Abstract
molA(HI1472) from H. influenzae encodes a periplasmic binding protein (PBP) that delivers substrate to the ABC transporter MolB2C2 (formerly HI1470/71). The structures of MolA with molybdate and tungstate in the binding pocket were solved to 1.6 and 1.7-Å resolution, respectively. The MolA binding protein binds molybdate and tungstate but not other oxyanions such as sulfate and phosphate, making it the first class III molybdate binding protein structurally solved. The ~100 μM binding affinity for tungstate and molybdate is significantly lower than observed for the class II ModA molybdate binding proteins that have nanomolar to low micromolar affinity for molybdate. The presence of two molybdate loci in H. influenzae suggests multiple transport systems for one substrate, with molABC constituting a low-affinity molybdate locus. (Word count 123/150)
INTRODUCTION
Molybdenum, found in trace amounts in the environment, is a necessary cofactor for redox enzymes that catalyze reactions at carbon, sulfur and nitrogen atoms, including nitrogenase and nitrate reductase, key enzymes in the nitrogen cycle (Imperial et al., 1985). Cellular uptake systems must be able to distinguish molybdate from other tetrahedral anions including sulfate, phosphate and tungstate; this ability has been attributed to differences in size, coordination number or protonation (Hu et al., 1997; Lawson et al., 1998; Ledvina et al., 1998; Luecke and Quiocho, 1990; Pflugrath and Quiocho, 1985; Rech et al., 1996). Bacterial cells have developed highly efficient, distinct transport systems to regulate intracellular concentrations of these ligands (Higgins, 1992). For example, molybdate and sulfate are transported by separate systems in bacteria that have both systems present; however, in the absence of a fully functioning molybdate transport system, the sulfate system can compensate, albeit with less efficiency (Fitzpatrick et al., 2008; Rosentel et al., 1995). One of the most studied systems that control the transport of specific substrates across the membrane is the ABC transporter (ATP-Binding Cassette) superfamily (Higgins, 1992). For prokaryotic importers, the system consists of three components: a periplasmic or substrate binding protein (PBP), two transmembrane domains (TMDs) and two cytoplasmic nucleotide-binding domains (NBDs) (Higgins, 1992). The ATP binds across the interface of the NBDs and this binding and hydrolysis is thought to cause the rearrangement of the TMDs (Davidson and Maloney, 2007; Hollenstein et al., 2007a; Procko et al., 2009). Through conformational changes powered by ATP binding and hydrolysis, the substrate is shuttled from the periplasmic side to the inside of the cell. While in gram-negative bacteria the periplasmic protein scavenges specific ligands from the periplasmic space, in gram-positive bacteria, substrate binds to a binding protein tethered to the surface of the cell membrane (Gilson et al., 1988). In both cases, the PBP attaches onto the periplasmic side of the TMDs to deliver the substrate to the transmembrane domain. ABC transporters are typically classified by what they transport, identified through the binding specificity of the PBP (Quiocho and Ledvina, 1996). Despite the wide variety of ligands bound and lack of sequence conservation, PBPs have a common architecture and may be assigned into six distinct structural classes (Berntsson et al., 2010; Quiocho and Ledvina, 1996). The core of all PBPs consists of two structurally conserved globular domains with a binding pocket located in the cleft formed between the two domains. The two most common clusters that bind to ABC transport proteins are class III (cluster A) and class II (cluster D) PBPs (Berntsson et al., 2010). Class II PBP’s typically undergo multiple conformational changes in the presence or absence of substrate. Substrate bound at the inter-domain interface is thought to stabilize the PBP in what is classified as a “closed conformation” (Berntsson et al., 2010). While the substrate-free PBP binding pocket is accessible to solvent, upon substrate binding, residues in both domains bind to the ligand through a series of hydrogen bonds. This results in conformational changes from the apo to substrate bound structure that have been characterized as a “Venus flytrap” model (Mao et al., 1982) In contrast, class III PBPs exhibit little overall conformational change upon the binding of substrate (Berntsson et al., 2010), perhaps as a consequence of an additional “hinge” helix, not present in class II proteins, that connects the two globular domains.
The most extensively studied molybdate transport system is in E. coli, where molybdate acquisition is controlled through the molybdate operon, modABCD (Maupin-Furlow et al., 1995; Shanmugam et al., 1992). In E. coli, molybdate enters the cell via the high-affinity transport system, ModABC. modA is the first gene in the molybdate operon coding for the molybdate periplasmic binding protein, followed by modB and modC, representing the TMDs and NBDs of the transporter (Rech et al., 1995; Shanmugam et al., 1992; Wang et al., 1993). The mod loci in E. coli, R. capsulatus and A. vinelandii all contain a modD gene, with no known function (Rech et al., 1995). The mod operon in these organisms also encodes a molybdate dependent transcriptional repressor, modE (Grunden et al., 1996; Pau RN, 2002; van Veen et al., 1994b). As molybdate enters the cytoplasm, it binds to ModE which in turn represses the transcription of the high-affinity transporter (Pau RN, 2002). Once molybdate enters into the cytoplasm, H. influenzae (mop), Azotobacter vinelandii (modG) and Rhodobacter capsulatus (mopB) contain cytoplasmic molybdate-binding proteins or molbindins, which participate in the storage and homeostasis of molybdate inside the cell (Masters et al., 2005; Wiethaus et al., 2009).
The crystal structures of molybdate PBP ModA from E. coli (Hu et al., 1997), A. vinelandii (Lawson et al., 1998) Xanthomonas citri (Balan et al., 2006) and the tungstate PBP WtpA and TupA from Archaeoglobus fulgidus (Hollenstein et al., 2007b), Pyrococcus horikoshii, Methanosarcina acetivorans, Pyrococcus furiosus, Methanococcus jannaschii (Hollenstein et al., 2009) and Geobacter sulfurreducens (PDB ID:3LR1) are currently available (Hagen, 2011). The tertiary structures of these proteins are similar although the sequence similarity among these binding proteins is low (less than 30% sequence similarity between ModA/WtpA in different organisms). These proteins are members of the well-defined class II (cluster D) with proteins that bind tetrahedral oxyanions, including molybdate-, sulfate-, and phosphate-binding proteins (Bern tsson et al., 2010). These proteins are structurally distinct from the class III (cluster A) PBPs that have been classified as metal ions or metal coordinated siderophore binding proteins and have not been reported to serve as binding proteins for any oxyanions.
We have determined crystal structures of HI1472, a PBP from H. influenzae, bound to tungstate and molybdate. HI1472 is the cognate PBP of ABC transporter encoded by the gene products of HI1471 and HI1470, respectively, for which the structure has been reported (Lewinson et al., 2010; Pinkett et al., 2007). Although the ligand specificity of this transport system was not known when the structure was solved, in view of the similarities between this system and the vitamin B12 importer BtuCD in both structure and the ability to form a stable complex in the absence of substrate with the binding protein (Lewinson et al., 2010; Pinkett et al., 2007), it was suggested that HI1470/71 transported some type of iron chelate species. Based on the ability of HI1472 to bind molybdate and tungstate, however, it now appears likely that this system transports molybdate and/or tungstate. Consequently, we have designated each component of this H. influenzae molybdate transport system as MolA (PBP, HI1472), MolB (TMD, HI1471) and MolC (NBD, HI1470), herein. The structure of MolA, the first example of a class III PBP bound to molybdate and tungstate, displays a binding cavity and coordination that is distinct from ModA-type molybdate binding proteins, and sheds new light on the classification of molybdate transport systems (Hagen, 2011) .
RESULTS
Crystallization and structural determination of MolA bound to tungstate and molybdate
The H. influenzae molA gene codes for a 351 amino acid precursor protein, from which a 21 amino acid signal peptide is cleaved to produce a 330 amino acid mature protein. Crystallization of the mature protein expressed in LB media was achieved in 24% polyethylene glycol monomethyl ether 2000, 0.1 M sodium acetate pH 4.5, 0.2 M ammonium sulfate and 12% glycerol. A native data set to 1.6-Å resolution was collected at SSRL from a crystal belonging to space group P3121. An isomorphous 2.5 Å resolution data set collected at the home source with crystals soaked in potassium iodide was used to determine the structure of MolA (Figure 1) using the single anomalous dispersion (SAD) technique described in the experimental section. During the structure determination, non-protein electron density was observed in the binding pocket for an electron dense ligand with tetrahedral geometry (Figure 2A). The model was initially modeled as molybdate (MolA-MoO42−) and was subsequently refined as molybdate once it was confirmed through biochemical assays. As the protein was not supplemented with an oxyanion, the binding protein presumably scavenged traces of ligand from the growth medium. Since most molybdate binding proteins also have a similar affinity for tungstate (Rech et al., 1996), MolA was subsequently expressed in M9 media supplemented with sodium tungstate. MolA expressed in the presence of tungstate was purified and crystallized in conditions identical to the MolA-MoO4 2− crystallization conditions. Native data sets collected to 1.7 Å and 2.0 Å resolutions at energies of 12.667 keV and 10.209 keV (near the tungsten edge), respectively, were isomorphous to the molybdate data set collected previously. A difference Fourier map calculated from the tungstate dataset revealed a prominent peak (Figure 2E) in the binding pocket, corresponding to the location of the molybdate ion. The structure of tungstate bound MolA was determined by molecular replacement using the original structure, omitting the molybdate from the model. The Fourier difference map clearly showed tetrahedron shaped electron density at 60 sigma (figure 2E), indicating that a tungstate is bound within the active site. The density above the molybdate/tungstate in MolA shows two peaks, which are modeled as water molecules (figure 2A and 2C). MolA-tungstate (MolA-WO42−) and MolA-molybate (MolA-MoO42−) retain identical structural features with a root mean square (rms) deviation of 0.124 Å, despite the smaller unit cell and lower R-factors.
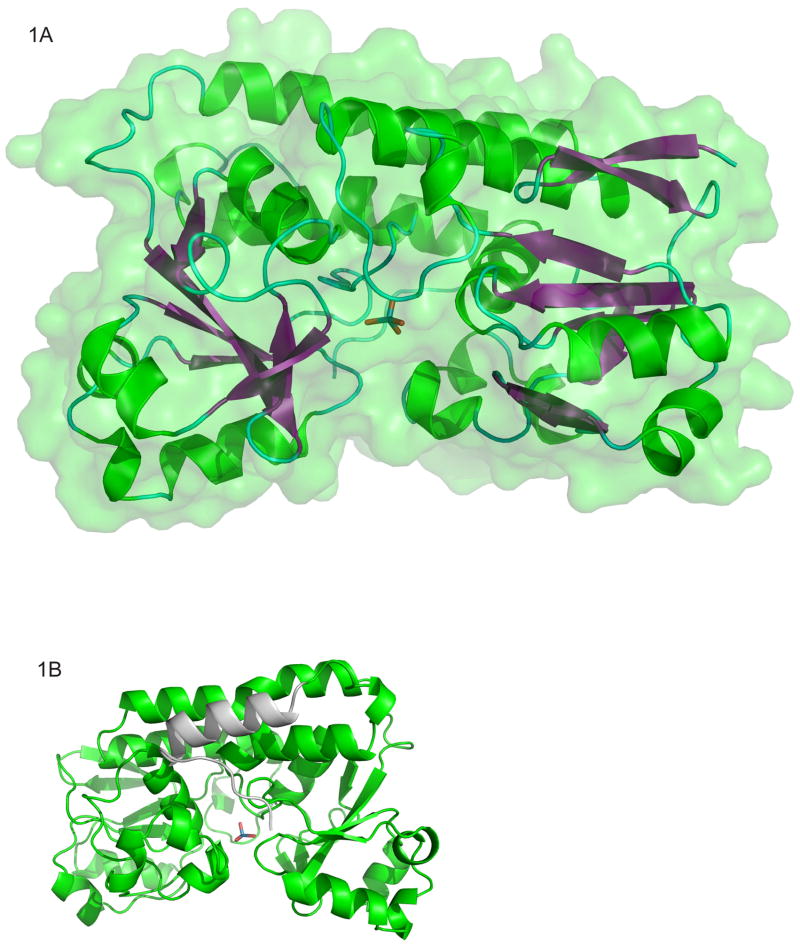
Figure 1.
The overall fold of molA (HI1472), a molybdate/tungstate binding protein. (A) The cartoon and space filling diagram of the molybdate bound conformation of molA (PDB code 3PSH). α-helices are shown in green, β-strands in purple and loop areas in cyan. The molybdate anion coordinated between the two domains is shown in a stick representation with the oxygen atoms in red and the molybdenum in cyan. (B) Ribbon diagram of molA. The structure resembles other class III substrate binding proteins with the exception of the extra helix shown in grey (residues 362–347) at the C-terminus. The graphics were made with PyMOL (W. L. DeLano, DeLano Scientific, San Carlos, CA).
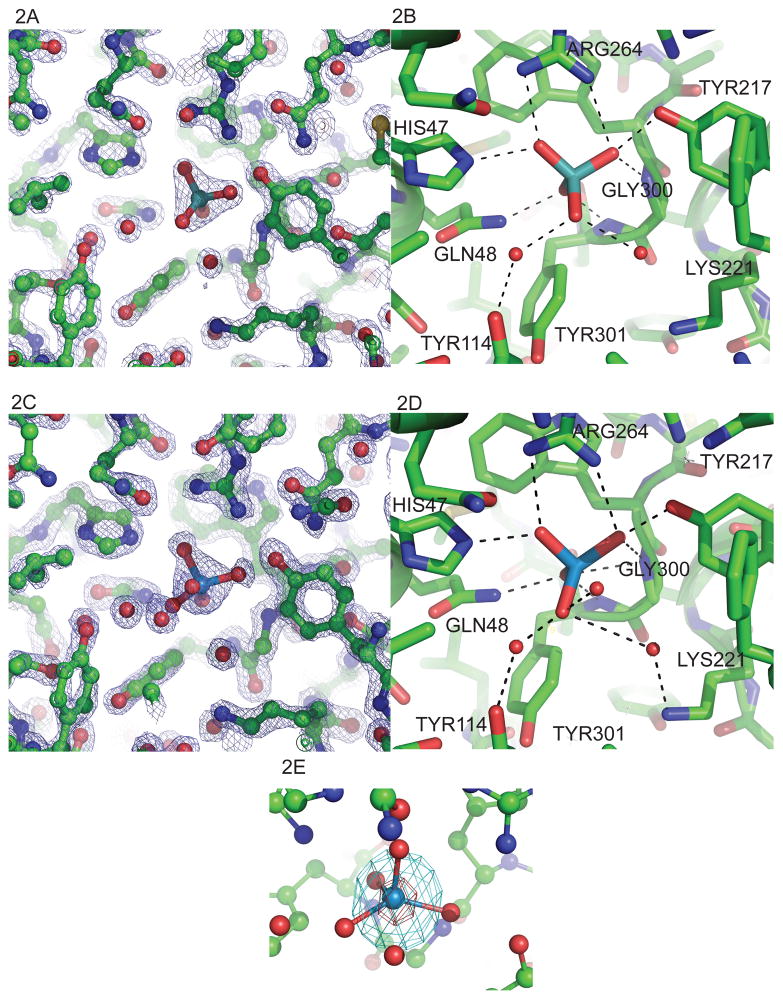
Figure 2.
The ligand binding site of HI1472 with substrate bound. (A) The binding pocket of molA with tungstate (PDB ID 3PSA) and (C) molybdate bound (PDB ID 3PSH). 2Fobs-Fcalc electron density is shown for the ligand binding site. (B) Hydrogen bonds between tungstate and (D) molybdate anion and molA. Residues shown in stick representation, bonds shown by dashed lines and water molecules are shown depicted as red spheres. 2Fobs-Fcalc electron density (contoured at 1.5 σ level) is shown for the tungstate and molybdate ligand binding site in figure 2A and 2C, respectively (also see Figure S5). (E) Anomolous difference map of the tungstate bound structure contoured at 5σ (cyan) and 60σ (red). The graphics were made with PyMOL (W. L. DeLano, DeLano Scientific, San Carlos, CA) .
MolA topology and binding coordination
The structure of MolA falls into the cluster A, class III periplasmic binding proteins with two topologically similar globular domains (domain I and II correspond to residues 1–175 and 197–322, respectively) with a signature hinge α-helix as an interdomain linker (residues 176–196) (Berntsson et al., 2010; Quiocho, 1990) (figure 1A). Domain I consists of 8 β-strands surrounded by 6 α-helices while domain II consists of 6 β-strands surrounded by 6 α-helices. Structural alignment with PDB3FOLD (Krissinel and Henrick, 2004) indicates that the MolA structure has a similar fold to the vitamin B12 PBP, BtuF (PDB 1N2Z-ligand bound) (Borths et al., 2002; Karpowich et al., 2003) and TroA (PDB 1TOA-ligand bound) (Lawrence et al., 1998), despite sequence identities of only 18% and 9% respectively. The C-terminal helix, α-12 (figure S4), at the tail end of MolA (residues 322–347 (figure 1B) is not present in the homologous BtuF and TroA. In addition to contacts with α-12, the hinge helix packs tightly against α-6 from domain I and α-11 from domain II, forming a hydrophobic core, which may result in a less flexible hinge region.
The electron density reveals a ligand site deep in the cleft between the loops of the two domains. Details of the interaction of both molybdate and tungstate with the protein are depicted in figure 2. The ligand is bound to MolA through favorable contacts with both domains of the protein, forming hydrogen bonds with His47 and Gln48 from domain I, and Tyr217, Arg264, Gly300 and Tyr301 from domain II (figure 2B/2D). The side chain groups of His47 and Gln48 form hydrogen bonds with the anion from domain I, while the side chain hydroxyl of Tyr217, the side chain guanidinium from Arg264 and the main chain amide of Gly300 from domain II all donate hydrogen bonds to the same oxygen of the atom (figure 2B, figure S.5). In addition, a second oxygen accepts a hydrogen bond from the backbone amide of Tyr301. Unlike the ModA molybdate bound PBPs, where all hydrogen bonds to the molybdate oxygens involve protein residues (Hagen, 2011),in MolA, one of the ligand oxygens is only hydrogen bonded to water. This water molecule in turn is hydrogen bonded to Lys221 in the MolA-WO42− (tungstate bound) structure (figure 2D). This differs from MolA-MoO42− (molybdate bound) structure where Tyr114 forms a hydrogen bond with a water molecule, forming a hydrogen-bonding network (figure 2B). The molybdate coordination seen in the ModA structures from E. coli (Hu et al., 1997) and A. vinelandii (Lawson et al., 1998) consists of seven hydrogen bonds between the protein and the molybdate, and the residues within the binding site are conserved in these two structures. A lack of sequence conservation is evident between the E. coli and X. citri ModAs, although all structures of MolA and ModA reveal a conserved tyrosine as part of the active site (Hagen, 2011).
Molybdenum content and binding affinity of MolA
To test the affinity for other anions, a ligand-dependent protein gel shift assay was performed with 4000-fold molar excess of sulfate, phosphate, molybdate and tungstate. The gel shift demonstrated that the protein binds to both molybdate and tungstate, but not sulfate or phosphate (figure S1). Native 12% acrylamide gels showed that MolA migrated in an electric field with an apparent overall negative charge. Addition of the oxyanions molybdate and tungstate resulted in increased MolA mobility, which indicated the protein had an increased negative charge and/or a smaller volume, while no mobility shift was evident in the presence of sulfate or phosphate.
In order to determine the occupancy of molybdate bound to MolA, the protein was expressed in Luria-Bertani (LB) media supplemented with sodium molybdate or tungstate and subsequently purified with affinity chromatography to remove unbound ligand. The protein concentration was measured with a bicinchoninic acid (BCA) assay (Pierce, Rockford, IL) and by measuring absorbance at 280 nm (A280). The metal content was determined by Inductively coupled plasma mass spectroscopy (ICP-MS) analysis (figure 3A and S2). At 0.117 μM MolA, there was 7.1 parts per billion (ppb) molybdenum, showing MolA had 0.071 μM of molybdate bound per MolA (figure 3). This suggests that after removal of the free ligand during purification, at least 61% of MolA binding sites remained occupied with molybdate. This experiment was repeated in the presence of protein expressed in 10 mM tungstate where at 0.117 μM MolA, there was 11.9 ppb tungstate, showing MolA had 0.064 μM of tungstate bound per MolA. This data shows that MolA is able to bind to both tungstate and molybdate with a slight preference for molybdate. As a control, the protein was also expressed in the presence of sulfate, where the occupancy of molybdate was 24%. MolA was also expressed in the presence of sulfate and molybdate to measure molybdate binding at high concentrations of an alternate substrate (S2). However, the levels of molybdate stayed constant, indicating that sulfate does not significantly compete for binding of MolA.
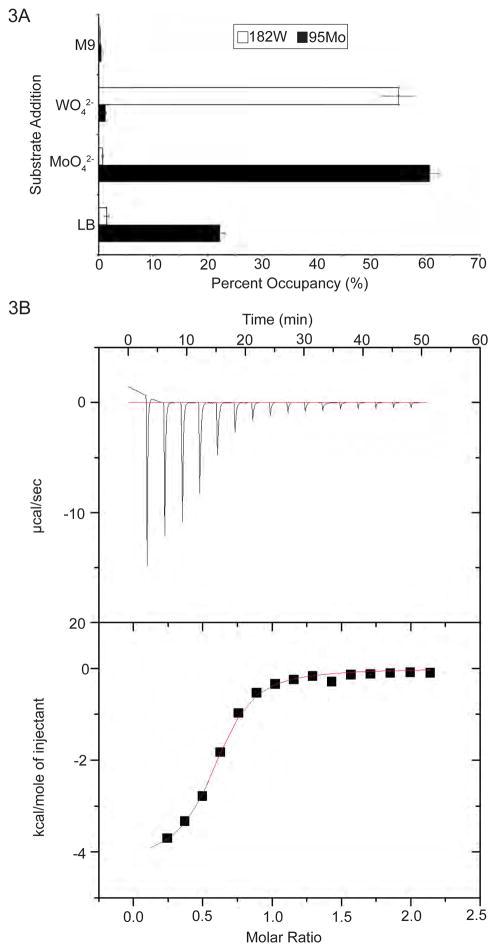
Figure 3.
Metal content analysis and molybdate binding affinity of MolA. (A) ITC of sodium molybdate binding into HI1472. ITC curves from the titration of sodium molybdate (19.9 mM) into MolA (1.98 mM). The Kd was estimated to be 67 uM. Raw data (top) and integration (bottom) (also see Figures S1 and S2). (B) ICP-MS quantitation of the molybdenum and tungsten content of HI1472. Protein was expressed in LB, minimal media (M9), and LB supplemented molybdate or tungstate, which significantly increases occupancy, while protein isolated from cells grown in minimal media exhibits reduced occupancies of these species, indicating that the LB contains Mo and W contaminants (also see Figure S3).
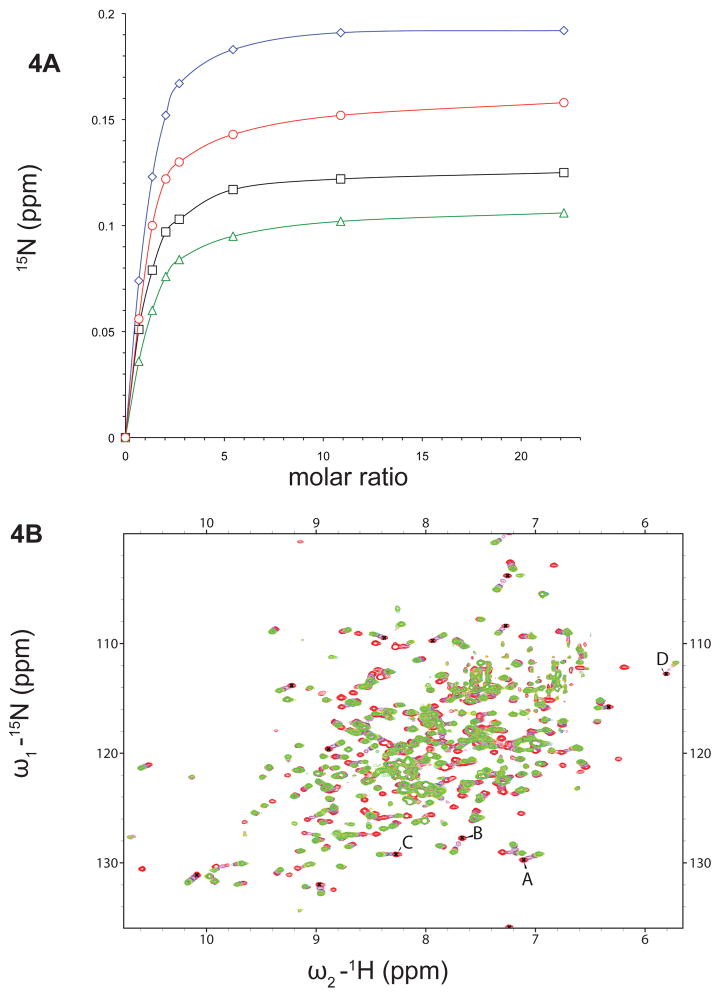
Since we were able to establish MolA binds molybdate and tungstate present in growth media, we next determined binding constants using Isothermal Titration Calorimetry (ITC) and Nuclear Magnetic Resonance (NMR) titrations. Using ITC, MolA was observed to bind to tungstate and molybdate with a stoichiometry of 0.667 ± 0.012 and 0.565 ± 0.010 mole oxyanion per mole of protein, respectively, as deduced from the heat consumption upon addition of tungstate or molybdate to the protein solution (figure 3B). From the binding curve for molybdate, the Kd value was determined to be 67 ± 9 μM. For the tungstate binding curve, it was not possible to significantly increase the protein concentration to obtain more data points, however the apparent Kd for tungstate when the protein is saturated was determined to be 84 ± 4 μM (Figure S.3.). Binding in the 100 micromolar range was confirmed with NMR titrations of 15N-labeled MolA with unlabeled molybdate (figure 4A). Titrations of molybdate with 15N-labeled MolA shifted a subset of resonances in the NMR spectrum to new positions (figure 4B), indicative of a specific complex with fast dissociation kinetics. Non-linear least-squares fitting of 13 independent binding isotherms yielded an average apparent KD of 134 ± 14 μM. Collectively, the ITC and NMR results confirm a direct interaction between MolA and molybdate with an affinity on the 100 micromolar scale.
Figure 4.
NMR Binding studies of MolA. A. Titration curves of HI1472 (280.8 mM) with Na2MoO2− 4 at variable ratios (from 1:0.7 to 1:22, HI1472:Na2 MoO2−4). B. Overlay of Transverse Relaxation Optimized Spectroscopy (TROSY) spectra of 15N labeled HI1472 before (red) and after (green) addition of 6.12 mM Na2MoO2−4 (ratio of 1:22 protein:substrate). Peaks chosen for the titration curve are labeled as A, B, C and D respectively.
DISCUSSION
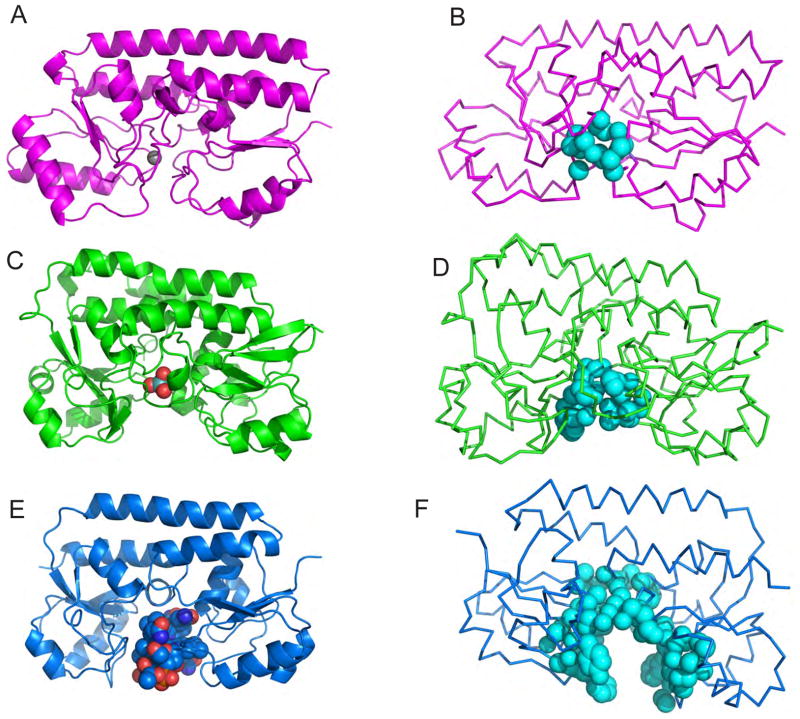
Despite classification of MolA as a putative iron-chelate binding protein, it is clear from the biochemical and structural studies presented here that MolA is a molybdate binding periplasmic protein. As mentioned, while class III PBPs exhibit a similar fold, they share little sequence similarity and bind to a wide variety of substrates. What is most intriguing is the size of the substrate does not seem to be associated with a drastic conformational change to the overall structure. As a representative set of class III proteins, we compared the MolA PBP to structures of class III PBP bound to either a large substrate (BtuF, 1N2Z) or a single ion (TroA, 1TOA). Vitamin B12 bound BtuF (PDB-1N2Z) (figure 5A) contains the two globular domains connected with the hinge helix as does TroA (pdb-1TOA), a zinc/manganese PBP (figure 5E) with a single zinc molecule bound in the active site. When compared using PDB3FOLD, the rms deviation between MolA and BtuF is 3.2 Å and between MolA and TroA is 3.5 Å. While the overall folds are similar, the largest differences are apparent in the size of the binding pocket. Utilizing the program CASTp to measure the binding pocket (Dundas et al., 2006), the binding pocket for BtuF is 2275 Å3 (figure 5F) compared to MolA with a binding pocket of 330 Å3 (figure 5D) and 105 Å3 for TroA bound to zinc (figure 5F) indicating a correlation between the sizes of the substrate and binding pocket. Reflecting the 10-fold difference in binding pocket volumes, but similar rms deviations of the overall fold, the principal variations are in the loops present in the binding pocket to accommodate large and small substrates without major conformational changes to the overall structure. When aligned with MolA, TroA provides a narrow cavity to which zinc is bound (figure 6A), while the loops present in the BtuF structure provide a larger opening to accommodate the binding of vitamin B12. With a slightly larger cavity than TroA and smaller than BtuF, the molybdate bound MolA is in an intermediate conformation (figure 6C). While many of these loops contain residues that make contact with the ligand, others play more of a structural role, making the cavity larger or smaller. Since there is little conformational change in class III PBPs upon ligand binding, we suspect that the selectivity is established by both specific residues and cavity accessibility.
Figure 5.
Measuring the size of binding pockets of class III PBPs. The cartoon diagrams of (A) Zn(II) bound TroA (magenta), (C) Molybdate-bound MolA (green) and (E) Vitamin B12 bound BtuF. Ribbon diagrams of (B) Zn(II) bound TroA (magenta), (D) Molybdate-bound MolA (green) and (F) Vitamin B12 bound BtuF, with cyan spheres to represent the binding pocket calculated by the program CASTp. Although TroA, MolA, and BtuF have the same overall fold, the variable size of binding pocket can accommodate the appropriate substrate.
Figure 6.
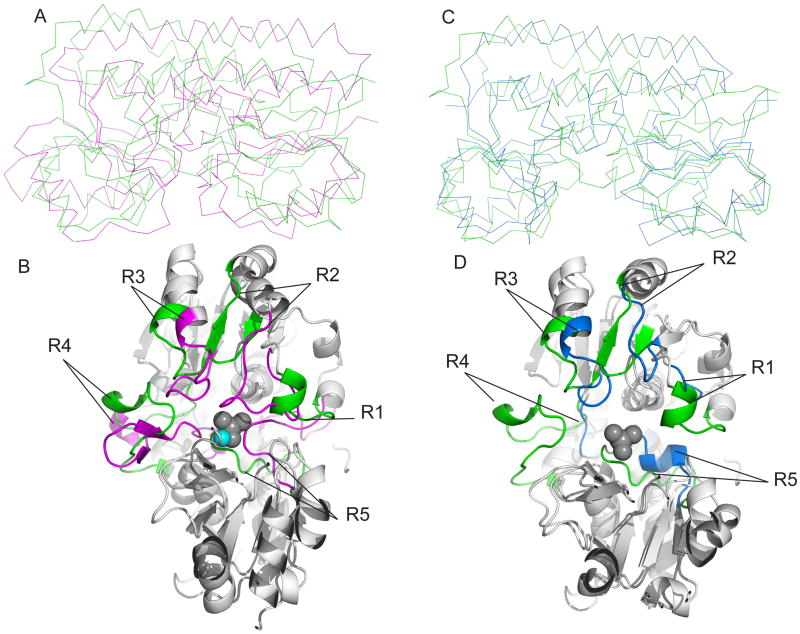
Structural alignment of class III PBPs. The structures of MolA, TroA and BtuF were superimposed by a secondary structure matching method, using PDB3Fold. (A) Superimposed ribbon diagrams of molybdate bound-MolA (green) and Zn (II)-bound TroA. (B) The view from the binding pocket of superimposed cartoon diagram of MolA and TroA. Loops that play a role in the conformation of the binding pocket for molybdate bound-MolA and Zn (II)-bound TroA are colored green and magenta, respectively. The loops that make up the binding site of Zn (II)-bound TroA are labeled R2 (residues 60–77), R3 (residues 88–98), R4 (residues119–137) and R5 (residues 273–291). The loops that make up the binding site of molybdate bound-MolA are labeled R1 (residues 69–77), R2 (residues 91–98), R3 (residues 112–119), R4 (resides 134–153) and R5 (292–304) (also see Figure S4).
(C) Superimposed ribbon diagrams of molybdate bound-MolA (green) and Vitamin B12 bound-BtuF (blue) (D) The view from the binding pocket of superimposed cartoon diagram of molybdate bound-MolA and Vitamin B12 bound-BtuF. Loops that play a role in the conformation of the binding pocket for molybdate bound-MolA and Vitamin B12 bound-BtuF are colored green and blue, respectively. The loops that make up the binding site of Vitamin B12 bound-BtuF are labeled R1 (residues 49–52), R2 (residues 64–71), R3 (residues 85–98), R4 (residues 107–110) and R5 (residues 239–249). The grey spheres represent molybdate bound in the active site of MolA, the cyan sphere represents zinc bound to TroA (vitamin B12 was omitted for clarity of the active site of BtuF). The graphics were made with PyMOL (W. L. DeLano, DeLano Scientific, San Carlos, CA) (also see Figure S4).
In case of TroA, BtuF and MolA, the majority of the variable length loops reside on domain I of the PBP. In the structural schematic in figure S4, the 5 “loop regions” that play a role in the size of the binding pocket are designated R1 to R5. R1, R2, R3 and R4 of MolA, BtuF and TroA reside on domain I before the hinge helix, while R5 is present on domain II. These regions are colored from the point of divergence in the aligned structures (figure 6B/6D). For clarity, the structures of TroA and MolA (figure 6B) are aligned with substrates zinc and molybdate, respectively, while MolA and BtuF (figure 6D) are aligned with molybdate in the absence of vitamin B12. The binding of substrates to the TroA and MolA structures seem to have a “sidedness”, where the substrate resides closer to domain II than in the center of the binding cavity, so that R1 through R4 reach across the binding pocket, to make contact with the substrate or play a role in reducing the size of the pocket. For TroA, the four loops that line the inner cavity, R1/R2, R4 and R5, make contacts with the zinc ion, while R3 is a 8 residue loop that extends inward, forming a narrow channel between domain I and II. For clarity, the longest loop in the binding pocket of TroA is classified as R1/R2 because it structurally aligns with the shorter R1 and R2 loops of MolA. For BtuF, loops R1 through R5 all make direct or water mediated contacts with vitamin B12, repositioning themselves to form a much larger binding pocket. This differs for MolA, where only R5 makes contact with the substrate, while R1–R4 extend into the cavity forming a smaller pocket. For TroA, the R1/R2 loop (residues 60–74) stretch from domain I across the binding pocket making contact with the zinc ion, and effectively reducing the size of the pocket. The R2 loop from MolA (residues 91–98) and BtuF (residues 64–71) is shorter in length and thus does not obstruct the binding pocket. This is also apparent in loop R4 of TroA (residues 119–137) that extends from the outside of domain I to the inside of the pocket, reducing the potential size. However, R4 loop in BtuF (residues 107–110) is a short 4-residue loop allowing for a larger binding pocket. R5 for MolA, TroA and BtuF, located on domain II, is part of the region closest to the hinge helix or “top” of the cavity. In all three structures, R5 extends from the hinge region to restrict the size of the pocket from above. While the aspartic acid on R5 in TroA makes contact with the zinc molecule, when aligned with MolA, the positioning of this loop would clash sterically with molybdate (figure 6B) and the larger vitamin B12 molecule. This is also apparent in MolA, where the positioning of the R1 loop would cause a steric clash with the vitamin B12 molecule, which sheds light on the roles of each of the loop regions in shaping the binding pocket.
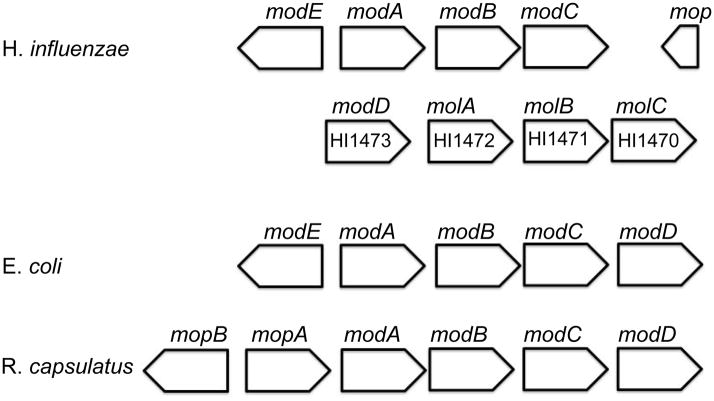
We have established through ITC, NMR and structural studies that MolA binds to molybdate and tungstate with ~100 μM affinity. Further support for the role of MolABC in molybdate transport comes from analysis of the organization of the molybdate transport genes in H. influenzae; while modABC genes are part of the same operon (as in E. coli), unlike E. coli, modD is located in the operon of molABC (figure 7). Comparative studies of the amino acid sequence of modD reveals that H. influenzae has 47% sequence identity to ModD ortholog in E. coli (blastp search)(Karlin and Altschul, 1990). Since modD is commonly found in the molybdate locus of mod genes, this would suggest that the molABC operon encodes for a second molybdate transport system.
Figure 7.
Organization of genes in the molybdate transport loci of H. influenzae, E. coli, and R. capsulatus. modA, modB and modC encode for the periplasmic binding protein, transmembrane domain and nucleotide-binding domain respectively. modE (mopB and mopA in R. capsulatus) code for transcriptional regulators while the gene classified as a molybdenum transport protein modD has no known function, but is typically found in operons with modABC.
Biological evidence for multiple molybdate uptake systems in bacteria has been accumulating. In E. coli mod mutants, molybdate is transported by a sulfate transport system (cysTWA) or by a non-specific anion transport system (Grunden and Shanmugam, 1997; Rosentel et al., 1995). In a sulfate transport-defective mutant of E. coli, the molybdate transporter is capable of transporting sulfate to partially suppress the Cys- phenotype, although this requires derepression of the modABC genes, which are expressed at low levels in a wildtype strain (Self et al., 2001). This would indicate that E. coli has multiple independent transport systems (ModABC, cysTWA and an unidentified anion transport system) capable of transporting molybdate (Lee et al., 1990; Rosentel et al., 1995). CysA has a Kd for molybdate in the milimolar range but can transport molybdate from media containing molybdate concentrations in the micromolar range. In the case of a double mod cysA mutant, formate dehydrogenase (FHL) activity is optimum at 100 μM molybdate (Self et al., 2001). When the double mutant strain is supplemented with selenate and sulfate, selenate but not sulfate also affect FHL activity, indicating an alternate system with a preference for selenate over molybdate (Lee et al., 1990).
Evidence for high and low affinity molybdate transport systems has also been noted for K. pneumoniae (Imperial et al., 1985). The K. pneumoniae high affinity system encoded by ModABC is involved in transport from 10 nM to a 1 μM saturation point. However, strains with modABC mutations show nitrate reductase activity when grown in more than 10 μM molybdate, suggesting two concentration dependent uptake systems in K. pneumoniae. This is also observed in A. vinelandii where mutations in the molybdenum transport operon show reductase activity at high concentrations (10 μM) of molybdate in comparison to wildtype, indicating both low and high-affinity molybdate uptake systems (Mouncey et al., 1995; Premakumar et al., 1996). Similar observations have been made in Bradyrhizobium japonicum (Maier et al., 1987), although the low affinity routes for molybdate uptake have yet to be fully characterized. It should be further noted that the presence of multiple transports systems is not restricted to molybdate and sulfate ions, but also to phosphate, as two transport systems for this oxyanion have been established in Acinetobacter johnsonii 210A (van Veen et al., 1994a). The structures presented here establish that MoIB2C2A (formerly HI1470/71/72) constitute a new class of molybdate/tungstate transporters. The presence of a second molybdate transport loci in H. influenzae, indicates that there are multiple ways in which molybdate is regulated in the H. influenzae system. With affinity in 100 μM range for molybdate and tungstate in an organism with the modABC molybdate transport system classified as a high-affinity transport system, we believe we have the structure of a PBP that is part of the low affinity transport system, molABC. The low affinity would suggest that this system would not be the primary route of transport for molybdate, but an alternate route. Since H. influenzae is highly adapted to its human host where quantities of molybdate are reported in the ~100 – 1000 nanomolar range in whole blood (Allaway et al., 1968), we speculate that in a system with two modes of transport for a specific substrate, each binding protein will take on a different role to maintain levels of substrate. With the structure of the periplasmic binding protein for the MolB2C2 ABC transporter now in hand, genetic experiments will be necessary to further explain the need for a high and low affinity molybdate transport system in H. influenzae.
EXPERIMENTAL PROCEDURE
Cloning, expression and purification of MolA
MolA was amplified from Haemophilus influenzae KW20 genomic DNA and subcloned into pET19b vector (EMD Biosciences, Madison, WI), with N-terminal 8xHistidine tag. The protein was expressed in E. coli BL21 (DE3) cells (EMD Biosciences, Madison, WI) at 37° C in LB medium with 100 μg/ml ampicillin. The cells were induced with 1mM IPTG for 3 hrs, harvested and stored at −80 °C. For purification of MolA, 10 gm cells were resuspended in 10ml of osmotic shock solution (40% sucrose, 1mM EDTA, pH 8.0, 10mM Tris-HCL, pH 7.5) and protease inhibitor, PMSF. After stirring and 1-hour incubation at 4°C, the solution was introduced to 1L of cold water for shock. MgCl2 and start buffer components were added for a final concentration of 2mM MgCl2, 17 mM imidazole, 250 mM NaCL, 25 mM Tris HCl pH 7.5 before centrifugion at 8,000 rpm. The supernatant was loaded onto a 3 mL equilibrated Ni-NTA affinity chromatography column (Qiagen, Valencia, CA), washed with 10 column volumes of wash buffer (17 mM imidazole, 250 mM NaCL, 25 mM Tris HCl pH 7.5) and eluted with 10 column volumes of elution buffer (150 mM imidazole, 250 mM NaCL, 25 mM Tris HCl pH 7.5). The eluate was dialyzed against 25 mM Tris HCl pH 7.5, 250 mM NaCl. The purified protein was concentrated using an Amicon Ultra concentrator (Millipore).
For protein expression of MolA supplemented with tungstate, the protein was expressed in E. coli BL21 (DE3) cells (EMD Biosciences, Madison, WI) at 37° C in M9 minimal media (Sambrook J, 2001) with 100 μg/ml ampicillin and grown to an OD of ~0.7, before supplementing the media with 0.1mM sodium tungstate. The cells were induced with 0.5 mM IPTG and grown for an additional 2 hrs, harvested and stored at −80 °C. Tungstate bound MolA was purified as described above.
Protein Crystallization and Data Collection
Crystals were grown using the sitting drop method at 4 °C by combining protein with a precipitant solution in a 1:1 ratio. The precipitant solution in the reservoir contained 24% PEG 2000 MME, 0.1 M sodium acetate pH 4.5, 0.2M ammonium sulfate and 12% glycerol. All crystals were frozen in liquid nitrogen prior to data collection. Data collection on tungstate-MolA was performed at the APS. Data collection on molybdate-MolA was performed at the SSRL. Data was indexed and integrated with HKL2000 and scaled with SCALEPACK(Borek et al., 2003).
Structure Determination and Refinement
To solve the molybdate bound structure of MolA, a 1.6 Å resolution native data and a 2.5 Å iodide-edge data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) and the home source respectively, and processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997). Protein phases were determined by the single wavelength anomalous dispersion method using a single wavelength data set from a potassium iodide soaked crystal. The program ShelX-CDE was used to determine the iodide sites and initial phases using SHARP (Bricogne et al., 2003). The initial partial structure was automatically built by Arp/wArp (Perrakis et al., 1999) and followed by manual rebuilding in COOT(Emsley and Cowtan, 2004). Crystallographic refinement using REFMAC (Murshudov et al., 1997) resulted in a final model with an R and R-free of 32% and 33%, respectively, at 1.6 Å resolution.
To solve the tungstate bound MolA structure, molecular replacement was attempted using the data of the MolA-molybdate. The structure was solved using the program PHASER(Read, 2001), followed by manual rebuilding in COOT(Emsley and Cowtan, 2004) (Emsley and Cowtan, 2004). Crystallographic refinement using REFMAC (Murshudov et al., 1997) resulted in a final model with an R and R-free of 21% and 23%, respectively, at 1.7 Å resolution. Data collection, phasing and refinement statistics are provided in Table 1.
Table I. Crystallographic data and refinement statistics.
Structure determination of molybdate and tungstate bound-bound MolA
| Data processing | SSRL 12–2 | APS 21-ID-F | Home source | APS 21-ID-G |
|---|---|---|---|---|
| MolA-MoO2−4 | MolA-WO42 | MolA-MoO2−4 | MolA-WO42 | |
| Native | Native | KI | Peak | |
| Unit Cell space group P3121 | a=123.30 Å, c=103.04 Å | a=122.71 Å, c=103.22 Å | a=124.06 Å, c=103.66 Å | a=122.82 Å, c=103.23 Å |
| Wavelength (Å) | 0.97856 | 0.97856 | 1.54015 | 1.21434 |
| Resolution (Å) | 30-1.6 | 30-1.7 | 30-3.0 | 30-2.0 |
| Unique reflections | 106,986 | 94,269 | 18,931 | 61,303 |
| Redundancy | 8.0 | 7.9 | 16.0 | 4.7 |
| a Completeness | 99.9%(100) | 99.9% (99.8) | 100%(100%) | 99.8% (100) |
| b Rmerge | 0.077 (0.333) | 0.074 (0.348) | 0.177 (0.103) | 0.093 (0.343) |
| Refinement | ||||
| Resolution (Å) | 30-1.6 | 30-1.7 | ||
| Reflections used | 99,137 | 94,269 | ||
| Test reflections | 11,884 | 4,868 | ||
| c RWork (%) | 32.25 | 21.48 | ||
| c Rfree (%) | 33.16 | 23.40 | ||
| Average B factor (Å2) | 16.6 | 14.5 | ||
| Rmsd bond length (Å) | 0.036 | 0.009 | ||
| Rmsd bond angle (°) | 1.45 | 2.61 |
Numbers in parentheses refer to the highest resolution shell only.
Rsym=σ|Ih-<I>h|/σ Ih, where <I>h is average intensity over symmetry equivalents.
R-factor= σ |Fobs-Fcalc|/σ Fobs.
Gel shift assay
The gel-shift experiments were done according to the method developed by Rech et al. (Rech et al., 1996). In brief, 18 μg of purified MolA protein in 25mM Tris, pH 7.5, 250 mM NaCl were incubated with 50 mM final concentration of sodium molybdate, sodium tungstate, sulfate, phosphate in the presence of 66 mM potassium acetate, pH 5. The samples were incubated overnight at 4 °C before running on a 12% native polyacrylamide gel. The protein was electrophoresed at 200 V on ice for 2.5 hrs.
Biochemical analysis of MolA with Inductively Coupled Plasma-Mass Spectrometry (ICP-MS)
Quantitation of Mo and W was accomplished using ICP-MS of acid digested samples. 0.117 μM of HI1472 was digested in Trace Metal Grade nitric acid (65–70%, Fisher), 3% v/v final concentration, overnight at 67 °C. (with the exception of the M9 grown HI1472 which required 0.237 μM for detectable levels of molybdenum/tungsten). A multi-element internal standard (Spex CertiPrep Inc) was then added to achieve a 5.0 ng/mL final concentration. The sample was diluted to a total volume of 3mL in Ultrapure H2O (18.2 MΩ.cm, Milli-Q Academic System, Millipore). Mixed molybdenum (Sigma-Aldrich) and tungsten (Sigma-Aldrich) elemental standards (0, 1.00, 5.00, 10.0, 25.0, 50.0, 100, and 250 ng/mL) were made up in a 3% nitric acid (v/v) solution also containing 5.0 ng/mL of internal standards in the total sample volume of 25 mL. ICP-MS was performed on an X Series II ICP-MS controlled with Thermo Plasmalab software (Thermo Fisher Scientific). Samples were introduced manually and scanned for 95Mo and 118W with 3 runs (100 sweeps). Each sample was analyzed in triplicates and the data were corrected for molybdenum levels found in the ICP-MS buffer (250mM NaCL, 25mM Tris pH 7.5).
Isothermal Titration Calorimetry (ITC)
Isothermal titration calorimetry experiments were carried out using an iTC200 calorimeter (MicroCal USA/General Electric Healthcare). Prior to all ITC experiments, MolA grown in M9 minimal media was dialyzed extensively against an ITC buffer (25 mM Tris, pH 7.5) at 4° C, following affinity purification as described in the purification section. The sodium molybdate and sodium tungstate (Sigma Aldrich, Inc) solutions were prepared in the same buffer for the titrations. MolA was equilibrated at 25°C in the calorimeter cell and sodium molybdate or sodium tungstate was titrated in. For the molybdate titration, the concentration of HI1472 in the cell was 1.98 mM and the concentration of the sodium molybdate in the syringe was 19.8 mM. In this case, sixteen 2.5 μL injections were performed for a molar ratio of 2.25:1. For the tungstate titration, the concentration of HI1472 in the cell was 0.3 mM and the concentration of the sodium tungstate in the syringe was 4.5 mM. In this case, twenty 1.5 μL injections were performed for a molar ratio of 2.25:1. Titrations were also performed into buffer alone to allow for subtraction of the baseline. The integrated heats were fitted with the single binding site model to obtain the Kd through curve fitting with Origin 5.0 software.
NMR Titrations
MolA uniformly labeled with 15N isotope was produced as described above, except that cells were grown in M9 minimal media containing 15N-ammonium sulfate (Spectra Stable Isotopes) and dialyzed in a desalting buffer at pH 6.5. NMR data were acquired on a Varian Inova 600 MHz spectrometer at 25°C. NMR data processing was performed using NMRPipe (Delaglio et al., 1995). Substrate-free MolA (281 μM) was prepared in 20 mM deuterated Tris buffer at pH 6.5. A measured amount (500 μL) of this solution was immediately transferred into an NMR tube and capped minimize the evaporation of the solvent. Sodium molybdate was titrated in at molar ratios ranging from ~0.5:1 to 22:1, substrate and protein, respectively, with seven additions. A 1H-15N correlated spectrum was recorded after each addition. The collected data was analyzed using Sparky (Goddard and Kneller) and fitting was done with xmgrace.
Supplementary Material
HIGHLIGHTS.
MolA is a class III molybdate periplasmic binding protein (PBP), which delivers substrate to MolB2C2, the ABC transporter from H. influenzae.
Structures of MolA were solved to 1.6 Å and 1.7 Å resolutions bound to molybdate and tungstate, respectively.
MolA is distinguished from the more common class II ModA PBP by the presence of a hinge helix connecting two globular domains and a significantly lower binding affinity for molybdate.
The presence of two molybdate loci indicates multiple molybdate transport systems for low and high affinity transport are present in H. influenzae.
Acknowledgments
We would like to thank Arabela Grigorescu and Ishwar Radhakrishnan for useful discussions. We would like to thank Keith MacRenaris and Rebecca Marvin for assistance with ICP-MS, Yongbo Zhang for assistance with NMR titration studies and Kaspar Locher for the initial cloning of HI1472. This work is based on research performed at SSRL, APS (21-ID-D-G/F), funded by the Department of Energy. Research at Caltech was supported in part by NIH GM045162, by the Gordon and Betty Moore for their support of the Molecular Observatory, and by the James Irvine Foundation and United Negro College Fund - Merck for postdoctoral fellowships (H.W.P). This work was supported in part by a grant from ACS-IRG 93-037-15. Support for L.T.L was provided by an NIH training grant T32 GM008382.
Each publication, press release or other document that cites results from NIH grant-supported research must include an acknowledgment of NIH grant support and disclaimer such as “The project described was supported by Grant Number T32GM008382 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health”. Award recipients are required to comply with the NIH Public Access Policy. This includes submission to PubMed Central (PMC), upon acceptance for publication, an electronic version of a final peer-reviewed, manuscript resulting from research supported in whole or in part, with direct costs from National Institutes of Health. The author’s final peer-reviewed manuscript is defined as the final version accepted for journal publication, and includes all modifications from the publishing peer review process. For additional information, please visit http://publicaccess.nih.gov/.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors of the MolA periplamic binding protein have been deposited in the Protein Data Bank under ID code 3PSH and 3PSA for MolA-MoO2−4 and MolA-WO42− respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allaway WH, Kubota J, Losee F, Roth M. Selenium, molybdenum, and vanadium in human blood. Arch Environ Health. 1968;16:342–348. doi: 10.1080/00039896.1968.10665069. [DOI] [PubMed] [Google Scholar]
- Balan A, Santacruz CP, Moutran A, Ferreira RC, Medrano FJ, Perez CA, Ramos CH, Ferreira LC. The molybdate-binding protein (ModA) of the plant pathogen Xanthomonas axonopodis pv. citri. Protein Expr Purif. 2006;50:215–222. doi: 10.1016/j.pep.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010 doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- Borek D, Minor W, Otwinowski Z. Measurement errors and their consequences in protein crystallography. Acta Crystallogr D Biol Crystallogr. 2003;59:2031–2038. doi: 10.1107/s0907444903020924. [DOI] [PubMed] [Google Scholar]
- Borths EL, Locher KP, Lee AT, Rees DC. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc Natl Acad Sci U S A. 2002;99:16642–16647. doi: 10.1073/pnas.262659699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta crystallographica. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Davidson AL, Maloney PC. ABC transporters: how small machines do a big job. Trends Microbiol. 2007;15:448–455. doi: 10.1016/j.tim.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick KL, Tyerman SD, Kaiser BN. Molybdate transport through the plant sulfate transporter SHST1. FEBS Lett. 2008;582:1508–1513. doi: 10.1016/j.febslet.2008.03.045. [DOI] [PubMed] [Google Scholar]
- Gilson E, Alloing G, Schmidt T, Claverys JP, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- Grunden AM, Ray RM, Rosentel JK, Healy FG, Shanmugam KT. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol. 1996;178:735–744. doi: 10.1128/jb.178.3.735-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunden AM, Shanmugam KT. Molybdate transport and regulation in bacteria. Arch Microbiol. 1997;168:345–354. doi: 10.1007/s002030050508. [DOI] [PubMed] [Google Scholar]
- Hagen WR. Cellular uptake of molybdenum and tungsten. Coordination Chemistry Reviews. 2011;255:1117–1128. [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Comellas-Bigler M, Bevers LE, Feiters MC, Meyer-Klaucke W, Hagedoorn PL, Locher KP. Distorted octahedral coordination of tungstate in a subfamily of specific binding proteins. J Biol Inorg Chem. 2009;14:663–672. doi: 10.1007/s00775-009-0479-7. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007a;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007b;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- Hu Y, Rech S, Gunsalus RP, Rees DC. Crystal structure of the molybdate binding protein ModA. Nat Struct Biol. 1997;4:703–707. doi: 10.1038/nsb0997-703. [DOI] [PubMed] [Google Scholar]
- Imperial J, Ugalde RA, Shah VK, Brill WJ. Mol- mutants of Klebsiella pneumoniae requiring high levels of molybdate for nitrogenase activity. J Bacteriol. 1985;163:1285–1287. doi: 10.1128/jb.163.3.1285-1287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowich NK, Huang HH, Smith PC, Hunt JF. Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J Biol Chem. 2003;278:8429–8434. doi: 10.1074/jbc.M212239200. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Pilling PA, Epa VC, Berry AM, Ogunniyi AD, Paton JC. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure. 1998;6:1553–1561. doi: 10.1016/s0969-2126(98)00153-1. [DOI] [PubMed] [Google Scholar]
- Lawson DM, Williams CE, Mitchenall LA, Pau RN. Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA. Structure. 1998;6:1529–1539. doi: 10.1016/s0969-2126(98)00151-8. [DOI] [PubMed] [Google Scholar]
- Ledvina PS, Tsai AL, Wang Z, Koehl E, Quiocho FA. Dominant role of local dipolar interactions in phosphate binding to a receptor cleft with an electronegative charge surface: equilibrium, kinetic, and crystallographic studies. Protein Sci. 1998;7:2550–2559. doi: 10.1002/pro.5560071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Wendt JC, Shanmugam KT. Identification of a new gene, molR, essential for utilization of molybdate by Escherichia coli. J Bacteriol. 1990;172:2079–2087. doi: 10.1128/jb.172.4.2079-2087.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinson O, Lee AT, Locher KP, Rees DC. A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation. Nat Struct Mol Biol. 2010;17:332–338. doi: 10.1038/nsmb.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke H, Quiocho FA. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature. 1990;347:402–406. doi: 10.1038/347402a0. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Graham L, Keefe RG, Pihl T, Smith E. Bradyrhizobium japonicum mutants defective in nitrogen fixation and molybdenum metabolism. J Bacteriol. 1987;169:2548–2554. doi: 10.1128/jb.169.6.2548-2554.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Pear MR, McCammon JA, Quiocho FA. Hinge-bending in L-arabinose-binding protein. The ”Venus’s-flytrap” model. J Biol Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- Masters SL, Howlett GJ, Pau RN. The molybdate binding protein Mop from Haemophilus influenzae--biochemical and thermodynamic characterisation. Arch Biochem Biophys. 2005;439:105–112. doi: 10.1016/j.abb.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Rosentel JK, Lee JH, Deppenmeier U, Gunsalus RP, Shanmugam KT. Genetic analysis of the modABCD (molybdate transport) operon of Escherichia coli. J Bacteriol. 1995;177:4851–4856. doi: 10.1128/jb.177.17.4851-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouncey NJ, Mitchenall LA, Pau RN. Mutational analysis of genes of the mod locus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J Bacteriol. 1995;177:5294–5302. doi: 10.1128/jb.177.18.5294-5302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- Pau RNLD. Molybdenum and tungsten: their roles in biological processes. New York, Basel: Marcel Decker, Inc; 2002. Transport, homeostasis, regulation, and binding of molybdate and tungstate to proteins; pp. 31–74. [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Mol Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Pflugrath JW, Quiocho FA. Sulphate sequestered in the sulphate-binding protein of Salmonella typhimurium is bound solely by hydrogen bonds. Nature. 1985;314:257–260. doi: 10.1038/314257a0. [DOI] [PubMed] [Google Scholar]
- Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315:373–377. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- Premakumar R, Jacobitz S, Ricke SC, Bishop PE. Phenotypic characterization of a tungsten-tolerant mutant of Azotobacter vinelandii. J Bacteriol. 1996;178:691–696. doi: 10.1128/jb.178.3.691-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko E, O’Mara ML, Bennett WF, Tieleman DP, Gaudet R. The mechanism of ABC transporters: general lessons from structural and functional studies of an antigenic peptide transporter. FASEB J. 2009;23:1287–1302. doi: 10.1096/fj.08-121855. [DOI] [PubMed] [Google Scholar]
- Quiocho FA. Atomic structures of periplasmic binding proteins and the high-affinity active transport systems in bacteria. Philos Trans R Soc Lond B Biol Sci. 1990;326:341–351. doi: 10.1098/rstb.1990.0016. discussion 351–342. [DOI] [PubMed] [Google Scholar]
- Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Rech S, Deppenmeier U, Gunsalus RP. Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J Bacteriol. 1995;177:1023–1029. doi: 10.1128/jb.177.4.1023-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech S, Wolin C, Gunsalus RP. Properties of the periplasmic ModA molybdate-binding protein of Escherichia coli. J Biol Chem. 1996;271:2557–2562. doi: 10.1074/jbc.271.5.2557. [DOI] [PubMed] [Google Scholar]
- Rosentel JK, Healy F, Maupin-Furlow JA, Lee JH, Shanmugam KT. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J Bacteriol. 1995;177:4857–4864. doi: 10.1128/jb.177.17.4857-4864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JRD. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY; 2001. [Google Scholar]
- Self WT, Grunden AM, Hasona A, Shanmugam KT. Molybdate transport. Res Microbiol. 2001;152:311–321. doi: 10.1016/s0923-2508(01)01202-5. [DOI] [PubMed] [Google Scholar]
- Shanmugam KT, Stewart V, Gunsalus RP, Boxer DH, Cole JA, Chippaux M, DeMoss JA, Giordano G, Lin EC, Rajagopalan KV. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- van Veen HW, Abee T, Kortstee GJ, Konings WN, Zehnder AJ. Substrate specificity of the two phosphate transport systems of Acinetobacter johnsonii 210A in relation to phosphate speciation in its aquatic environment. J Biol Chem. 1994a;269:16212–16216. [PubMed] [Google Scholar]
- van Veen HW, Abee T, Kortstee GJ, Pereira H, Konings WN, Zehnder AJ. Generation of a proton motive force by the excretion of metal-phosphate in the polyphosphate-accumulating Acinetobacter johnsonii strain 210A. J Biol Chem. 1994b;269:29509–29514. [PubMed] [Google Scholar]
- Wang G, Angermuller S, Klipp W. Characterization of Rhodobacter capsulatus genes encoding a molybdenum transport system and putative molybdenum-pterin-binding proteins. J Bacteriol. 1993;175:3031–3042. doi: 10.1128/jb.175.10.3031-3042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethaus J, Muller A, Neumann M, Neumann S, Leimkuhler S, Narberhaus F, Masepohl B. Specific interactions between four molybdenum-binding proteins contribute to Mo-dependent gene regulation in Rhodobacter capsulatus. J Bacteriol. 2009;191:5205–5215. doi: 10.1128/JB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.