Abstract
Methylation profile was analyzed in ninety-five patients with childhood acute lymphoblastic leukemia (ALL). Methylation of both MGMT and p16 genes were associated with higher age (p=0.01 and p=0.03, respectively). Methylation of both p15 and SHP1 genes occurred more frequently in T-ALL than in precursor B-ALL (p=0.02 and p=0.01, respectively). In contrast, methylation of the DAPK gene was more frequent in precursor B-ALL (p=0.01). Patients with methylation of multiple genes more likely had T cell phenotype, and are classified as medium/high risk (p=0.004 and p=0.03, respectively). These results suggest that methylation status is associated with clinicopathological features in childhood ALL.
Keywords: methylation, multiple genes, ALL
Introduction
Childhood ALL is a heterozygous disease with a widely variable outcome. Differences in the behavior and prognosis of the leukemia suggest that ALL can be divided into several biologic subgroups. Many investigators have tried to identify useful clinical markers that could divide the ALL patients into several groups. However, useful markers have been difficult to identify.
Like other tumors, ALL results from the accumulation of genomic abnormalities that affect normal control of cellular growth (1). Inactivation of tumor suppressor genes is a pivotal pathway of leukemogenesis in ALL. DNA methylation at CpG sites in the promoter region can cause gene silencing, which may provide an alternate pathway to gene inactivation in addition to their deletions or mutations. Methylation in the promoter region is a frequent, acquired epigenetic event involved in the pathogenesis of many kinds of human malignancies including leukemia (2-4). Studies of methylation of ALL samples were mostly examined at single candidate genes, and only limited data are available on the extent of concurrent methylation (5-7). Moreover, little data are available concerning the association between methylation pattern and clinocopathological features. To understand the frequency and clinical relevance of aberrant methylation, we analyzed DNA methylation patterns of 14 genes in 95 patients with childhood ALL.
Materials and Methods
Clinical Sample
Ninety-five primary DNA samples of childhood ALL were obtained from individuals who participated in the ongoing Multicenter Trial (ALL-BFM 2000) of the German Berlin-Frankfurt-Münster (BFM) group (diagnosis between October 1999 and September 2002)(8). These patients were selected by the availability of cryopreserved pre-treatment cell samples in addition to the needs for routine programs. Informed consent was obtained from the patients, their parents, or both, as appropriate. The percentage of blast cells was at least 80 %, and usually more than 90 % in the cell samples; and DNA was extracted from them. Clinical information at the initial diagnosis was available for all of the 95 children in this study and is summarized in Table 1. Immunophenotypic subgroups were defined as follows: common ALL: TdT+, CD19+, CD10+, cyIgM−, surface Ig−; pre-B ALL: TdT+, CD19+, CD10+/−, cyIgM+, surface Ig−; T-ALL: TdT+, cyCD3+, CD7+. Precursor-B ALL includes pre-B ALL and common-ALL. Control normal DNA was extracted from peripheral blood of 10 healthy individuals.
Table 1.
Clinicopathological characteristics of 95 childhood ALL patients by methylation profile
| Clinical features at diagnosis |
Total 95 (%) |
patients with 0-1 gene methylated 57 (%) |
patients with multiple genes methylated 38 (%) |
p |
|---|---|---|---|---|
| Age (years) | ||||
| <10 | 63 (69) | 40 (73) | 23 (64) | |
| ≥10 | 28 (31) | 15 (27) | 13 (36) | 0.37 |
| Sex | ||||
| Male | 41 (45) | 23 (42) | 18 (50) | |
| Female | 50 (55) | 32 (58) | 18 (50) | 0.44 |
| WBC counts (103/μl) | ||||
| <20 | 54 (59) | 33 (60) | 21 (58) | |
| ≥20 | 37 (41) | 22 (40) | 15 (42) | 0.87 |
| CNS involvement | ||||
| no | 72 (83) | 43 (81) | 29 (85) | |
| yes | 15 (17) | 10 (19) | 5 (15) | 0.62 |
| Immunophenotype | ||||
| T-ALL | 29 (33) | 11 (21) | 18 (50) | |
| precursor-B ALL* | 60 (67) | 42 (79) | 18 (50) | 0.004 |
| t (4;11) | ||||
| negative | 76 (97) | 44 (98) | 32 (97) | |
| positive | 2 (3) | 1 (2) | 1 (3) | 0.82 |
| TEL/AML1 | ||||
| negative | 66 (86) | 36 (80) | 30 (94) | |
| positive | 11 (14) | 9 (20) | 2 (6) | 0.09 |
| Response to chemotherapy# | ||||
| good | 73 (84) | 45 (83) | 28 (85) | |
| poor | 14 (16) | 9 (17) | 5 (15) | 0.85 |
| BFM risk | ||||
| low | 24 (26) | 19 (35) | 5 (14) | |
| medium/high | 67 (74) | 36 (65) | 32 (86) | 0.03 |
, Precursor-B ALL includes pre-B and common-ALL;
, Poor response was defined as the presence of more than 1000/μl peripheral blood blast cells at day 8 of therapy. ( ) numbers in parenthesis represent percentage.
Primer. and methylation-specific PCR
A total of 14 genes, including cell cycle control genes (p14, p15, p16, Rb, p21, p27, APC, cyclin D2, and FHIT), DNA repair genes (MGMT, and hMLH1), apoptosis-related genes (DAPK), a differentiation-associated gene (RARβ), and a negative regulator of the Jak/STAT signaling pathway (SHP1) were analyzed in this study.
Genomic DNA was modified using the sodium bisulfite method as described (9). Briefly, one μg of genomic DNA was treated with 0.2 M NaOH for 10 min at 37°C. Aliquots of 10 mM hydroquinone (30 μl) and 3 M sodium bisufite (Sigma Chemical Co., St. Louis. Mo) were added, and the solution was incubated at 50°C for 16 h. Treated DNA was purified by the use of a Wizard DNA purification system (Promega Corporation, Madisn WI). Modified DNA was stored at −80°C until used. Bisulfite treatment converts unmethylated cytosines to uracils while leaving the methylated cytosines unaffected.
PCR was performed using specific primers for each of the genes. After amplification, 15 μL of the PCR product was separated on a 3 % agarose gel containing 0.3 mg/mL ethidium bromide. For each MSP reaction, a positive control [CpGenome™, DNA universally methylated DNA (Intergen, Purchase, NY, USA.)] and negative control (normal lymphocyte DNA) was used. Water blank was used as a negative control in each set of PCR reactions.
Statistical analysis
Difference in the distribution among patients either with or without methylation was analyzed using χ2 test or two tailed Fisher’s exact test. An effect was considered statistically significant if the P value was 0.05 or less.
Results
Methylation profile was analyzed in ninety-five patients with childhood ALL. The genes analyzed includes p14, p15, p16, Rb, MGMT, MLH1, FHIT, p21, p27, RARβ, APC, DAPK, cyclin D2, and SHP1. None of these 14 genes were methylated in white blood cell DNA from 10 healthy individuals (data not shown), suggesting that methylation of these genes was associated with leukemia, not merely changes found in lymphocytes.
Most of these genes demonstrated methylation in a proportion of patients (Fig. 1). RARβ, FHIT, p15, and DAPK showed methylation in a significant proportion of cases (33, 28, 25, and 13 %, respectively). Frequency of methylation in other gene was: SHP1, 9 %; MLH1, 8 %; MGMT, 6 %, p16, 4 %; and APC, 4%. By contrast, p27, cyclin D2, and Rb were rarely altered (2, 2, and 1 %) (Table 2). None of the p14 or the p21 genes were hypermethylated (Fig. 2).
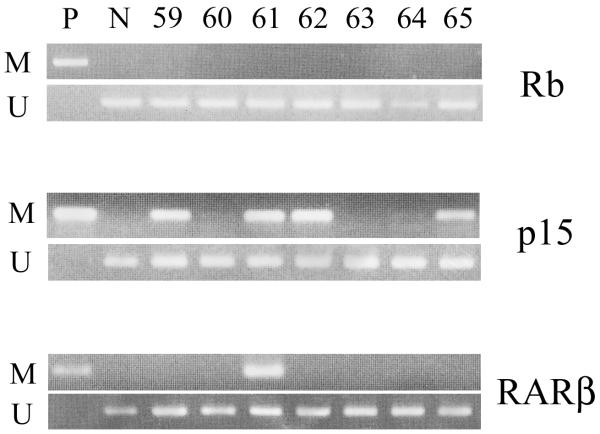
Figure 1. Methylation analysis in childhood ALL patients.
Examples are shown for both Methylated and Unmethylated primers. Patients No. 59, 61, 62, and 65 showed methylation in the p15 promoter. Patient No. 61 had methylation in the RARβ promoter. None of the patients acquired methylation in the Rb gene. P, positive control (universally methylated DNA); N, negative control (normal lymphocyte DNA).
Table 2.
Methylation profile in childhood ALL
| Features | frequency, % |
|---|---|
| Methylated genes | |
| RARβ | 33 |
| FHIT | 28 |
| P15 | 25 |
| DAPK | 13 |
| SHP1 | 9 |
| MLH1 | 8 |
| MGMT | 6 |
| P16 | 4 |
| APC | 4 |
| P27 | 2 |
| cyclin D2 | 2 |
| Rb | 1 |
| P14 | 0 |
| P21 | 0 |
| No. methylated genes | |
| 0 | 29 |
| 1 | 28 |
| 2 | 21 |
| 3 | 10 |
| 4 | 5 |
| 5 | 2 |
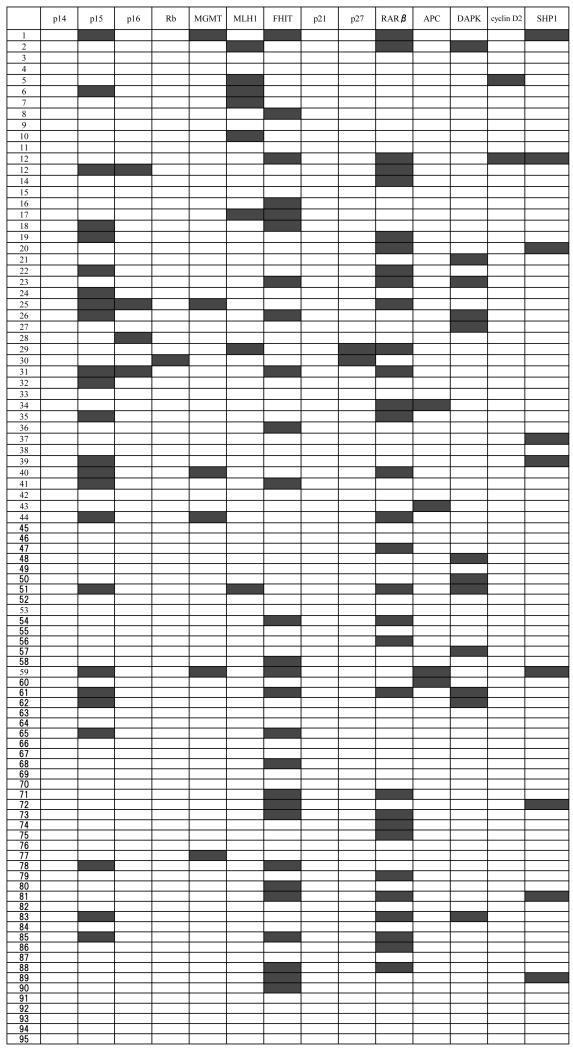
Figure 2. Methylation profile of 14 genes in 95 childhood ALL patients.
We analyzed methylation status of the 14 genes in 95 patients with childhood ALL. Black boxes represent methylated cases, white boxes denote nonmethylated cases.
Sixty-six patients (69 %) showed methylation of at least one gene, indicating that methylation is a frequent event in the development of childhood ALL. Twenty-eight patients (29 %) had methylation in one gene, 21 individuals (22 %) had methylation in two genes, 10 children (11 %) had methylation in three genes, five patients (5 %) had methylation in four genes, and two individuals (2 %) had methylation in five genes. In total, thirty-eight (40 %) patients showed methylation of multiple genes (Table 2).
In order to analyze the relationship of methylation in different genes in childhood ALL, we performed statistical analysis. We found that methylation of several genes occurred concurrently. Methylation of p15 gene was associated with methylation of p16, MGMT, and RARβ (p=0.049, p=0.004, and p=0.013, respectively). Methylation of FHIT was coordinated with methylation of SHP1 (p=0.015). However, methylation of p16 did not coexist with methylation of MGMT.
We analyzed the relationship between methylation status and clinicopathological features at presentation, including age, sex, WBC, CNS involvement, immunophenotype, t (4; 11), TEL/AML1, response to prednisolone, BFM risk classification, and prognosis. We found that methylation of several genes was correlated with standard prognostic factors, including immunophenotype, and age. Methylation of both MGMT and p16 genes were associated with higher age (p=0.01 and p=0.03, respectively). Methylation of both p15 and SHP1 genes occurred more frequently in T-ALL than in precursor B-ALL (p=0.02 and p=0.01, respectively). In contrast, methylation of the DAPK gene was more frequent in precursor B ALL (p=0.01). In addition to these statistically significant associations, we found some interesting observations. Methylation of both MGMT and RARβ genes occurred more frequently in T-ALL. Methylation of the DAPK gene was associated with CNS involvement. Methylation of the FHIT gene was more frequent in children with t (4;11), and with good response to prednisolone. However, these associations were not statistically significant. We did not find any significant relationship between methylation status of each gene and prognosis.
We then correlated the relationship between the numbers of methylated genes and clinicopathological features (Table 1). We found that patients with methylation of multiple gene more likely had T cell phenotype, and are classified as medium/high risk (p=0.004 and p=0.03, respectively). In addition to these statistically significant associations, we identified that TEL-AML1 [ETV6-RUNX1] fusion positive patients are less likely to have methylation of multiple genes. However, this association was not statistically significant. We did not find any significant relationship between number of methylated gene and prognosis.
Discussion
DNA methylation at CpG sites in the promoter region plays an important role in the development of hematological malignancies. Several methylation studies have been performed in childhood ALL (6, 7, 10, 11). In this study, we have analyzed the methylation profile in 95 cases of childhood ALL. Sixty-six patients (69 %) showed methylation of at least one gene, and thirty-eight (40 %) patients showed methylation of multiple genes. These results suggest that methylation is a frequent event in childhood ALL, and is involved in the development of this disease.
We found that the RARβ (33 %), the FHIT (28 %), and the p15 (25 %) genes were frequently methylated in patients with childhood ALL. Frequent methylation of the both FHIT and p15 genes has been reported by other investigators (6, 7, 10, 12, 13). Taken together, our results confirm that methylation of these genes is important in the development of childhood ALL. However, methylation of the RARβ gene has not been reported.
We have recently found frequent methylation of the RARβ gene (89 %) in myelofibrosis with myeloid metaplasia (14). This gene was methylated in 21 % patients with acute promyelocytic leukemia (15). A recent study revealed that acute promyelocytic leukemia having the PML-RAR fusion protein is associated with hypermethylation and silencing of the RARβ gene (16). These results imply a mechanistic link between genetic and epigenetic changes during transformation and suggest that hypermethylation contributes to the early steps of leukemogenesis.
During statistical analysis of concomitant methylation, p15 gene was associated with p16 (p=0.05). Both of the genes compose cell cycle regulatory pathway, and work at the same point. Therefore, inactivation of either of the genes alleviates the need for inactivation of the other gene. However, inactivation of both genes may collaborate to allow cell cycle to proceed and prevent apoptosis. These results suggest that methylation is not randomly distributed, but discrete and unique methylation phenotype is established in childhood ALL.
The main clinical value of this methylation analysis lies in its ability to divide ALL patients into several biologic subgroups. We found that methylation of several genes was correlated with standard prognostic factors, including immunophenotype and age. Methylation of p15 occurred more frequently in T-ALL than in precursor B-ALL (p=0.02). Tsellou et al, reported frequent methylation of the p15 gene in T-ALL (17). Taken together, hypermethylation of p15 may be involved in the pathogenesis of T-cell origin ALL, but not in that of B-cell origin ALL. In contrast, methylation of the DAPK gene was more frequent in precursor B ALL (p=0.01). Methylation of both MGMT and p16 genes were associated with higher age (p=0.01 and p=0.03, respectively). We did not find any significant relationship between methylation status of each gene and prognosis. Mirebeau et al, reported similar findings that methylation of p15 and p16 did not influence the patients’ outcome in childhood B-lineage ALL (18). In contrast, Milani et al, identified 20 genes with DNA methylation levels that predicted relapse of leukemia (19). However, none of these genes were included in this study.
We found that patients with methylation of multiple genes more likely have T cell phenotype, and are classified as medium/high risk (p=0.004 and p=0.03, respectively). Roman-Gomez et al, reported that methylation of multiple genes is an independent prognostic factor in ALL (10, 20). However, no significant relationship occurred between number of methylated genes and prognosis in our series. Treatment of childhood ALL has been improved, and prognosis of our ALL-BFM 2000 cohort is remarkable making the identification of a significant association exceedingly difficullt. Further methylation studies with longer follow up are necessary to clarify this issue.
We previously performed methylation analysis in relapsed childhood ALL (21). Frequency of methylation in each gene was: MGMT, 56%; RARβ, 44%; and p16, 22%, respectively. None of the p14, p15, Rb, APC, hMLH1, DAPK, and FHIT genes were hypermethylated. We compared these results with results obtained in this study. RARβ was methylated in both initial diagnosis and relapse. The three genes (FHIT, p15, and DAPK) were methylated only at the initial presentation. MGMT and p16 acquired methylation at relapse. Four genes (MLH1, APC, Rb, and p14) were not methylated at any stage of this disease. These results suggest that hypermethylation is involved in both development and relapse of childhood ALL. Interestingly, several genes which had been methylated at initial presentation lost their methylation at relapse. These differences in the methylation profile between initial diagnosis and relapse reflect molecular differences between these two stages, and suggest that different genetic alterations are involved in each event. A larger study is necessary to clarify this issue.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pui C-H, Robison LL, Look AT. Acute Lymphoblastic Leukemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Uehara E, Takeuchi S, Tasaka T, Matsuhashi Y, Yang Y, Fujita M, Tamura T, Nagai M, Koeffler HP. Aberrant methylation in promoter-associated CpG islands of multiple genes in therapy-related leukemia. Int J Oncol. 2003;23:693–696. [PubMed] [Google Scholar]
- 3.Kuang SQ, Tong WG, Yang H, Lin W, Lee MK, Fang ZH, Wei Y, Jelinek J, Issa JP, Garcia-Manero G. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22:1529–1538. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- 4.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Research. 2007;67:2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Manero G, Daniel J, Smith TL, Kornblau SM, Lee M-S, Kantarjian HM, Issa J-PJ. DNA methylation of multiple promoter-associated CpG islands in adult acute lymphocytic leukemia. Clin. Cancer Res. 2002;8:2217–2224. [PubMed] [Google Scholar]
- 6.Garcia-Manero G, Jeha S, Daniel J, Williamson J, Albitar M, Kantarjian HM, Issa J-PJ. Aberrant DNA methylation in pediatric patients with acute lymphoblastic leukemia. Cancer. 2003;97:695–702. doi: 10.1002/cncr.11090. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez MI, Siraj AK, Bhargava M, Ozbek U, Banavali S, Chaudhary MA, Solh HEI, Bhatia K. Concurrent methylation of multiple genes in childhood ALL: Correlation with phenotype and molecular subgroup. Leukemia. 2003;17:1845–1850. doi: 10.1038/sj.leu.2403060. [DOI] [PubMed] [Google Scholar]
- 8.Stanulla M, Schaeffeler E, Flohr T, Cario G, Schrauder A, Zimmermann M, Welte K, Ludwig WD, Bartram CR, Zanger UM, Eichelbaum M, Schrappe M, Schwab M. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. JAMA. 2005;293:1485–1489. doi: 10.1001/jama.293.12.1485. [DOI] [PubMed] [Google Scholar]
- 9.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U.S.A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roman-Gomez J, Jimenez-Velasco A, Castillejo JA, Agirre X, Barrios M, Navarro G, Molina FJ, Calasanz MJ, Prosper F, Heiniger A, Torres A. Promoter hypermethylation of cancer-related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104:2492–2498. doi: 10.1182/blood-2004-03-0954. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Takeuchi S, Hofmann WK, Ikezoe T, van Dongen JJ, Szczepański T, Bartram CR, Yoshino N, Taguchi H, Koeffler HP. Aberrant methylation in promoter-associated CpG islands of multiple genes in acute lymphoblastic leukemia. Leukemia Research. 2006;30:98–102. doi: 10.1016/j.leukres.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Zheng S, Ma X, Zhang L, Gunn L, Smith MT, Wiemels JL, Leung K, Buffler PA, Wiencke JK. Hypermethylation of the 5′ CpG island of the FHIT gene is associated with hyperdiploid and translocation-negative subtypes of pediatric leukemia. Cancer Research. 2004;64:2000–2006. doi: 10.1158/0008-5472.can-03-2387. [DOI] [PubMed] [Google Scholar]
- 13.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Calasanz MJ, Garate L, San Jose-Eneriz E, Cordeu L, Prosper F, Heiniger A, Torres A. CpG island methylator phenotype redefines the prognostic effect of t(12;21) in childhood acute lymphoblastic leukemia. Clin. Cancer Res. 2006;12:4845–4850. doi: 10.1158/1078-0432.CCR-05-2592. [DOI] [PubMed] [Google Scholar]
- 14.Jones LC, Tefferi A, Idos GE, Kumagai T, Hofmann WK, Koeffler HP. RARβ2 is a candidate tumor suppressor gene in myelofibrosis with myeloid metaplasia. Oncogene. 2004;23:7846–7853. doi: 10.1038/sj.onc.1207510. [DOI] [PubMed] [Google Scholar]
- 15.Chim CS, Wong SY, Kwong YL. Aberant gene promoter methylation in acute promyelocytic leukaemia: profile and prognostic significance. British Journal of Haematology. 2003;122:571–578. doi: 10.1046/j.1365-2141.2003.04462.x. [DOI] [PubMed] [Google Scholar]
- 16.Croce LD, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Coco FL, Kouzarides T, Nervi C, Minucci S, Pelicci PG. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 17.Tsellou E, Troungos C, Moschovi M, Athanasiadou-Piperopoulou F, Polychronopoulou S, Kosmidis H, Kalmanti M, Hatzakis A, Dessypris N, Kalofoutis A, Petridou E. Hypermethylation of CpG islands in the promoter region of the p15INK4B gene in childhood acute leukaemia. Eur. J. Cancer. 2005;41:584–589. doi: 10.1016/j.ejca.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Mirebeau D, Acquaviva C, Suciu S, Bertin R, Dastugue N, Robert A, Boutard P, Méchinaud F, Plouvier E, Otten J, Vilmer E, Cavé H. EORTC-CLG: The prognostic significance of CDKN2A, CDKN2B and MTAP inactivation in B-lineage acute lymphoblastic leukemia of childhood. Results of the EORTC studies 58881 and 58951. Haematologica. 2006;91:881–885. [PubMed] [Google Scholar]
- 19.Milani L, Lundmark A, Kiialainen A, Nordlund J, Flaegstad T, Forestier E, Heyman M, Jonmundsson G, Kanerva J, Schmiegelow K, Söderhäll S, Gustafsson MG, Lönnerholm G, Syvänen AC. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood. 2010;115:1214–1225. doi: 10.1182/blood-2009-04-214668. [DOI] [PubMed] [Google Scholar]
- 20.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Garate L, Jose-Eneriz ES, Cordeu L, Barrios M, Prosper F, Heiniger A, Torres A. Promoter hypermethylation and global hypomethylation are independent epigenetic events in lymphoid leukemogenesis with opposing effects on clinical outcome. Leukemia. 2006;20:1445–1448. doi: 10.1038/sj.leu.2404257. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita C, Yang Y, Takeuchi S, Matsushita M, van Dongen JJM, Szczepański T, Bartram CR, Seo H, Koeffler HP, Taguchi H. Aberrant methylation in promoter-associated CpG islands of multiple genes in relapsed childhood acute lymphoblastic leukemia. Oncol Rep. 2004;12:97–99. [PubMed] [Google Scholar]