constant and was not silenced through random differentiation by embryoid body (EB) formation and directed differentiation toward neural stem cells (NSCs). However, when cells were further differentiated into more specialized neural cell types, such as dopaminergic neurons, expression of transgenes was partially or largely shut down as assessed by decreased GFP signal.
To solve this problem and assure that our retargeting system can be applied to various mature cell types of different lineages, we improved the vector construction strategy by incorporating chromatin insulator sequences. Chromatin insulators (reviewed in Refs. [9–11]) are cis-acting barrier elements that protect promoter fragments from DNA methylation-mediated silencing. Insulators may also enhance expression driven by a weak promoter [12]. Several types of insulators have been identified in both invertebrates and vertebrates. One of the widely used insulator sequences, cHS4, is derived from the constitutive DNase I hypersensitive site at the 5′ end of the chicken β-globin locus [13]. cHS4 and other insulator elements have previously been described in multiple cell lines, and have been used to stabilize expression of virally transduced transgenes in human cell types, including hESCs [14–16]. However, data on transgene silencing in hESCs upon differentiation to more mature cell types by genetic engineering strategy involving insulators have not been reported so far.
Here, we tested the utility of insulator sequences in engineering of hESCs for stable transgene expression. Our results support the conclusion that the dual strategy of integrating insulator sequences into retargeting platforms at the chromosome 13 locus ensures ubiquitous expression of 1 or more transgenes in various lineages derived from hESCs, including dopaminergic neurons, pancreatic endoderm, and mesodermal population. This work provides a reliable, yet easy-to-access protocol on genetic engineering of hPSCs, which can be extended to broader applications in the stem cell field.
Materials and Methods
Vector construction
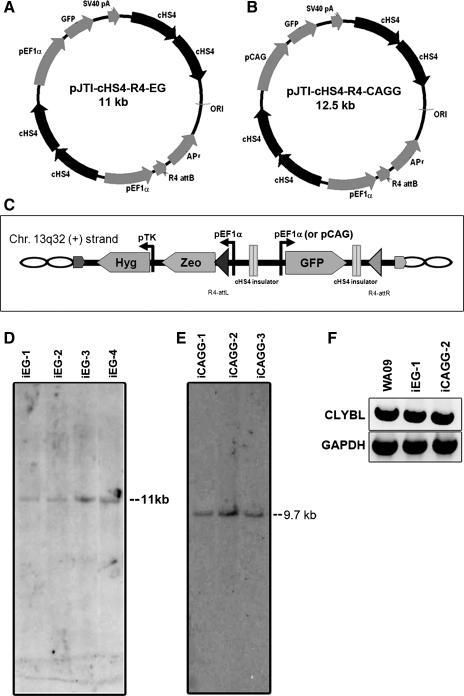
The plasmid pJTI/Zeo was cloned and used to generate platform hESC lines as described [7]. The insertion of the plasmid to chromosome 13q32 at the second intron of the CLYBL gene was identified by plasmid rescue, which was described in [8]. The uninsulated retargeting plasmid pJTI-R4-EG that contained pEF1α-EmGFP was cloned as described [7]. The double-insulated plasmids pJTI-cHS4-R4-EG (iEG, Fig. 1A) and pJTI-cHS4-R4-CAGG (iCAGG, Fig. 1B) that contained 2 pairs of 2.5-kilobase (kb) cHS4 double-insulator elements (each full-length cHS4 element is 1.25 kb long) and pEF1α-EmGFP or pCAG-EmGFP, respectively, were cloned into the Multisite Gateway system by 2 rounds of 3-way LR reactions to assemble the cHS4 double insulators and the promoter-reporter cassettes (Life Technologies). After cotransfection with pJTI R4 Int, a plasmid that expressed R4 integrase, the retargeting vectors inserted into the chromosome 13q32 site through recombination of R4 attB (constructed in the retargeting plasmid) and attP (constructed in the platform hESC line at chromosome 13). The zeocin resistance gene, activated by the constitutive elongation factor 1 alpha (EF1α) promoter, was used for selection of transfected clones. Expression of EmGFP (ie, the gene of interest) was driven by either the EF1α promoter (for iEG) or the CAG promoter (for iCAGG). The built-in R4 attP site then became R4-attL and R4-attR after the recombination (Fig. 1C). Primers for examining CLYBL gene expression are 5′-GAAGATGGCGCTACGTCTGC-3′ and 5′-CCCGCGGTGTTTGTCTAAGA-3′. The PCR product was 1,293 bp and covered all 9 exons of the CLYBL gene. GAPDH was amplified as an internal control for which the primers 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and 5′-CATGTGGGCCATGAGGTCCACCAC-3′ were used.
FIG. 1.
Retargeting plasmids used in this study and identification of retargeting events. The platform line generated in WA09 contained a docking site inserted at chromosome 13q32.3, which harbored a wild-type R4 attP site upstream of a Zeocin-resistance gene lacking a eukaryotic promoter. Insulated retargeting vectors pJTI-cHS4-R4-EG (ie, iEG, A) and pJTI-cHS4-R4-CAGG (ie, iCAGG, B) had 2 double-cHS4 insulator sequences flanking the EF1α or CAG promoter-driven GFP cassette. A wild-type R4 attB site was included to mediate recombinations between the plasmids and the platform line, which had an attP site already inserted. In the retargeting vector pJTI-cHS4-R4-CAGG (B), the EF1α promoter was replaced by the CAG promoter (a combination of the cytomegalovirus early enhancer element and chicken β-actin promoter) to obtain higher GFP expression levels in hESCs and their differentiated progeny. The retargeted hESC lines had double-insulator sequences flanking the expression cassette as indicated in (C) to prevent potential silencing events from interfering with the expression of inserted genes. Correct recombination events were screened by PCR as described in [7] and verified by Southern blot analysis (D). A predicted 11 kb single band was detected in representative clones obtained from retargeting with the pJTI-cHS4-R4-EG (iEG) vector (D), and a predicated 9.7 kb single band was shown in clones retargeted with the pJTI-cHS4-R4-CAGG (iCAGG) vector (E). To ensure that the generation of the platform at chromosome 13 did not cause any disruption of expression of endogenous genes, and that the CLYBL gene remained intact, RT-PCR was performed in the parental WA09 line and the retargeted iEG and iCAGG lines. GAPDH was used as an internal control. CLYBL expression was similar in all 3 lines examined (F). EF1α, elongation factor 1 alpha; GFP, green fluorescent protein; hESC, human embryonic stem cell.
Cell culture
WA09 (WiCell Research Institute) and the platform line derived from WA09 [7] (46, XX) were maintained as described [7,17]. Briefly, cells were cultured on a layer of mitomycin C (Sigma)-inactivated mouse embryonic fibroblast cells (MitC-MEFs) in hESC medium containing DMEM-F12 with glutamax, 20% knockout serum replacement, 1% nonessential amino acid, and 55 μM 2-mercaptoethanol, supplemented with 4 ng/mL basic fibroblast growth factor (bFGF). Cells were passaged using collagenase at a ratio of 1:4 every 4–5 days. hESCs were also maintained in a feeder-free fashion on Geltrex-coated dishes as needed, in hESC medium conditioned by MitC-MEF or StemPro hESC medium (all from Life Technologies). Cells were fed everyday and routine karyotype examination was done every 10–15 passages.
Transfection, retargeting, clone selection, and maintenance
Generation of the platform lines and the retargeted hESC lines was described previously [7,8]. Briefly, R4 cells were harvested using TrypLE and 1×106 cells were electroporated using a Neon microporator (Life Technologies) at 850 V, 30 ms, 1 pulse or ECM830 electroporator (BTX, 200 V at 10 ms, 2 pulses) with 10 μg of retargeting constructs. In the present work, either insulated or uninsulated EF1α-GFP (pJTI-cHS4-R4-EG) or CAG-GFP (pJTI-cHS4-R4-CAGG) was used. When the retargeting vectors integrated in a site-specific manner to the predetermined chromosome 13q32.3 locus, an EF1α promoter was placed upstream of the Zeocin resistance gene Sh ble. Ten micrograms of codon-optimized pCMV-R4 integrase expression plasmid was cotransfected along with the retargeting vectors. Transfected cells were seeded on MEFs and allowed to recover for 48 to 72 h. R4 integration-mediated retargeted clones were selected using 1.5–2.5 μg/mL zeocin (Life Technologies). Colonies were picked and expanded for further analysis.
Southern blot verification
Genomic DNA from individual clones was isolated using either the ChargeSwitch gDNA Mini Tissue Kit (Life Technologies) or DNAzol Reagent (Life Technologies). Southern blots were performed as described [7]. Briefly, genomic DNA (20 μg) from parent clones was digested with BamHI and separated overnight by electrophoresis on a 0.8% agarose gel. The DNA was transferred from the gel onto a Nytron SuPerCharge nylon membrane (Schleicher and Schuell) using a TurboBlotter (Schleicher and Schuell) according to the manufacturer's instructions. The genomic DNA was digested with SpeI, and the probe was obtained by amplification of the 1.4 kb GFP-SV40 polyA fragment from the plasmid pJTI-R4-EG by the following primers: 5′-ATGGTGAGCAAGGGCGAGGA-3′ and 5′-GATCCAGACATGATAAGATACATTGATGAG-3′. This probe was labeled with α-32P-dCTP (GE-Amersham or Perkin-Elmer) using the High-Prime DNA Labeling kit (Roche Applied Sciences). The membrane was incubated overnight with the labeled probe in QuickHyb Hybridization Solution (Strategene), washed twice at room temperature in a solution containing 2× SSC and 0.1% SDS, and once at 60°C in a solution containing 0.2× SSC and 0.1% SDS. The membrane was then exposed to a storage phosphor screen (GE-Amersham) and scanned using a Typhoon scanner (GE-Amersham).
Methylation analysis of endogenous and exogenous EF1α promoters
EF1α promoter region was analyzed for cytosine methylation of CpG dinucleotides in hESCs and their neural derivatives using the MethylMiner kit (Life Technologies) according to the manufacturer's instruction. DNA was processed into 100 bp to 1 kb fragments by sonication, and methylated DNA was captured on MBD-Dynabeads. Differentially methylated DNAs were collected with multifraction elution with a step-wise NaCl gradient. DNA fragments were precipitated with ethanol, and their methylation status was determined by quantitative PCR using 2 sets of primers (for endogenous EF1α promoter (designated as EE1AP1 and EE1AP2) and 1 set of primer for the exogenous EF1α promoters driving GFP (designated as EE1a GFP P2, to amply the overlapping region of 3′ end of the exogenous EF1α promoter and 5′ end of GFP). Primer sequences are EE1AP1 forward: 5′-GTGGAGAAGAGCATGCGTGA-3′; EE1AP1 reverse: 5′-CACGACATCACTTTCCCAGTT-3′; EE1AP2 forward: 5′-TGGTTCATTCTCAAGCCTCA-3′; EE1AP2 reverse: 5′-CCCGAATCTACGTGTCCAAT-3′; EE1aGFP P2 forward: 5′-TGGTTCATTCTCAAGCCTCA-3′; and EE1aGFP P2 reverse: 5′-CACCCCGGTGAACAGCTC-3′.
Random differentiation of hESCs
Undifferentiated hESCs were harvested using collagenase to generate EBs and were cultured for 4 days in suspension in differentiation medium containing DMEM-F12 with glutamax, 20% Knockout Serum Replacement, 1% nonessential amino acid, and 55 μM 2-mercaptoethanol. On day 5, EBs were seeded on Geltrex-coated plates for an additional 17 days of differentiation in the same medium. Then, the cells were subjected to flow cytometric analysis and immunocytochemistry.
Directed neural and dopaminergic differentiation
NSC was derived as previously described [18]. Briefly, hESC colonies were harvested and cultured in suspension as EBs for 8 days in hESC medium minus bFGF. EBs were then cultured for additional 2–3 days in suspension in neural induction media containing DMEM/F12 with glutamax, 1× NEAA, 1× N2, and bFGF (20 ng/mL) before attachment on cell culture plates. Neural rosettes formed 2–3 days after adherent culture were manually isolated and dissociated into single cells and replated onto culture dishes. The NSC population was expanded in Neurobasal media containing 1× NEAA, L-Glutamine 2 mM, 1× B27, and bFGF. Dopaminergic differentiation of NSCs was obtained by culturing NSCs in medium conditioned on PA6 cells for 4 weeks as previously described [18].
Differentiation into pancreatic endoderm
Differentiation of hESCs into pancreatic endoderm was performed following a published protocol with slight modifications [19]. Before differentiation, the hESCs were passaged onto gelatin-coated coverslips in 12-well plates (BD Falcon), and grown for 3 days. To generate definitive endoderm, the hESCs were washed with PBS and incubated for 24 h in DMEM:F12 with glutamax, 2 mg/mL bovine serum albumin fraction V (Sigma), 0.5× N2, 0.5× B27 (Invitrogen), 100 ng/mL Activin A, 50 ng/mL Wnt3a (R&D Systems), and 100 nM wortmannin (Sigma). Subsequently, the differentiating cells were incubated for 48 h in the above medium minus wortmannin and Wnt3a. To generate primitive gut tube-like tissue, the heterogeneous definitive endoderm population was incubated for 72 h in RPMI (Invitrogen) with 2% fetal calf serum (VWR Hyclone) and 50 ng/mL keratinocyte growth factor. For the generation of posterior foregut tissue, cells at the primitive gut tube stage were incubated for 72 h in DMEM:F12 with 1× N2, 300 nM (−)-indolactam V (Calbiochem), 2 mg/mL bovine serum albumin fraction V, and 10 ng/mL FGF10 (R&D Systems). To generate pancreatic endoderm, the posterior foregut cells were incubated for 72 h in DMEM (Invitrogen) with 1× B27. Then, the cells were incubated for another 72 h in CMRL (Invitrogen) with 50 ng/mL exendin-4 (Sigma-Aldrich), 50 ng/mL insulin-like growth factor 1 (IGF1; Sigma-Aldrich), 50 ng/mL hepatocyte growth factor (Peprotech), and 1× B27.
Induced mesoderm differentiation
Mesodermal lineage cell populations from insulated clones were obtained using a directed differentiation protocol modified from a published work [20]. Briefly, iCAGG cells were adapted to feeder-free conditions and were grown in StemPro hESC medium (Invitrogen). Cells were then harvested by Accutase and plated in a chemically defined medium consisting of IMDM:F12, Activin A (100 ng/mL; R&D systems), bFGF (20 ng/mL; Stemgent), BMP4 (10 ng/mL; R&D systems), and LY294002 (10 μM; Tocris) for 2 days. On the third day, Activin A was withdrawn from culture and starting from day 4, the medium was changed to 10% fetal bovine serum (FBS) in DMEM and cells were continued to be induced for an additional 8–10 days.
Flow cytometric analysis and immunocytochemistry
Retargeted clones, their parental nonengineered lines, or differentiated cells were harvested using TrypLE; cell debris was excluded from analysis by gating based on forward and side scatter. Data were collected using BD FACS CantoII or FACS Calibur Flow Cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Immunocytochemistry was carried out as described [21,22]. Briefly, undifferentiated hESC or differentiated cells were fixed with 2% paraformaldehyde and incubated with blocking buffer for 30 min. Primary antibodies were added to the cells and incubated at 4°C overnight followed by addition of appropriate secondary antibodies. The following primary antibodies were used: Oct4 (1:500; Abcam), SSEA4 (1:500; Life Technologies), Tra-1-60 (1:100; Millipore), Tra-1-81 (1:100; Millipore), β3 tubulin (Tuj1, 1:4,000; Sigma), PAX6 (1:50; Developmental Studies Hybridoma Bank), Nestin (1:500; BD Biosciences), smooth muscle actin (SMA, 1:200; Sigma), CD31 (R&D systems; 1:200), Brachyury (1:200; Santa Cruz), PDGFRα (1:200; BD), α-fetoprotein (1:500; Sigma), tyrosine hydroxylase (TH; 1:1,000), rabbit anti-HNF1b (1:100; Santa Cruz), goat anti-SOX17 (1:1,000; R&D Systems), goat anti-PDX1 (1:10,000; Abcam), and mouse anti-NKX6.1 (1:50; Hybridoma F55A10-c). The following secondary antibodies were used: Alexa Fluor 594- or 488-conjugated anti-mouse IgG (1:1,000), Alexa 594- or 488-conjugated anti-rabbit IgG, (1:1,000), Alexa Fluor-conjugated donkey anti-goat IgG, and allophycocyanin (1:1,000; all from Life Technologies), Cy3-conjugated donkey anti-mouse, Cy3-conjugated donkey anti-goat (1:200), Cy3-conjugated donkey anti-rabbit (1:200; Jackson ImmunoResearch) (Invitrogen). DAPI or ToPro3 was used for counter nuclei staining. Images were captured using a Zeiss Axiovision microscope with z-stack split view function or Leica SL and SP2 confocal laser scanning microscopes, and images were processed using AdobePhotoshop CS or Paint Shop Pro XI. Quantification of coexpression of GFP with lineage markers was done using ImageJ software.
Results
Construction and retargeting of insulator vectors in R4 platform hESC lines
We have previously established platform lines from a normal hESC line WA09 and an abnormal hESC line BG01V at the chromosome 13q32.3 locus by PhiC31 integrase-mediated recombination and created several reporter lines driven by an EF1α promoter [7,8,23]. Expression of reporters in these hESC lines was maintained in long-term culture at undifferentiated state. However, when complicated and prolonged differentiation protocols were applied, at least 50% of the cells became dark and stopped expressing GFP, indicating transgene silencing. To circumvent this problem, we engineered 2 copies of the paradigm insulator sequence cHS4 in tandem to flank an EF1α-EmGFP or CAG-EmGFP cassette in the R4 retargeting vector (Fig. 1) [7]. After electroporation and zeocin selection (1.0–2.5 μg/mL), clones of both insulated EF1α-EmGFP (iEG) and CAG-EmGFP (iCAGG) were obtained and verified to be correctly inserted into the platform locus by PCR and Southern blot analysis (Fig. 1).
Characterization of clones retargeted by insulator vectors
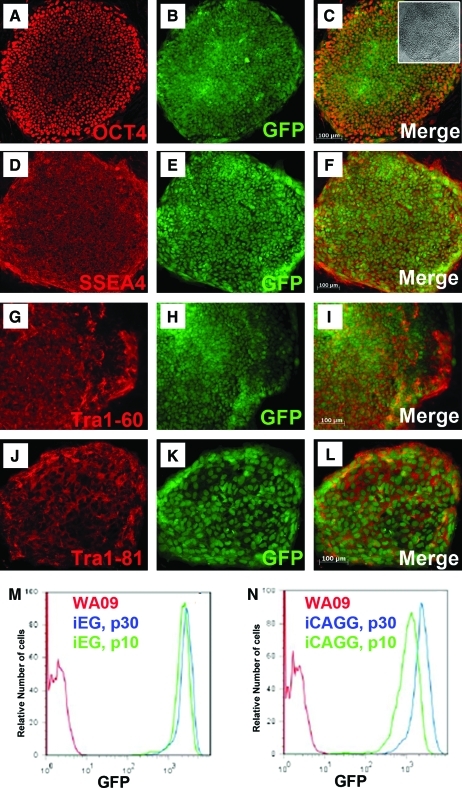
To ensure that the retargeted cells with insulators maintained their normal ESC characteristics, we examined the pluripotency markers OCT4, SSEA4, Tra1-60, and Tra1-81 (Fig. 2). Consistent with their parental WA09 line, the insulated iEG and iCAGG clones maintained a normal karyotype, and coexpressed GFP uniformly in addition to all of the pluripotent markers. Similar to the uninsulated clones [7], GFP expression was retained for more than 30 passages continuously over a period of 4 months without any reduction (Fig. 2M, N).
FIG. 2.
Double-insulated pJTI-cHS4-R4-EG (iEG) and pJTI-cHS4-R4-CAGG (iCAGG) vectors were successfully retargeted to the chromosome 13q32.3 site, and expressed GFP. Immunocytochemistry results of clones from both vectors are similar, and only those of iCAGG clones are shown here. A representative clone displayed the typical human hESC colony morphology (C inset), expressed GFP, and coexpressed characteristic hESC markers, including OCT4 (A–C), SSEA4 (D–F), Tra1-60 (G–I), and Tra1-81 (J–L). The GFP expression profile was identical in prolonged culture for over 20 passages for both iEG (M) and iCAGG (N). x-axis represents the GFP intensity; y-axis represents percentage of Max, an indicator of the number of cells. Red curves represent the parental cell line WA09, which did not express GFP. Green curves represent clones at passage 10. Blue curves represent clones at passage 30. Scale bar, 100 μm. Color images available online at www.liebertonline.com/scd
The platform was built via PhiC31-mediated integration at an attP pseudosite of the hESC genome. One of the advantages of PhiC31 integrase-mediated integrations over random insertions is that attP pseudosites are almost always located at intergenic regions or introns of a gene; therefore, the odds of disrupting or interfering with normal gene function are lower [24], although they cannot be completely ruled out. It was determined that, in our platform lines, insertion of retargeting the platform was located at the second intron of the CLYBL gene, which encodes citrate lyase subunit β-like protein, an enzyme in the tricarboxylic acid cycle [25]. The CLYBL gene does not appear to be developmentally regulated or involved in any diseases, although polymorphisms have been identified in 3 amino acids in the CLYBL protein (http://bioinf.umbc.edu/DMDM). We also searched for the possibilities that the introns of CLYBL might be active to encode functional microRNAs or other noncoding RNAs and did not find any evidence that these introns might be involved in any of these activities (www.genome.ucsc.edu). Therefore, the intron 2 of CLYBL where our platform site was inserted seemed to be a constitutive genomic locus, which was permissive for further genetic engineering. Nevertheless, we examined the retargeted clones by RT-PCR and showed that its expression and the size of the mRNA remained unchanged in both iEG and iCAGG clones compared with the parental WA09 cells (Fig. 1F).
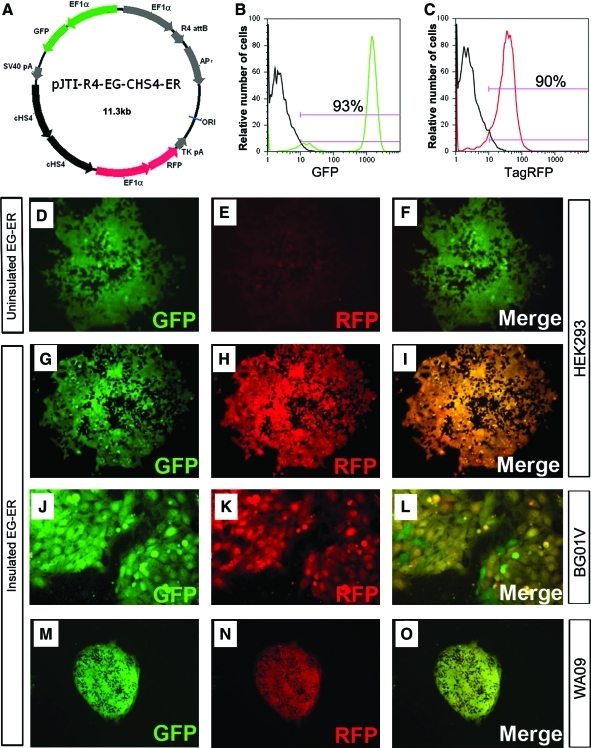
Our engineered lines were derived from 2 hESC lines, WA09 and BG01V. Please note that all data presented in this article are from the normal hESC line WA09 and its engineered derivatives. The parental lines and the insulated and uninsulated clones were karyotyped routinely, which showed the maintenance of a normal karyotype. Data from BG01V, a karyotypically abnormal line, and its engineered clones showed similar results but were not used in the preparation of this article, except for Figure 7J–L, where a retargeting vector EF1αGFP-Insulator (cHS4)2-EF1αRFP was used to show the effect of insulator in balancing the expression of 2 promoter reporters.
FIG. 7.
Double-insulator elements prevented unbalanced expression of transgenes in cis. Uninsulated and insulated dual-reporter plasmids pJTI-R4-EF1α-GFP-EF1α-RFP and pJTI-R4-EF1α-GFP-(cHS4)2-EF1α-RFP were used to transfect HEK293 cells and the expression level of GFP and RFP was compared (A). Expression of GFP and RFP was biased with GFP being expressed at a much higher level (D–F) in uninsulated clones. However, when 2 copies of cHS4 were placed between these 2 cassettes, expression of GFP and RFP was almost identical as revealed by direct fluorescence microscopy of the native signals (G–I). This was also confirmed by retargeting to the platform hESC line generated in an abnormal hESC line BG01V (J–L) and a widely used normal hESC line WA09 using an episomal expression backbone (M–O). In addition to immunocytochemistry assays, flow cytometric analysis also confirmed that similar percentages of cells expressed both reporters (∼93% for GFP+, as shown in B, ∼90% for TagRFP+, shown in C). x-axis represents the intensity of GFP or TagRFP; y-axis represents the relative number of cells. Nonengineered ESCs were used as negative controls (black curves in B and C). EG:EF1α-EmGFP, ER: EF1α-RFP. EmGFP, emerald green fluorescent protein. Color images available online at www.liebertonline.com/scd
Directed differentiation toward neural and dopaminergic lineages
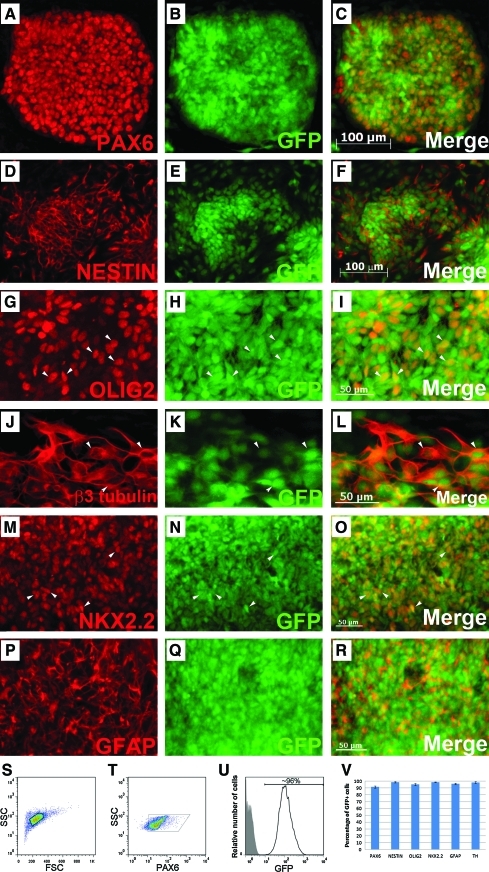
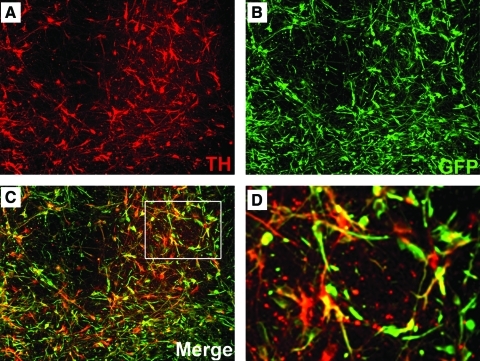
One of the original goals of this work was to generate hESC lines expressing a GFP reporter in undifferentiated hESCs and all of their progeny for cell tracking purposes in transplantation studies. However, GFP expression was silenced in the previously reported uninsulated EF1α-GFP line when it was differentiated into mature dopaminergic neurons. To evaluate whether the insulated lines maintained transgene expression constitutively, we performed directed differentiation of iEG and iCAGG to an array of neural cell types, including dopaminergic lineage. Throughout NSC and the subsequent neural subtype specification, cells continued to express GFP; no dark cells were observed with fluorescence microscopy. Almost every NSC that expressed PAX6 or NESTIN maintained robust GFP expression (about 92%–96% of PAX6+ cells and 99% of NESTIN+ cells coexpressed GFP, Fig. 3A–F, S–U, bar graph 3V). In addition, β3-tubulin+ neurons, OLIG2+ or NKX2.2+ glial progenitors, as well as GFAP+ astrocytes coexpressed GFP (about 95% of OLIG2+ cells, 98.5% of NKX2.2+ cells, and 96% of GFAP+ cells coexpressed GFP, Fig. 3G–R, bar graph in Fig. 3V). Most importantly, in these double-insulated iEG or iCAGG clones, robust GFP expression was retained in all cells constantly even at the terminal differentiated stage when cells started to express TH, indicative of dopaminergic neuron formation (about 98% of TH+ cells expressed GFP, Figs. 4 and 3V). These results suggest that the GFP reporter flanked by double insulators at the constitutive chromosome 13 site maintained its expression throughout the differentiation process to mature neural cell types.
FIG. 3.
iEG and iCAGG clones differentiated into NSCs, neurons, and glial cells without any silencing of GFP expression. The iEG and iCAGG clones were specified toward NSCs. Representative iCAGG clones showed robust and constitutive GFP expression along the time course of differentiation, together with NSC markers PAX6 (A–C) and NESTIN (D–F), neuronal marker β3 tubulin (J–L), glial markers OLIG2 (G–I) and NKX2.2 (M–O), and astrocyte marker GFAP (P–R). Flow cytometric analysis was applied to determine the percentage of cells that retained GFP expression during differentiation (S–U). Cells differentiated after the protocol described in Materials and Methods were harvested, fixed, and stained using a Pax6 antibody that did not stain undifferentiated ESCs. Samples were gated on the basis of the appropriate forward scatter (FSC) and side scatter (SSC) to exclude cell debris (S). The majority of the cells underwent directed neural differentiation were Pax6+ (T), and 96% of which continued to express GFP (U). The shaded gray area represented the negative control cells. Quantification of coexpressed cells of GFP and neural lineage markers is shown in (V). Note that in all experiments, the GFP shown was from a native signal without any additional immunostaining using GFP antibodies, indicating the robustness of the transgene expression in our system. Arrowheads indicated representative double-labeled cells. Scale bars, 100 μm (A–F), 50 μm (G–R). NSCs, neural stem cells. Color images available online at www.liebertonline.com/scd
FIG. 4.
iEG and iCAGG clones differentiated into dopaminergic neurons, while maintaining robust GFP expression. TH staining (A) revealed the relatively high differentiation efficiency of iCAGG cells toward dopaminergic lineage. These cells continued to express GFP (B, C). (D) Higher magnification of inset in (C). Please refer to Fig. 3V for quantification of coexpression of TH and GFP. TH, tyrosine hydroxylase. Color images available online at www.liebertonline.com/scd
Directed differentiation into pancreatic endocrine precursors
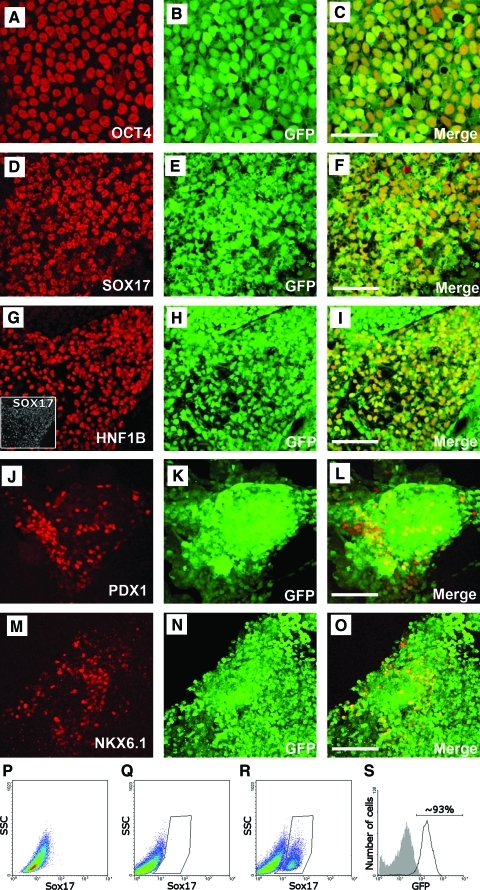
To further examine the GFP expression of insulated clones after differentiation, the cells were differentiated into pancreatic endoderm cells after defined stages [19]. When cells transitioned from a pluripotent hESC (Fig. 5A–C) state to definitive endoderm and primitive gut tube, which were characterized by expression of transcription factors SOX17 (Fig. 5D–F) and HNF1B (TCF2; Fig. 5G–I), respectively, almost all cells retained GFP expression. Moreover, when these cells commenced to posterior foregut and pancreatic endoderm lineages, as defined by PDX1 (Fig. 5J–L) and NKX6.1 (Fig. 5M–O) expression, respectively, GFP expression was sustained at high levels. Quantification by flow cytometry analysis confirmed that ∼93% of SOX17+ definitive endoderm cells expressed GFP (Fig. 5P–S). In addition, cells expressing other endodermal lineage markers, such as alpha fetal protein (AFP), retained GFP expression as well (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/scd). Data presented were from a representative iCAGG clone, and no detectable difference in GFP expression or differentiation efficiency was observed during the entire differentiation period of iEG and iCAGG clones.
FIG. 5.
Double-insulated retargeted clone iCAGG differentiated into pancreatic endoderm retained expression of GFP throughout the entire differentiation process. iCAGG hESCs were subjected to a multistep differentiation protocol, and analyzed for expression of GFP (native signal, B, E, H, K, N) and coexpression of the stage-specific transcription factors (red, C, F, I, L, O) OCT4 (A) for undifferentiated cells, SOX17 (D) for definitive endoderm, HNF1B (G) and SOX17 (G, inset) for primitive gut tube, PDX1 (J) for posterior foregut, and NKX6.1 (M) for pancreatic endocrine precursors. The individual images were merged (right-hand column) to show colocalization. Flow cytometric analysis was applied to determine the percentage of cells that retained GFP expression during differentiation (P–S). No SOX17+ cells were detected in undifferentiated cells (P) or cells that were allowed to differentiate without growth factors (Q). Three days after treatment with growth factors, 17% of the cells had differentiated into SOX17+ definitive endoderm (R), ∼93% of which were positive for GFP (S). Scale bars 100 μm. Color images available online at www.liebertonline.com/scd
Differentiation into mesodermal cells
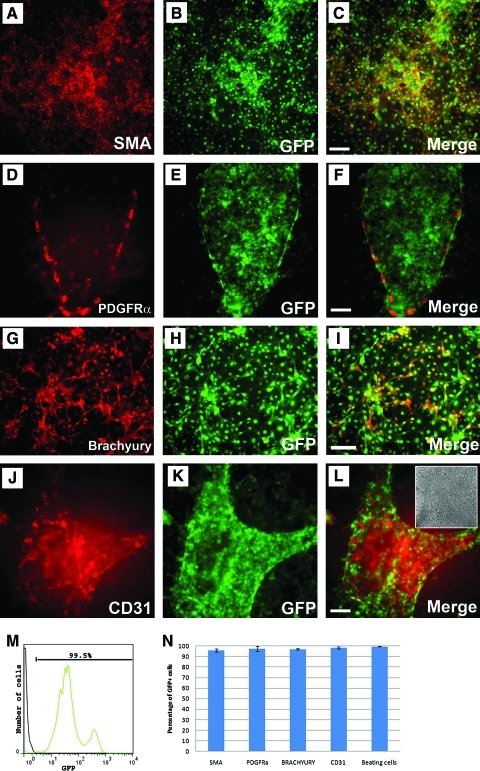
To assess whether GFP expression was maintained in the insulated clones during differentiation into mesodermal progeny, we differentiated the iCAGG clone using a directed mesodermal differentiation protocol [20,26]. At the end of the 14-day protocol, all cells retained GFP expression (Fig. 6). In particular, by day 11 of differentiation, multiple GFP-expressing beating clusters were readily to be detected in culture, indicating that reporter expression was maintained upon formation of cardiomyocytes (Supplementary Fig. S2 and Supplementary Movie S1). In addition, when the beating clusters were manually dissected and dissociated into single cells by TrypLE, almost all cells (average 99.5%) showed robust GFP expression as identified by flow cytometric analysis (Fig. 6M, N). We also observed vasculature-like cell patterns that appeared to be endothelial cells lining-up the structure (Fig. 6L, inset). Immunocytochemistry revealed that these cells expressed platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31, about 98% of CD31+ cells coexpressed GFP, Fig. 6J–L, N), supporting their mesodermal identity. Several other commonly used mesodermal markers were also examined, including cardiac mesoderm marker platelet-derived growth factor receptor alpha (PDGFRα) [27,28], Brachyury, and SMA (Fig. 6). Cells expressing these markers retained GFP expression at a high level; about 97.6% of PDGFRα+ cells, 97% of Brachyury+ cells, and 96% SMA+ cells coexpressed GFP (Fig. 6N), which was comparable to undifferentiated ES cells. These findings combined support that insulated clones maintained GFP expression during extensive differentiation processes toward various somatic cell types.
FIG. 6.
GFP expression was retained in insulated clones after directed differentiation toward mesodermal lineages. Markers that characterized mesodermal lineages such as SMA (A–C) colabeled with GFP after differentiation. Cells expressing cardiovascular markers PDGFRα (D–F) and Brachyury (G–I) also retained robust GFP expression. PECAM1 (CD31), a common marker for endothelial cells, was detected to be coexpressed with GFP (J–L, inset in L shows the phase image of J–L). In addition, single cells dissociated from beating clusters were analyzed by flow cytometry and showed coexpression with GFP (M). Black curve represents a control cell line that does not have GFP expression; green curve represents the iCAGG-derived beating cells. Quantification of coexpressed cells of GFP and mesodermal lineage markers is shown in (N). Scale bar, 100 μm. Color images available online at www.liebertonline.com/scd
Double-insulator elements prevented unbalanced expression of transgenes in cis
Our data showed that when transgenes were flanked by insulator elements at the chromosome 13 site, their expression was protected from silencing during complicated differentiation process. To further test whether insulators possess other beneficial functions, we compared dual reporter plasmids in uninsulated (pJTI-R4-EF1α-GFP-EF1α-RFP) and insulated (pJTI-R4-EF1α-GFP-[cHS4]2-EF1α-RFP) formats. Promoter interference has been reported when 2 strong promoters are placed in proximity in a vector. Our aim is to test whether the insulator sequence in between would eliminate or reduce such interference. As shown in Fig. 7, expression of GFP and red fluorescent protein (RFP) was unbalanced with GFP expressing at a much higher level in uninsulated clones. However, when insulator sequences were placed between these 2 cassettes, expression of GFP and RFP was almost identical as revealed by direct fluorescence microscopy of the native signals. Similar results were obtained in multiple hESC lines (WA09, and a karyotypically abnormal line BG01V), when using a different version of insulator elements (such as EHD1), as well as using episomal vectors that did not integrate into the hESC genome but were able to replicate during cell cycle (Fig. 7).
DNA methylation of the transgene promoters is reduced in insulated clones compared with uninsulated clones after differentiation
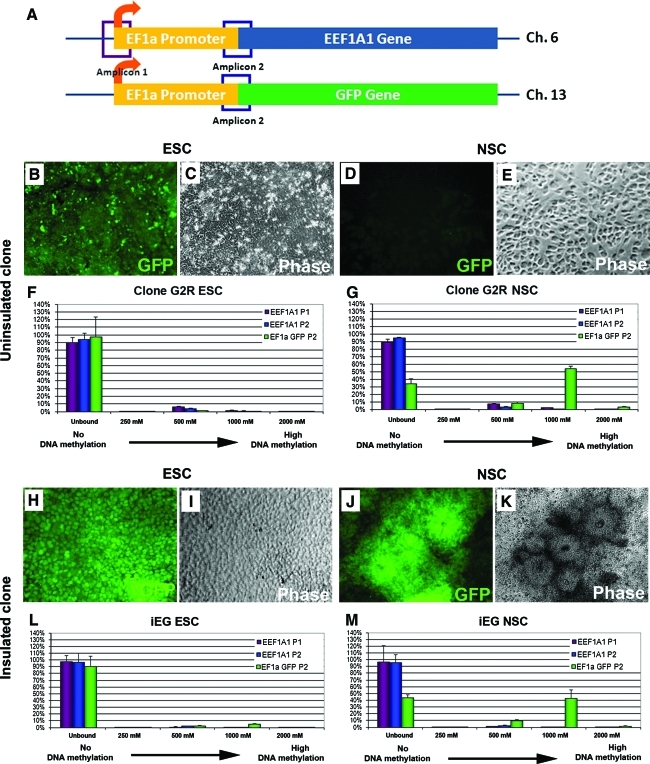
Our data show that chromatin insulators prevent gene silencing. To further investigate whether epigenetic modifications such as DNA methylation are involved in effects elicited by insulator elements in this scenario, we assessed DNA methylation status of both the endogenous and exogenous EF1α promoter (which drove GFP expression) using MethylMiner, a well-established methylation enrichment and fractionation method based on binding of methylated fragments with MBD protein. Quantification of DNA methylation at the promoter regions was determined by subsequent quantitative PCR using primers specifically designed for endogenous and exogenous EF1α promoter regions (Fig. 8A). At the undifferentiated ESC stage, all GFP reporter lines tested showed no or very little DNA methylation at both the endogenous proximal EF1α promoter (Fig. 8F, H, purple and blue bars) and the exogenous EF1α promoter (Fig. 8F, H, green bars). However, when these clones were differentiated in culture toward neural lineages, DNA methylation was detected at the exogenous promoter amplicon (Fig. 8G, green bars), whereas the endogenous promoter remained unmethylated (Fig. 8G, purple and blue bars). There is a clear correlation between high DNA methylation of the exogenous EF1α promoter and the reduction of GFP expression (compare Fig. 8B and D, and green bars in 8F and G). Similar to uninsulated clones, the insulated clone iEG showed no methylation at the undifferentiated stage (Fig. 8L). When it was differentiated, the exogenous EF1α promoter was methylated but to a lesser degree by a reduction of 10%–15% (compare the green bars in Fig. 8G and M), indicating that in this insulated clone, cHS4 elements acted partially through reducing DNA methylation of transgene promoters.
FIG. 8.
DNA methylation status of EF1α promoters. Location of PCR amplicons for the endogenous EF1α promoter at chromosome 6 and the exogenous EF1α promoter at the chromosome 13 locus is shown in (A). GFP and phase images of a representative uninsulated EG clone named G2R and an insulated EG clone (iEG) at undifferentiated (B, C; H, I) and differentiated NSC stage (D, E; J, K) are also shown. While endogenous EF1α promoter remained unmethylated at both ESC stage and after neural differentiation, the exogenous EF1α that drove GFP had higher methylation status (F–G). An increase of DNA methylation after differentiation was also observed in insulated iEG clone, but to a lesser degree when compared with G2R (L, M). Primer sets EEF1A1 P1 and EEF1A1 P2 were used to detect methylation status of endogenous EF1α promoter. The primer set EF1α GFP P2 was used to detect methylation status of the exogenous EF1α promoter. x-axis represents elution fractions with differential methylation levels. There was a direct correlation between salt concentration in elution buffer and DNA methylation. y-axis represents percentage of total amplified DNA signal from all fractions collected. Higher bars in higher salt concentrations represent higher DNA methylation. Bars in the unbound section represented fractions that did not have any DNA methylation. Color images available online at www.liebertonline.com/scd
Discussion
Previously, we created EF1α-EmGFP hESC lines integrated at chromosome 13q32.3, a predetermined genomic locus safe for transgene integration [7]. Although GFP expression was maintained in various early lineage precursors, gene silencing was observed when these lines were terminally differentiated into somatic cell types, such as dopaminergic neurons. To circumvent this problem, we flanked the genes of interest by double-insulator sequences, providing a shelter for exogenous promoters from possible epigenetic modifications that might interfere with transgene expression. In these double-insulated reporter lines, GFP expression remained robust throughout the courses of directed differentiation into NSCs, dopaminergic neurons, pancreatic endoderm, and mesodermal lineages.
That GFP expression was maintained without any reduction through dopaminergic, pancreatic, and mesodermal differentiation not only shows that our double-insulated retargeting system is constitutive in various differentiated cell types, but also provides a powerful tool to further study in these lineages. A GFP-labeled, hESC-derived dopaminergic neuron population is highly desirable for investigating dopaminergic lineage development in human. Although TH+ cells derived from mouse ESCs or abnormal hESCs that are genetically tagged with GFP (driven by CMV or EF1α promoter) retain GFP expression [29,30], such hESC lines with normal karyotype have not been created. Here, our double-insulated clone is able to be differentiated into TH+ GFP-expressing dopaminergic cells (Fig. 4). These cells have the potential to facilitate in vitro dopaminergic differentiation experiments and in vivo tracking after transplantation, making it feasible to directly monitor survival, integration, and striatal circuitry reconstruction of grafted cells, which may lead to identification of critical factors that modulate the degenerative brain environment, hence providing clues for hPSC-based therapy for Parkinson's patients.
Similar to dopaminergic derivation, the differentiation of hESCs toward pancreatic lineages is a lengthy in vitro process, which might alter transgenes genetically or epigenetically, causing gene silencing and preventing the application of genetic tags in such lineages. The data presented here offer a GFP-labeled population of pancreatic endoderm cells and provide a defined genomic locus and an effective engineering strategy for potential generation of pancreatic lineage-specific reporters in hPSCs in future investigations. Such reporters will be applied to optimization of differentiation protocols, obtaining purified insulin-producing cells from heterogeneous populations, as well as identification of key factors important for pancreatic endocrine differentiation in vitro.
Clues provided in previous reports suggest that insulators function through blocking de novo DNA methylation to ensure stable and consistent transgene expression over extended culture [31]. To pinpoint the mechanism that cHS4 insulator elements act in our hESC clones in particular, we compared the DNA methylation status of both endogenous and exogenous EF1α promoter in uninsulated and insulated EF1α-GFP clones. Our results indicate that prevention or reduction of DNA methylation of transgene promoters only contributes partially to the protection of insulator elements in this system (Fig. 8); other possible mechanisms such as changes of status in histone acetylation between the insulated and uninsulated clones after differentiation will be examined. Our system, with the ability to target only one copy of transgene at a defined genomic locus, has provided a platform allowing for further investigation along this line, since uncontrolled transgene copy number has also been reported to interfere with epigenetic modifications [32]. Additional experiments to investigate the comprehensive mechanisms that different types of chromatin insulators establish to shield transgenes from silencing in hPSCs are of interest.
There are 2 major components of the integrating strategy that ensure appropriate regulation of transgenes in the retargeted line: (1) the identification of the safe integration locus at chromosome 13q32.3 (within the intron of the CLYBL gene), and (2) the use of chromatin insulator sequences to flank the gene of interest, supplying another layer of protection from undesired regulation that might cause gene silencing. These data indicate that chromatin insulators facilitate the appropriate transgene expression in hESCs and reduce the interference caused by adjacent strong promoters.
Of further interest is whether these chromatin insulator sequences function in a broader context, including hiPSC lines or other vector constructs for different genetic engineering purposes. The chromosome 13 site is the only genomic locus we have tested for insulating ability, although we have tested the same locus in at least 2 different hESC lines, H9 and BG01V (an abnormal hESC line with a karyotype of 48XY, +12, +17, but otherwise shares all of the hESC characteristics). Based on the data from episomal vectors (Fig. 7) and previous reports from other groups, who used lentiviral [14] or adenoviral transduction [12], it would be reasonable to predict that the insulating strategy could be extended to other loci and various genetic engineering systems.
In conclusion, our current work and previous work [7,8] show detailed and extensive characterization of a genetic manipulation system where a safe docking site is identified in the hESC genome and an optimized double-insulator retargeting strategy is provided. The system has the capacity to accept large, multigenic elements, and can be combined with Cre-Lox and Flp-FRT system to make versatile tools for additional genetic manipulations. The system ensures appropriate regulation of transgene expression and significantly reduces possible gene silencing as shown for a variety of hESC derivatives.
Supplementary Material
Acknowledgments
This work was supported by Life Technologies Corporation, California Institute for Regenerative Medicine Tools and Technology Award RT1-011071 to Y.L., the Juvenile Diabetes Research Foundation (Grant awards 3-2008-477 and 35-2008-628), California Institute for Regenerative Medicine Early Translation Award TR-1250 to J.F.L., and Hartwell Individual Biomedical Investigator Award and NIH/NICHD 2K12HD001259-11 WRHR Career Development Award to L.C.L.
Author Disclosure Statement
All authors declare that they have no potential conflict of interest in connection with this article.
References
- 1.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Irion S. Luche H. Gadue P. Fehling HJ. Kennedy M. Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 5.Costa M. Dottori M. Ng E. Hawes SM. Sourris K. Jamshidi P. Pera MF. Elefanty AG. Stanley EG. The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nat Methods. 2005;2:259–260. doi: 10.1038/nmeth748. [DOI] [PubMed] [Google Scholar]
- 6.Du ZW. Hu BY. Ayala M. Sauer B. Zhang SC. Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells. 2009;27:1032–1041. doi: 10.1002/stem.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y. Thyagarajan B. Lakshmipathy U. Xue H. Lieu P. Fontes A. MacArthur CC. Scheyhing K. Rao MS. Chesnut JD. Generation of platform human embryonic stem cell lines that allow efficient targeting at a predetermined genomic location. Stem Cells Dev. 2009;18:1459–1472. doi: 10.1089/scd.2009.0047. [DOI] [PubMed] [Google Scholar]
- 8.Thyagarajan B. Liu Y. Shin S. Lakshmipathy U. Scheyhing K. Xue H. Ellerstrom C. Strehl R. Hyllner J. Rao MS. Chesnut JD. Creation of engineered human embryonic stem cell lines using phiC31 integrase. Stem Cells. 2008;26:119–126. doi: 10.1634/stemcells.2007-0283. [DOI] [PubMed] [Google Scholar]
- 9.Bushey AM. Dorman ER. Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaszner M. Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 11.West AG. Gaszner M. Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 12.Mori-Uchino M. Takeuchi T. Murakami I. Yano T. Yasugi T. Taketani Y. Nakagawa K. Kanda T. Enhanced transgene expression in the mouse skeletal muscle infected by the adeno-associated viral vector with the human elongation factor 1alpha promoter and a human chromatin insulator. J Gene Med. 2009;11:598–604. doi: 10.1002/jgm.1337. [DOI] [PubMed] [Google Scholar]
- 13.Pikaart MJ. Recillas-Targa F. Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y. Ramezani A. Lewis R. Hawley RG. Thomson JA. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells. 2003;21:111–117. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 15.Puthenveetil G. Scholes J. Carbonell D. Qureshi N. Xia P. Zeng L. Li S. Yu Y. Hiti AL. Yee JK. Malik P. Successful correction of the human beta-thalassemia major phenotype using a lentiviral vector. Blood. 2004;104:3445–3453. doi: 10.1182/blood-2004-04-1427. [DOI] [PubMed] [Google Scholar]
- 16.Vieyra DS. Goodell MA. Pluripotentiality and conditional transgene regulation in human embryonic stem cells expressing insulated tetracycline-ON transactivator. Stem Cells. 2007;25:2559–2566. doi: 10.1634/stemcells.2007-0248. [DOI] [PubMed] [Google Scholar]
- 17.Xue H. Wu S. Papadeas ST. Spusta S. Swistowska AM. MacArthur CC. Mattson MP. Maragakis NJ. Capecchi MR, et al. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27:1836–1846. doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swistowski A. Peng J. Han Y. Swistowska AM. Rao MS. Zeng X. Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS One. 2009;4:e6233. doi: 10.1371/journal.pone.0006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Amour KA. Bang AG. Eliazer S. Kelly OG. Agulnick AD. Smart NG. Moorman MA. Kroon E. Carpenter MK. Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 20.Vallier L. Touboul T. Chng Z. Brimpari M. Hannan N. Millan E. Smithers LE. Trotter M. Rugg-Gunn P. Weber A. Pedersen RA. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y. Han SS. Wu Y. Tuohy TM. Xue H. Cai J. Back SA. Sherman LS. Fischer I. Rao MS. CD44 expression identifies astrocyte-restricted precursor cells. Dev Biol. 2004;276:31–46. doi: 10.1016/j.ydbio.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Van Hoof D. Munoz J. Braam SR. Pinkse MW. Linding R. Heck AJ. Mummery CL. Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Lieu PT. Machleidt T. Thyagarajan B. Fontes A. Frey E. Fuerstenau-Sharp M. Thompson DV. Swamilingiah GM. Derebail SS. Piper D. Chesnut JD. Generation of site-specific retargeting platform cell lines for drug discovery using phiC31 and R4 integrases. J Biomol Screen. 2009;14:1207–1215. doi: 10.1177/1087057109348941. [DOI] [PubMed] [Google Scholar]
- 24.Beard C. Hochedlinger K. Plath K. Wutz A. Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 25.Morikawa J. Nishimura Y. Uchida A. Tanaka T. Molecular cloning of novel mouse and human putative citrate lyase beta-subunit. Biochem Biophys Res Commun. 2001;289:1282–1286. doi: 10.1006/bbrc.2001.6125. [DOI] [PubMed] [Google Scholar]
- 26.Touboul T. Hannan NR. Corbineau S. Martinez A. Martinet C. Branchereau S. Mainot S. Strick-Marchand H. Pedersen R, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 27.Tada S. Era T. Furusawa C. Sakurai H. Nishikawa S. Kinoshita M. Nakao K. Chiba T. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 28.Kattman SJ. Witty AD. Gagliardi M. Dubois NC. Niapour M. Hotta A. Ellis J. Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Chen J. Sai SY. Vazin T. Coggiano M. Freed WJ. Human embryonic stem cells which express hrGFP in the undifferentiated state and during dopaminergic differentiation. Restor Neurol Neurosci. 2009;27:359–370. doi: 10.3233/RNN-2009-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng X. Chen J. Sanchez JF. Coggiano M. Dillon-Carter O. Petersen J. Freed WJ. Stable expression of hrGFP by mouse embryonic stem cells: promoter activity in the undifferentiated state and during dopaminergic neural differentiation. Stem Cells. 2003;21:647–653. doi: 10.1634/stemcells.21-6-647. [DOI] [PubMed] [Google Scholar]
- 31.Dickson J. Gowher H. Strogantsev R. Gaszner M. Hair A. Felsenfeld G. West AG. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010;6:e1000804. doi: 10.1371/journal.pgen.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osterlehner A. Simmeth S. Gopfert U. Promoter methylation and transgene copy numbers predict unstable protein production in recombinant Chinese hamster ovary cell lines. Biotechnol Bioeng. 2011 2011 May 26; doi: 10.1002/bit.23216. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.