Abstract
Common themes are emerging in the molecular mechanisms of long non-coding RNA-mediated gene repression. Long non-coding RNAs (lncRNAs) participate in targeted gene silencing through chromatin remodelling, nuclear reorganisation, formation of a silencing domain and precise control over the entry of genes into silent compartments. The similarities suggest that these are fundamental processes of transcription regulation governed by lncRNAs. These findings have paved the way for analogous investigations on other lncRNAs and chromatin remodelling enzymes. Here we discuss these common mechanisms and provide our view on other molecules that warrant similar investigations. We also present our concepts on the possible mechanisms that may facilitate the exit of genes from the silencing domains and their potential therapeutic applications. Finally, we point to future areas of research and put forward our recommendations for improvements in resources and applications of existing technologies towards targeted outcomes in this active area of research.
Keywords: airn/Kcnq1ot1, enhancers, epigenetic modifications, non-coding RNA, Xist/HOTAIR, imprinting
Introduction
Long non-coding RNAs (lncRNAs) are molecules often longer than 2 kb in length with a coding potential of less than 100 amino acids [1–3]. The number of lncRNAs exceeds that of protein coding genes [4–6] and their discovery has revolutionised the field of molecular biology.
Since their sequence provides no obvious clues regarding their function and the fact that they are poorly conserved across species, ncRNAs were viewed as non-functional and their presence and significance is still being debated [7–9]. However, as new studies identify the functions of individual ncRNAs, it is now apparent that many ncRNAs are the key regulators of transcriptional and translational output and therefore of cell fate and function [10–12]. While most mRNAs are exported to the cytoplasm for translation, many lncRNAs are now known to be retained in various sub-nuclear compartments [6, 13–15] suggesting that such RNAs may have a potential function in the compartment where they are localised.
Several nuclear lncRNAs have been studied in detail and investigations into the molecular functions of lncRNAs reveal more unexpected similarities in molecular functions than previously anticipated. Here, we will focus on some examples that provide important paradigms for gene regulation by lncRNAs through interaction with chromatin remodelling complexes. We believe that we have discovered only the tip of the iceberg of a hitherto unknown network of nuclear non-coding RNA/chromatin interaction.
Studies on four lncRNAs (Kcnq1ot1, Airn, Xist and HOTAIR) investigated individually by independent laboratories reveal that their function is to regulate transcription of multiple target genes through epigenetic modifications. These investigations have established fundamental principles of lncRNA function with broad implications. On the mouse X-chromosome, expression of lncRNA X-inactive specific transcript (Xist) from the designated inactive X-chromosome is essential for the silencing of the inactive X-chromosome [16–19]. On the Insulin like growth factor 2 receptor (Igf2r) imprinted cluster, located on mouse chromosome 17, the expression of paternal-specific non-coding transcript antisense Igf2r RNA non-coding (Airn, 108 kb), is required for the silencing of three genes on the paternal allele. These genes are spread over a large genomic region spanning 400 kb [20]. On mouse chromosome 7, the potassium voltage-gated channel subfamily Q member 1 (Kcnq1) imprinted cluster, spread over a 1 Mb genomic region in embryos, contains multiple genes and is silenced on the paternal allele by the un-spliced lncRNA Kcnq1 overlapping transcript 1 (Kcnq1ot1, 91 kb) in cis [21, 22]. Some genes on the Homeobox D (HOXD) cluster, located over a 40 kb genomic region on human chromosome 2, are silenced by lncRNA HOTAIR, which originates from the HOXC cluster on chromosome 12 [23]. The elucidation of the molecular mechanisms of such long-range regulation reveals at least three common themes in the silencing process (Table 1).
Table 1.
List of investigated long non-coding RNAs and their protein partners
| lncRNA | Size (kb) | Spliced | Cover | Regulated genes | Escaped genes | Chromatin remodelling complex | Ref. |
|---|---|---|---|---|---|---|---|
| Airn | 108 | Yes | Yes | Multiple genes, in cis clusters | Yes, development specific escape | G9a | [20, 27] |
| Kcnq1ot1 | 94 | No | Yes | Multiple genes in cis clusters | Yes, tissue specific escape | G9a | [22, 50] |
| PRC2 | |||||||
| PRC1 | |||||||
| Xist | 17 | Yes | Yes | Multiple genes in cis clusters on X chromosome | Yes, development specific escape | PRC2 | [31, 67–69] |
| HOTAIR | 2 | Yes | Not known | Multiple genes in trans at HOXD locus, individual targets all over the genome | N/A | PRC2 | [23, 29] |
| LSD1 |
Silencing mediated by lncRNAs is imposed via recruitment of chromatin remodelling complexes
The involvement of RNAs in epigenetic silencing was proposed by various investigators [24, 25] based on the observation that while many enzymatic members of the chromatin remodelling complexes did not have DNA binding domains, they possessed RNA binding domains. Molecular investigations revealed the association between lncRNAs such as Kcnq1ot1, Airn, Xist, HOTAIR and chromatin remodelling complexes such as Polycomb repressive complexes 1 and 2, (PRC1 and PRC2) [22, 23, 26–32] which mediate mono-ubiquitinylation of Lysine 119 of Histone 2A (H2AK119ub)[33] and di- and tri-methylation of Histone 3 lysine 27 (H3K27me2 and H3K27me3)[34, 35], respectively; Lysine Specific Demethylase 1 (LSD1)/CoREST which demethylates mono- and di-methylated Histone 3 at Lysine 4 (H3K4) [36] and G9a histone methyl transferase which catalyses Histone 3 Lysine 9 di- and tri-methylation (H3K9me2 and H3K9me3) [37, 38].
At the Kcnq1 imprinted locus, Kcnq1ot1 lncRNA interacts with histone methyltransferase G9a and members of the PRC2 complex [22]. In addition, Terranova et al. reported close proximity between Kcnq1ot1 lncRNA and members of PRC2 and PRC1 complex [28]. At the Igf2r imprinted locus, Airn also associates with G9a [27]. The imprinted genes in the Igf2r and Kcnq1 clusters show repressive histone marks of K3K9me3 and H3K27me3 most likely induced by G9a and PRC2-remodelling complexes, respectively [22, 27]. It should be noted that these studies were performed in extra embryonic placental tissue in mouse and the mechanisms of imprinting in embryonic tissues may be different. On the X-chromosome, Xist lncRNA interacts with Ezh2 and Suz12 components of the PRC2 complex via a repeat A region (RepA) and the recruitment of PRC2 to the inactive X-chromosome induces the repressive epigenetic mark of H3K27me3 [31]. At the HOXD locus, HOTAIR also recruits PRC2 complex to induce silencing of specific genes [23]. It is noteworthy that at some of the loci mentioned above, the target genes fail to be silenced in the absence of the lncRNA [20, 23, 31, 39] thus implying that lncRNAs are essential for steering chromatin remodelling complexes to distinct target sites in order to induce silencing. Since these complexes interact with multiple lncRNAs, it appears that association with lncRNAs defines their target specificity. For example the repressive complex G9a, in concert with lncRNA Airn, targets the Igf2r imprinted locus, while in association with Kcnq1ot1, G9a represses genes in the Kcnq1 locus [22, 27]. Similarly PRC2 in association with HOTAIR, targets the HOXD locus [23]; with Kcnq1ot1 it targets the Kcnq1 cluster [22], and while associated with RepA/Xist, it modifies histones on the X-chromosome [31]. Thus each protein complex is capable of being directed by multiple lncRNAs (Fig. 1 A–D). However, it is not clear if members of chromatin remodelling protein complexes have distinct domains for binding with specific ncRNAs or whether they bind in general to ncRNA molecules presenting certain secondary structures as seen in Xist [31, 40]. Indeed, it has recently been reported that short RNAs (50–200 nt), transcribed from repressed loci by stalled RNA polymerase II, interact with the PRC2 complex through their stem loop secondary structure and mediate gene repression through epigenetic modification [41]. It is not known whether the other ncRNAs such as Promoter associated short and long RNAs (PASRs, and PALRs) and promoter upstream transcripts (PROMPTs) generated around promoters as well as the vast numbers of small RNAs now known to be retained in the nucleus [6, 15, 42–44] possess distinct secondary structures and participate in local epigenetic regulation through interaction with chromatin remodelling complexes.
Figure 1.
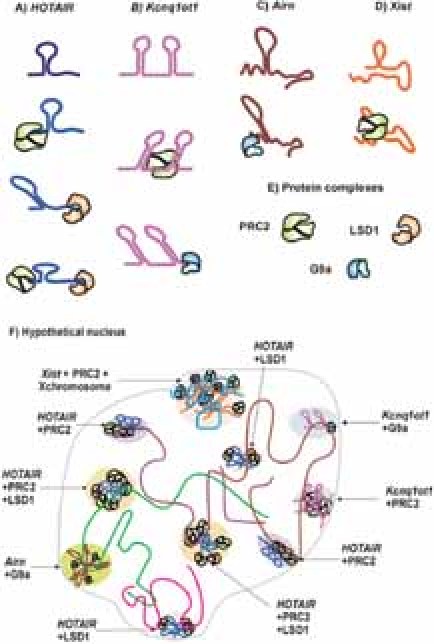
Complexity in lncRNA-chromatin modifying complex interactions. Chromatin remodelling complexes are capable of interacting with multiple lncRNAs. Similarly, lncRNAs may interact with more than one protein complex. A to D: Protein lncRNA interactions may result in conformational changes, which may help distinguish target specificity. E: Chromatin remodelling complexes F: Hypothetical nucleus depicting lncRNA–protein complexes and their silencing compartments. Xist domain is shown at the nuclear periphery with inactive X chromatin. Airn and Kcnq1ot1 are shown to silence specific genes on their respective imprinted loci while HOTAIR is seen to target loci genome wide in concert with different protein complexes. Note that mouse Hotair does not participate in silencing the HoxD cluster and is not reported to interact with chromatin remodelling complexes [55]. Human HOTAIR is depicted in this schema.
Interestingly, some lncRNAs also appear to interact with more than one chromatin-modifying complex. For example HOTAIR is known to interact with both PRC2 and LSD1/CoREST/REST complexes [29] and Kcnq1ot1 interacts with G9a as well as the PRC2 complex [22, 28, 30]. Recent reports identify ANRIL (antisense non-coding RNA in the INK4 locus) as another candidate lncRNA which interacts with more than one chromatin remodelling complex to induce silencing in cis [45, 46]. ANRIL (3.8 kb) originates close to the INK4A gene on chromosome 9 in humans and interacts with the CBX7 component of the PRC1 complex to induce silencing of the INK4A and INK4B loci [46] and with SUZ12 component of the PRC2 complex to mediate epigenetic silencing of the p15INK4B gene [45]. Thus the interaction of a single ncRNA with multiple chromatin modifying complexes to target specific genes may be a widespread phenomenon.
Indeed, in a high throughput RIP-Chip analysis, Khalil et al. found that 40% of long intergenic ncRNAs (lincRNAs) associated with the CoREST complex were also associated with the PRC2 complex, indicating that lincRNAs can have shared and independent targets [26]. Genome wide ChIP-Chip analysis of human promoters reveals 4,740 and 2,116 gene promoters occupied by PRC2 and LSD1, respectively, while 721 promoters are occupied by both complexes, suggesting shared and individual targets of each complex [29]. It is likely that repression at shared targets is mediated by ncRNAs capable of binding more than two complexes as seen with HOTAIR [29]. Thus it is now apparent that target specificity of lncRNAs can also be altered depending on the interacting chromatin modifying complex. These multiple interactions between chromatin complexes and lncRNAs may be sequence dependent as in the case of HOTAIR which has distinct domains for interaction with PRC2 and LSD1 [29]. However, the target distinction of the lncRNA-chromatin remodelling complexes is most likely mediated by conformational changes induced by these interactions (Fig. 1). It should be noted that while the ncRNAS mentioned above are certainly required for the initiation of silencing at their respective targets, it is not yet clear if like Xist, they are dispensable for the maintenance of silent epigenetic state at their target loci.
Additional mechanisms of ncRNA mediated silencing may exist in a gene or tissue specific manner. Certainly, Airn utilises gene specific silencing mechanisms even within the placenta, the Slc22a3 gene is silenced through recruitment of G9a, however silencing of the neighbouring Igf2r gene does not require G9a since its imprinted status is not affected in G9a KO mice [27]. In mouse ES cells, the Igf2r gene is persistently expressed at low levels from the paternal imprinted allele despite DNA methylation at its promoter [47], indicating that Airn transcription itself may interfere with transcription initiation at Igf2r [48, 49]. At the Kcnq1 locus, ubiquitously imprinted genes (genes imprinted in placenta, embryo and adult tissues) are silenced in the placenta and liver by recruitment of Dnmt1 by Kcnq1ot1 [50]. The observation of imprinting and X inactivation phenomena, despite the poor conservation of ncRNAs such as Airn in opossum and dog [51, 52] and Xist in marsupials [53, 54] and the presence of a dysfunctional, poorly conserved Hotair in mouse [55], suggests the existence of compensatory layers of gene regulation in such species. It is likely that other modes of silencing may also emerge as common mechanisms for ncRNA-mediated silencing. Alternatively, the functional module of the lncRNAs in such species may be much shorter and dependent on secondary structure rather than length or primary sequence.
The promoters of genes silenced by lncRNAs are covered with lncRNAs
A physical association between lncRNAs and the chromatin of their target loci is emerging as a common theme for very long ncRNAs that silence genes in clusters. Xist was first shown by RNA-FISH studies to coat the chromatin at the inactive X chromosome in a non-uniform manner, where euchromatic regions on the inactive X remained devoid of Xist coating in the initial stages of X-chromosome inactivation [56–58]. The physical association of Xist with chromatin was further confirmed by immunoprecipitation with antibodies against macroH2A1, a histone H2A variant enriched on the inactive X chromosome [59] (reviewed in [60]). On the Kcnq1 locus, RNA-DNA-FISH studies reveal that Kcnq1ot1 also associates with imprinted genes on the Kcnq1 imprinted chromatin region [22, 61, 62] and at the Igf2r locus. Nagano et al. demonstrated through RNA-DNA-FISH a cloud of ‘Airn’ over the imprinted Slc22a3 at E11.5 [27]. Such a ncRNA-cloud has not yet been reported for HOTAIR, which mediates silencing in trans and is only 2 kb long. Current data are insufficient to conclude whether this covering of targets is the exclusive property of long ncRNAs acting in cis over clustered targets.
The coating of lncRNA over targeted genomic loci is postulated to create a ‘silent nuclear compartment’ resulting in the recruitment of chromatin remodelling complexes to maintain silent chromatin marks and restrict access to the transcriptional machinery [63]. At the sub-nuclear level, such silent compartments created by Xist and Kcnq1ot1 cover have been reported to localise to the perinucleolar region [22, 64, 65] suggesting that lncRNAs may induce changes in the spatial organisation of chromatin in the nucleus. It is notable that the lncRNAs involved in the formation of these silent domains are cis acting, particularly long (over 10 kb) and known to silence genes spread over large genomic loci on single chromosomes. It is not known if the many small RNAs retained in the nucleus participate in the formation of such clouds, since it is difficult to visualise small RNAs with current techniques such as FISH. Chow et al. recently reported the presence of small siRNAs arising from the young LINE1 elements in the Huwe-1 gene, which facilitate its silencing by inclusion into the Xist domain [66]. Kanhere et al. reported sRNAs arising from PRC2 target genes that participate in the recruitment of PRC2 to their promoters [41]. It will be of interest to investigate if these short RNAs remain in the vicinity of their target loci and participate in the formation of a cover.
Silenced genes enter while active genes remain outside the lncRNA silencing compartment
The location of genes under the cover of lncRNAs appears to be dynamic. Detailed analyses reveal that the genes that undergo X-inactivation gradually relocate deep inside the Xist-covered silent compartment as they are silenced [63]. Genes that escape X-inactivation remain outside this silent domain [63]. It was recently shown that inclusion of genes within the Xist silencing compartment is dependent on the density and proximity of young full length long interspersed nuclear elements (LINEs) to the genes [66]. However, the evidence from genes such as Jarid1c (Kdm5c/Smcx), Shroom2 and Mid1, which undergo tissue or development stage specific inactivation [67–69], suggests that in addition to the abundance and proximity of LINEs, other tissue/development-specific factors may also play a role in facilitating inclusion into the silencing compartment. Jarid1c is a LINE poor gene [66], which initially undergoes X-inactivation but is activated at later stages of development [67]. Jarid1c is expressed at equal levels in males and females in neonatal brains and adult liver [70] but escapes X-inactivation in adult female brains [70]. RNA-DNA-FISH studies reveal that Jarid1c remains at the inner edge of the Xist silent compartment in cells where it is inactive and it is located outside the Xist compartment in cells where it is active [63]. Another gene Shroom2, undergoes X-inactivation and displays PRC2-mediated H3K27me3 mark of repressive chromatin in a tissue specific manner; while Mid1 gene shows H3K27me3 enrichment in female embryos but not in adult liver, indicating that it undergoes X-inactivation in embryonic tissues and later escapes X-inactivation [69]. Although RNA-DNA-FISH data for Mid1 and Shroom2 are not available, it is plausible that these genes are also located inside the Xist silencing compartment when they undergo X-inactivation. To escape inactivation Mid1 and Shroom2 would have to exit the silencing compartment, since the transcription machinery is located outside this compartment.
The exit of genes from lncRNA silencing domains is also seen at the imprinted loci. The Slc22a3 gene is imprinted in embryos at E11.5 but shows biallelic expression at E15.5 [27]. Using RNA-DNA-FISH techniques, Nagano et al. demonstrated Slc22a3 inside the silencing compartment under cover of Airn at E11.5 when it is imprinted and a reduction in Slc22a3 loci covered by Airn at E15.5 after escape from imprinting [27]. The interaction between Airn and the Slc22a3 promoter demonstrated by RNA TRAP experiments at E11.5 was reduced at E15.5 [27]. On the Kcnq1 cluster, genes are differentially regulated in placenta and embryos at E12.5 and studies of the lncRNA cover index over such genes suggests that only silenced genes remain inside the inactivation domain [61]. Interestingly, the imprinted loci are not particularly abundant in LINEs [71] suggesting that factors other than LINEs regulate the inclusion of genes in the silencing compartment at the imprinted loci. The fact that on the X-chromosome, as well as the imprinted loci, genes can escape from the silencing compartment into the transcriptionally active domains, despite the presence of the perpetrating lncRNA and repressive chromatin complexes in the vicinity, also suggests an additional layer of regulatory control that governs exit from the silencing compartment. Furthermore, tissue/development stage-specific silencing of X-linked and imprinted genes [67, 69, 70, 72] also argues against genomic features as key regulators of entry into silencing domains and suggests that at certain loci inclusion into lncRNA silencing compartment may be regulated by other factors responsive to development stage or tissue specific molecular signals. Intriguingly, the abundant expression and retrotransposition of LINE-1 in neuronal precursor cells is postulated to create gene disruption and diversity in the genome [73].
It will be of interest to investigate if such LINEs also play an active role in gene silencing by facilitating the influence of lncRNAs.
Possible role for enhancers in escaping epigenetic regulation mediated by lncRNAs
It is now apparent that genes once silenced by inclusion into the silent domains of the lncRNAs are capable of reactivation in a tissue or development stage specific manner.
This reactivation most likely requires the genes to escape from the silent compartment. What regulates the exit of genes from the silencing domains created by lncRNAs? For such regulation to be effective, the controlling mechanism must remain outside the influence of the silencing compartment mediated by lncRNAs and be able to respond to developmental cues. Genomic regions called enhancers meet both requirements and are likely candidates for such regulation. Enhancers are DNA elements which provide binding sites for sequence specific transcription factors and induce transcription by facilitating the recruitment of RNA pol II to promoters (reviewed in [74]). FISH and 3C studies have shown that enhancers activate transcription in cis and in trans and that transcription activation by enhancers requires physical contact with the promoters via chromatin looping [75]. It is now known that distal elements bound by p300 with a chromatin signature of high H3K4me1 and H3K27ac and low H3K4me3 marks enhancers while core promoters are nucleosome free regions flanked by high H3K4me3 and bound by RNA polymerase II [76, 77]. About 25% of enhancers are also bound by RNA polymerase II [78] and it was recently reported that a fraction of extra-genic and intra-genic enhancers are actively transcribed from the H3K4me1 domain, giving rise to non-polyadenylated bidirectional transcripts called enhancer RNAs (eRNAs) [78, 79].
Although the function of eRNAs is as yet unclear, their expression levels are reportedly concordant with the expression levels of their target promoter transcripts [78] and their induction is reported to be a precise indication of the physical contact between enhancers and their target promoters [80]. Enhancers show developmental and activity dependent plasticity and tissue specificity [78, 81–83] indicating that they are responsive to cellular signals. Due to their physical distance from the target promoters, upon silencing of target genes via inclusion into ncRNA silencing compartments, the distal enhancers are likely to remain outside the repressive domains. Such regulatory genomic regions may be involved in mediating the escape from lncRNA mediated silencing. In particular their ability to contact promoter regions through looping of chromatin may play a role in rescuing genes out of the silent domains (Fig. 2).
Figure 2.
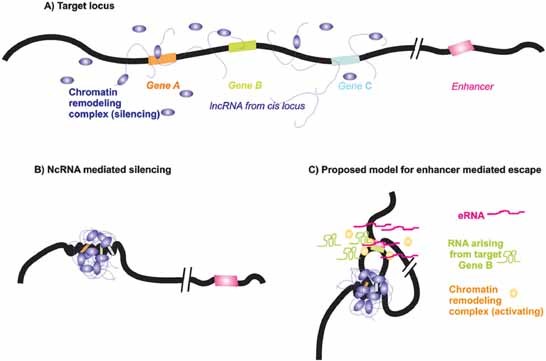
Schematic representation of lncRNA mediated silencing and proposed mechanism for escape. A: Hypothetical genomic locus with three genes (as labelled) regulated by a long non-coding RNA (blue) and a chromatin repressive enzyme complex (blue ovals) is shown. A distal enhancer for Gene B is represented in magenta. B: At the appropriate stage the lncRNA accumulates over the locus to form an lncRNA cloud. The cloud covers the genes and compacts chromatin via chromatin remodelling complexes. C: Proposed model for reactivation of silenced genes. A single gene is shown to escape silencing through exiting the silencing domain by looping out of the repressive compartment and physical contact with the distal enhancer. The chromatin looping may be mediated by other ncRNAs arising from the gene itself (green) in concert with eRNAs (magenta). We propose that enhancers may initiate the reactivation process through eRNAs via recruitment of chromatin activating complexes (orange spheres) or through competition for repressive complexes at the locus (not shown).
An area of future research could be to investigate if genes exit the lncRNA covered silent compartment with the help of eRNAs. The fact that epigenetic modification is precisely executed at specific promoters implies that a mechanism exists within cells to facilitate recognition of specific promoters. Recent evidence indicates that double stranded short synthetic RNAs targeted at promoter regions (agRNAs) can mediate silencing as well as activation of the targeted promoters [84–87], indicating that sRNAs can recognise genomic promoters of their origin. agRNAs were shown to form a complex with AGO protein and a locally arising antisense ncRNA at the Progesterone receptor (PR) locus [88], suggesting that promoter specific RNAs may require other ncRNA mediators to execute their function. Non coding RNAs have been shown to be transcribed from active [1, 6, 15, 44, 89] as well as repressed promoters [41]. It is probable that some of these promoter specific RNAs utilise sense or antisense eRNAs as the ncRNA mediators to bring into physical proximity, silenced target genes and their enhancers via chromatin looping. These eRNA complexes may then compete with lncRNAs for the chromatin repressor complexes and facilitate reactivation of silenced genes, thus mediating exit from the repressive compartment. Alternatively, eRNAs may interact with chromatin activating complexes, and upon close proximity with target genes, induce activation through epigenetic remodelling thus mediating exit from the silencing domain (Fig. 2).
Therapeutic applications of manipulating lncRNA mediated silencing
From the discussion above it is apparent that lncRNA mediated repression of genes is an intricate process involving chromatin remodelling enzymes and spatial reorganisation. The involvement of multiple factors suggests that the process is open to experimental manipulation at multiple levels. To be inactivated by Xist, genes on the X-chromosome require an abundance of LINEs in the genomic region and transcription of young LINE-1 from their vicinity [66]. In mammalian genomes, both full-length and truncated LINEs can be transcribed [90]. It is not clear what marks the imprinted genes must carry to distinguish them from non-imprinted genes within the cluster. Nevertheless the silencing and escape of genes appear to be tightly regulated and the nature of such regulation warrants investigation as it may have therapeutic applications in some disorders such as Rett Syndrome.
Rett Syndrome is an X-linked dominant neuro-developmental disorder where mutations in the MECP2 gene cause arrest of neurodevelopment in girls [91]. Girls with Rett syndrome possess one normal and one mutant copy of the MECP2 gene. Since MECP2 gene undergoes X-inactivation [63], in Rett patients, the normal copy of MECP2 gene is active only in 50% of cells while the other 50% cells express the mutant gene, which results in the phenotype. Indeed, phenotypic variations seen in Rett Syndrome are presumably dependent on the X-inactivation status of the patient [92, 93]. However, recent studies also implicate other factors [94, 95]. The MECP2 locus on the X-chromosome is drawn inside the Xist silencing compartment at day 4 after differentiation [63]. Activation of the inactive non-mutant MECP2 gene has long been proposed as a therapeutic avenue for Rett Syndrome [96]. The Mecp2 KO male mice display striking phenotypic similarities to female patients with MECP2 mutations [97].
Recently, Guy et al. reported a surprising reversal of the Rett phenotype seen in an experimental mouse model of Rett Syndrome by reactivation of the Mecp2 gene in a transgenic mutant mouse. This provided the proof of principle that reactivation of the normal copy of Mecp2 may provide therapeutic benefits in patients with loss of function MECP2 mutations [98]. Thus a strategy of preventing the inclusion of MECP2 in the silencing domain at early stages of differentiation or enforcing the exit of MECP2 gene from the XIST silencing domain in differentiated cells may have therapeutic applications.
It will be interesting to investigate if a combined experimental approach of targeted down regulation of allele specific MECP2 related LINE elements and allele specific over expression of MECP2 enhancer eRNAs prevents inclusion into the silencing compartment and facilitates activation of the MECP2 gene. It is noteworthy that the MeCP2 protein is known to repress LINE-1 transcription [99–101] and LINE-1 expression and retrotransposition is reported to be significantly higher in the adult Mecp2 KO mouse brain as seen with genomic DNA and RNA analysis [100, 101].
In addition to Rett syndrome, manipulation of lncRNA mediated silencing may also be beneficial in preventing cancer progression, recurrence and metastasis. Gupta et al. recently reported that HOTAIR was over expressed up to 2,000-fold in metastatic breast tumours [102]. They demonstrated a combined role of HOTAIR and PRC2 complex in breast cancer invasiveness via overexpression and knock down of HOTAIR and PRC2 components through in vitro and in vivo studies [102]. This study indicates that HOTAIR and PRC2 complex specifically act through silencing of metastasis suppressor genes and alteration of the epigenetic program of breast cancer cells to promote cancer progression and metastasis. Thus selective activation of key HOTAIR targets, through agRNAs for example, may be beneficial in preventing invasiveness of tumours. In addition, recent studies indicate that lncRNAs, which are highly expressed in solid tumours, may be involved in cancer progression and metastasis via other mechanisms. The Metastasis Associated Lung Adenocarcinoma Transcript-1 (MALAT-1 aka NEAT-2) is sequestered in nuclear speckles and is believed to alter the transcription program of cells through alternate splicing of target genes [103]. Identifying other mechanisms of ncRNA function may outline unforeseen strategies for cancer therapy.
Future studies
To unravel the molecules involved in epigenetic regulation by lncRNAs, it is important to first identify genes regulated by lncRNAs. Global investigation of chromatin-RNA interactions at different developmental stages is essential. Combined sequencing of RNA and DNA molecules in close proximity on a genomic scale will aid the discovery of chromatin-RNA associations at different development stages. Although chromatin associated RNAs (CARs) were recently sequenced on a genome wide scale, the exact region of their association with chromatin has not been investigated [104]. The identification of the region of chromatin interaction is essential for the discovery of targets since ncRNAs do not always associate with genomic regions of sequence homology. Gene expression analysis using high throughput quantitative techniques such as CAGE [105] conducted in parallel will identify ncRNA chromatin interactions resulting in activation or repression of genes. Thus new candidate ncRNAs likely to create silencing domains or participate in the activation of genes can be identified and individually investigated.
It is also necessary to identify the distal enhancers of genes regulated by ncRNAs. Although some recent studies have identified enhancers in neurons [78] and cardiomyocytes [83], since enhancers are tissue and development stage specific, there is a need to perform ChIP sequencing using antibodies against specific markers such as P300, H3K4me1, H3K27ac and histone variant H2A.Z in various tissues to identify tissue specific enhancers on a global scale. In addition, technologies such as HiC and ChIA-PET, which can identify genome wide chromatin-chromatin associations, have the capability to identify distal enhancers in physical contact with promoters [106, 107]. The HiC technique is based on proximity ligation and provides unbiased genome-wide maps of chromatin-chromatin association [106]. The CHIA-PET technique, based on immunoprecipitation, proximity ligation and paired end tag sequencing [107] will be especially useful in the identification of enhancers, if performed with antibodies against P300, H3K4me1 and H3K27ac.
It is also important to catalogue lncRNAs and permit their search on public genome browsers. Although some lncRNAs are viewable on public genome browsers and lncRNAdb, thousands of human lncRNAs and expressed retrotransposons identified in FANTOM3 [1, 90], lincRNAs identified in other projects using the k4-36 signature [108] and later with the RIP-seq assay [26] are not clearly annotated on the public browsers.
Thus we remain unaware of the regulatory lncRNAs expressed from the genomic vicinity of our genes of interest. Given the role of lncRNAs in gene regulation, the availability of dedicated tracks of full-length lncRNAs derived from FANTOM3, FANTOM5, ENCODE [1, 109, 110] and other similar projects would be of immense benefit to biologists seeking answers to gene regulation. Since most regulatory lncRNAs are likely to be nuclear, as major transcriptome sequencing projects such as ENCODE and FANTOM5 progress, additional tracks in public browsers based on the distribution of these lncRNAs such as nuclear, cytoplasmic, nucleoplasm or chromatin associated etc will speed up experimental validation of lncRNA related hypotheses.
Conclusions and perspectives
The common themes in the mechanism of silencing mediated by lncRNAs, such as Xist, HOTAIR, Kcnq1ot1 and Airn, have provided a sound template for the investigation of other similar molecules in cells, whose function remains unknown. Although in this review we have focused on the silencing aspect of lncRNA function, evidence is now emerging of lncRNAs participating in gene activation during chromatin looping [111]. Furthermore, novel gene specific mechanisms of silencing are also being uncovered [50].
Thus, it is clear that much remains to be learned in the field of lncRNA function. In addition, another layer of regulation appears to exist at the cellular level, which dictates the transcriptional program, by specifying lncRNA targets. This regulatory layer appears to be tissue and development stage specific and concerted efforts are needed to decipher this next level of control. Just as the lncRNAs have similarities in their modes of action, it is likely that the additional layer of regulatory control over lncRNA mediated silencing may also have common mechanisms. Whether distal enhancers, ncRNAs or other protein complexes exercise this control remains to be investigated. In the near future we may unravel universal techniques to reverse or enforce epigenetic silencing of specific targets mediated by lncRNAs, providing novel therapeutic avenues for some disorders.
Acknowledgments
A.S. is a visiting Scientist at the RIKEN Omics Science Center and is supported by a Japan Society for Promotion of Science (JSPS) long-term fellowship P09745. This work is supported by JSPS grant-in-aid to A.S., a grant of the 7th Framework to P.C. (Dopaminet), a Grant-in-Aids for Scientific Research (A) No.20241047 to P.C., the National Human Genome Research Institute grants U54 HG004557 to P.C., a grant from the Japan Society for the Promotion of Science (JSPS) through the ‘Funding Program for Next Generation World-Leading Researchers (NEXT Program)’, initiated by the Council for Science and Technology Policy (CSTP) and a Research Grant for RIKEN Omics Science Center from MEXT.
Glossary
Abbreviations:
- agRNA
antigene RNAs
- lincRNAs
long intergenic non-coding RNAs
- lncRNAs
long non-coding RNAs
- LINEs
long interspersed nuclear elements
- ncRNAs
non-coding RNAs
- siRNA
short interfering RNA
References
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and non-coding RNA: Challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frith MC, Bailey TL, Kasukawa T, Mignone F, et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3:40–8. doi: 10.4161/rna.3.1.2789. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P, Hayashizaki Y. Non-coding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17:139–44. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, Nix DA, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Bakel H, Hughes TR. Establishing legitimacy and function in the new transcriptome. Brief Funct Genomic Proteomic. 2009;8:424–36. doi: 10.1093/bfgp/elp037. [DOI] [PubMed] [Google Scholar]
- 9.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinger ME, Amaral PP, Mercer TR, Pang KC, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loewer S, Cabili MN, Guttman M, Loh YH, et al. Large intergenic noncoding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, et al. Long non-coding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapranov P, St Laurent G, Raz T, Ozsolak F, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ unannotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by non-coding RNAs. Nat Cell Biol. 2010;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockdorff N, Ashworth A, Kay GF, Cooper P, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–31. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 17.Brockdorff N, Ashworth A, Kay GF, McCabe VM, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 18.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 19.Penny GD, Kay GF, Sheardown SA, Rastan S, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 20.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 21.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, et al. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey RR, Mondal T, Mohammad F, Enroth S, et al. Kcnq1ot1 antisense non-coding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 23.Rinn JL, Kertesz M, Wang JK, Squazzo SL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Zhang H. A unified mode of epigenetic gene silencing: RNA meets polycomb group proteins. RNA Biol. 2005;2:8–10. doi: 10.4161/rna.2.1.1465. [DOI] [PubMed] [Google Scholar]
- 26.Khalil AM, Guttman M, Huarte M, Garber M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagano T, Mitchell JA, Sanz LA, Pauler FM, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 28.Terranova R, Yokobayashi S, Stadler MB, Otte AP, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–79. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Tsai MC, Manor O, Wan Y, Mosammaparast N, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umlauf D, Goto Y, Cao R, Cerqueira F, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of polycomb group complexes. Nat Genet. 2004;36:1296–300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Sun BK, Erwin JA, Song JJ, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko S, Li G, Son J, Xu CF, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Wang L, Erdjument-Bromage H, Vidal M, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 34.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Brown JL, Cao R, Zhang Y, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–46. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Lan F, Matson C, Mulligan P, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–17. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 38.Tachibana M, Sugimoto K, Nozaki M, Ueda J, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin JY, Fitzpatrick GV, Higgins MJ. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 2008;27:168–78. doi: 10.1038/sj.emboj.7601960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–74. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 41.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapranov P, Ozsolak F, Kim SW, Foissac S, et al. New class of genetermini- associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466:642–6. doi: 10.1038/nature09190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 44.Seila AC, Calabrese JM, Levine SS, Yeo GW, et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap KL, Li S, Munoz-Cabello AM, Raguz S, et al. Molecular interplay of the non-coding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Latos PA, Stricker SH, Steenpass L, Pauler FM, et al. An in vitro ES cell imprinting model shows that imprinted expression of the Igf2r gene arises from an allele-specific expression bias. Development. 2009;136:437–48. doi: 10.1242/dev.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted non-coding RNAs: Is transcription the answer? Trends Genet. 2007;23:284–92. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–83. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammad F, Mondal T, Guseva N, Pandey GK, et al. Kcnq1ot1 non-coding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–9. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 51.Weidman JR, Dolinoy DC, Maloney KA, Cheng JF, et al. Imprinting of opossum Igf2r in the absence of differential methylation and air. Epigenetics. 2006;1:49–54. doi: 10.4161/epi.1.1.2592. [DOI] [PubMed] [Google Scholar]
- 52.O'Sullivan FM, Murphy SK, Simel LR, McCann A, et al. Imprinted expression of the canine IGF2R, in the absence of an anti-sense transcript or promoter methylation. Evol Dev. 2007;9:579–89. doi: 10.1111/j.1525-142X.2007.00198.x. [DOI] [PubMed] [Google Scholar]
- 53.Duret L, Chureau C, Samain S, Weissenbach J, et al. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–5. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 54.Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–69. doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- 55.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hot air in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clemson CM, Chow JC, Brown CJ, Lawrence JB. Stabilization and localization of Xist RNA are controlled by separate mechanisms and are not sufficient for X inactivation. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, et al. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- 58.Murakami K, Ohhira T, Oshiro E, Qi D, et al. Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet Genome Res. 2009;125:19–25. doi: 10.1159/000207514. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert SL, Pehrson JR, Sharp PA. XIST RNA associates with specific regions of the inactive X chromatin. J Biol Chem. 2000;275:36491–4. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- 60.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–78. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 61.Redrup L, Branco MR, Perdeaux ER, Krueger C, et al. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–30. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murakami K, Oshimura M, Kugoh H. Suggestive evidence for chromosomal localization of non-coding RNA from imprinted LIT1. J Hum Genet. 2007;52:926–33. doi: 10.1007/s10038-007-0196-4. [DOI] [PubMed] [Google Scholar]
- 63.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–37. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 65.Mohammad F, Pandey RR, Nagano T, Chakalova L, et al. Kcnq1ot1/Lit1 non-coding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–28. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chow JC, Ciaudo C, Fazzari MJ, Mise N, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–69. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 67.Lingenfelter PA, Adler DA, Poslinski D, Thomas S, et al. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet. 1998;18:212–3. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 68.Prothero KE, Stahl JM, Carrel L. Dosage compensation and gene expression on the mammalian X chromosome: One plus one does not always equal two. Chromosome Res. 2009;17:637–48. doi: 10.1007/s10577-009-9063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–22. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One. 2008;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cowley M, de Burca A, McCole RB, Chahal M, et al. Short interspersed element (SINE) depletion and long interspersed element (LINE) abundance are not features universally required for imprinting. PLoS One. 2011;6:e18953. doi: 10.1371/journal.pone.0018953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim KP, Thurston A, Mummery C, Ward-van Oostwaard D, et al. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–42. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muotri AR, Chu VT, Marchetto MC, Deng W, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 74.Ong CT, Corces VG. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–93. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–57. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, et al. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–92. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim TK, Hemberg M, Gray JM, Costa AM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Santa F, Barozzi I, Mietton F, Ghisletti S, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang D, Garcia-Bassets I, Benner C, Li W, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lettice LA, Heaney SJ, Purdie LA, Li L, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–35. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 82.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 83.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108:5632–7. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janowski BA, Huffman KE, Schwartz JC, Ram R, et al. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol. 2005;1:216–22. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 85.Janowski BA, Younger ST, Hardy DB, Ram R, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 86.Li LC, Okino ST, Zhao H, Pookot D, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–10. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taft RJ, Glazov EA, Cloonan N, Simons C, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–8. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 90.Faulkner GJ, Kimura Y, Daub CO, Wani S, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 91.Amir RE, Van den Veyver IB, Wan M, Tran CQ, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 92.Huppke P, Maier EM, Warnke A, Brendel C, et al. Very mild cases of Rett syndrome with skewed X inactivation. J Med Genet. 2006;43:814–6. doi: 10.1136/jmg.2006.042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weaving LS, Williamson SL, Bennetts B, Davis M, et al. Effects of MECP2 mutation type, location and X-inactivation in modulating Rett syndrome phenotype. Am J Med Genet A. 2003;118A:103–14. doi: 10.1002/ajmg.a.10053. [DOI] [PubMed] [Google Scholar]
- 94.Zhu X, Li M, Pan H, Bao X, et al. Analysis of the parental origin of de novo MECP2 mutations and X chromosome inactivation in 24 sporadic patients with Rett syndrome in China. J Child Neurol. 2010;25:842–8. doi: 10.1177/0883073809350722. [DOI] [PubMed] [Google Scholar]
- 95.Bao X, Jiang S, Song F, Pan H, et al. X chromosome inactivation in Rett syndrome and its correlations with MeCP2 mutations and phenotype. J Child Neurol. 2008;23:22–5. doi: 10.1177/0883073807307077. [DOI] [PubMed] [Google Scholar]
- 96.Nan X, Bird A. The biological functions of the methyl-CpG-binding protein MeCP2 and its implication in Rett syndrome. Brain Dev. 2001;23(Suppl 1):S32–7. doi: 10.1016/s0387-7604(01)00333-3. [DOI] [PubMed] [Google Scholar]
- 97.Guy J, Hendrich B, Holmes M, Martin JE, et al. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–6. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 98.Guy J, Gan J, Selfridge J, Cobb S, et al. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–7. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retro-transposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Skene PJ, Illingworth RS, Webb S, Kerr AR, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–68. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muotri AR, Marchetto MC, Coufal NG, Oefner R, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gupta RA, Shah N, Wang KC, Kim J, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tripathi V, Ellis JD, Shen Z, Song DY, et al. The nuclear-retained non-coding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mondal T, Rasmussen M, Pandey GK, Isaksson A, et al. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Valen E, Pascarella G, Chalk A, Maeda N, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–65. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li G, Fullwood MJ, Xu H, Mulawadi FH, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guttman M, Amit I, Garber M, French C, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kanamori-Katayama M, Itoh M, Kawaji H, Lassmann T, et al. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 2011;21:1150–9. doi: 10.1101/gr.115469.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang KC, Yang YW, Liu B, et al. A long non-coding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]


