Two broad classes of models have been proposed to explain the patterning of the proximal-distal (PD) axis of the vertebrate limb (from the shoulder to the digit tips). Differentiating between them, we demonstrate that early limb mesenchyme in the chick is initially maintained in a state capable of generating all limb segments through exposure to a combination of proximal and distal signals. As the limb bud grows, the proximal limb is established through continued exposure to flank-derived signal(s), whereas the developmental program determining the medial and distal segments is initiated in domains that grow beyond proximal influence. In addition, the system we have developed, combining in vitro and in vivo culture, opens the door to a new level of analysis of patterning mechanisms in the limb.
The mechanisms that pattern the vertebrate limb mesenchyme so that the correct size, shape, and number of elements condense at precise locations have been argued in the literature for decades. Broadly, models of PD patterning can be divided into two general classes. One, exemplified by the progress zone model (1) posits that progressive distalization of limb pattern is based on an autonomous clocklike mechanism inherent to the undifferentiated mesenchymal cells. The second postulates that instructive cues from surrounding tissues are responsible for specifying the PD segments (2, 3). It has proven surprisingly difficult to differentiate between the autonomous and nonautonomous models experimentally. Here we try to address this issue by focusing on the establishment of the most proximal segment, the stylopod, as distinct from the more distal limb.
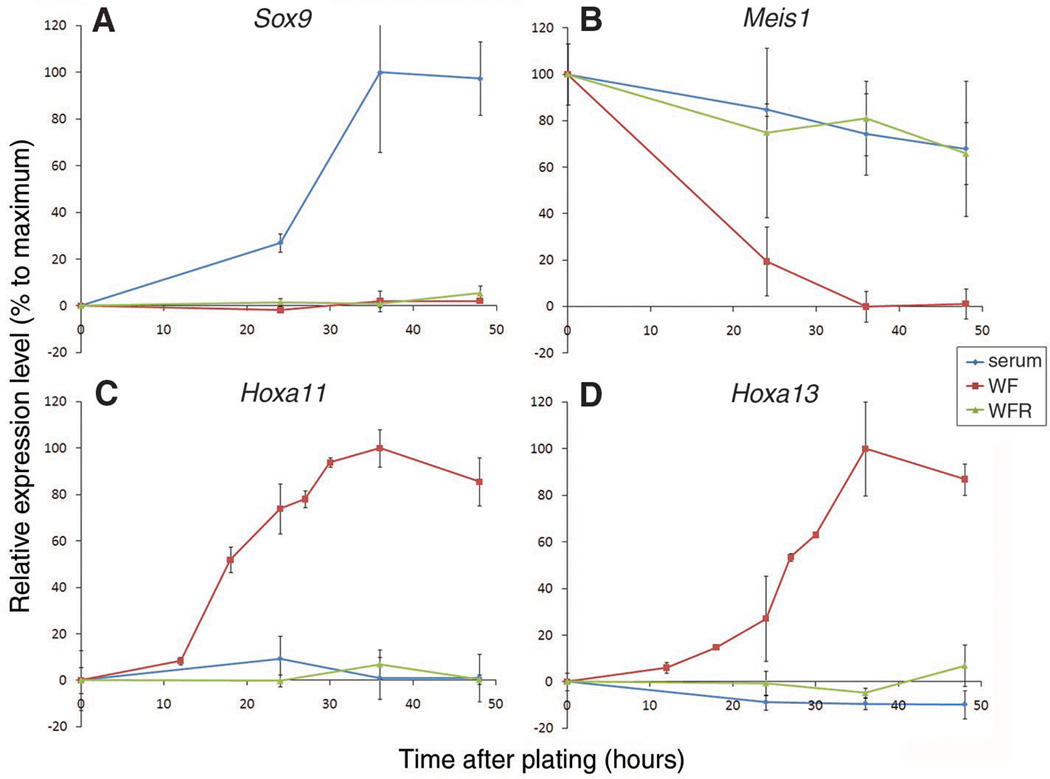
In the early vertebrate limb bud, mesenchymal cells encounter members of the fibroblast growth factor (FGF) family produced by the distal ectoderm and retinoic acid (RA) produced in the flank (2, 4). To clarify the roles these signals play in PD patterning, we have taken advantage of recently described conditions that allow limb bud cells to be maintained and manipulated in an undifferentiated state in vitro. When primary limb bud cells from Hamburger and Hamilton (5) stage 18 (HH18) chick embryos are cultured at high density, they quickly differentiate into chondrocytes (6). However, in the presence of Wnt3a and FGF8 proteins, both of which are normally secreted from the ectoderm, the cells remain proliferative and undifferentiated (7). As these cells are cultured, they continue to express markers, such as Axin2, Dusp6, and Msx1, which are characteristic of undifferentiated limb mesenchyme (7). The expression of PD markers in these cultured cells has not been examined. Whereas none of the known segmental markers are themselves required for PD specification [see discussion in Tabin and Wolpert (3)], at later stages during development in vivo, Meis1, Hoxa11, and Hoxa13 domains are congruent with the eventual stylopod, zeugopod, and autopod limb segments, respectively. We used quantitative reverse transcription polymerase chain reaction (RT-PCR) to detect these segmentally expressed limb markers in cells cultured in vitro (8). It has been proposed that cells falling out of range of distal signals in the limb bud become fixed in their PD pattern as they begin to differentiate—the so-called “differentiation front” (3). Consistent with this, dissociated primary distal HH18 mesenchymal cells expressing Meis1, but not Hoxa11 or Hoxa13, when first placed in culture with serum alone, maintained this profile at the onset of differentiation as Sox9 was up-regulated, before the formation of cartilage nodules. In contrast, we found that over time cells cultured with Wnt3a and FGF8 lost expression of the proximal marker, Meis1, and up-regulated expression of Hoxa11, a marker of the middle limb segment, followed by a distal marker, Hoxa13, an expression profile similar to distal cells of an intact limb bud (Fig. 1).
Fig. 1.
Wnt3a, FGF8, and RA act together to maintain markers of early limb mesenchyme in culture. Dissociated fresh HH18 distal limb bud cells were cultured with serum only (serum), serum + Wnt3a and FGF8 (WF), or serum + Wnt3a, FGF8, and RA (WFR) for increasing amounts of time. (A) Sox9, (B) Meis1, (C) Hoxa11, and (D) Hoxa13 expression levels were measured by quantitative PCR and normalized to β-actin expression.
In vivo, early limb bud cells are also exposed to RA from the flank in addition to FGF and Wnt activity. RA was previously shown to induce Meis1 expression, and it has been proposed to act as a proximal patterning signal (2, 3). Although this endogenous role of RA has recently been challenged, at least in the developing mouse limb bud (9, 10), the chick results indicate that RA, at minimum, may mimic or share redundancy with additional factor(s). As such, exogenous RA may act as a proxy for endogenous factors with analogous proximalizing activity (2, 11). Therefore, we next added all-trans retinoic acid (RA) at physiological concentrations (12) to the cultures with Wnt3a and FGF8. When primary limb cells were cultured with all three factors and, hence, exposed to a signaling milieu comparable to what is seen by the early limb bud mesenchyme, Meis1 expression was maintained, and Hoxa11 and Hoxa13 were not up-regulated (Fig. 1). Although this expression profile is similar to that of primary mesenchymal cells cultured in serum alone, the latter rapidly differentiate. In contrast, cells cultured with Wnt3a, FGF8, and RA remain undifferentiated while the expression of genes characteristic of the early limb mesenchyme is maintained. At higher doses, FGF8 appears to overcome the effect of RA to a limited extent, which results in a partial decrease in Meis1 expression and a concomitant increase in Hoxa11 expression (fig. S1).
To directly assess the developmental potential of cultured primary cells after exposure to various combinations of signals, we made use of a classic technique referred to as constructing a “recombinant limb.” Dissociated mesenchymal cells are reaggregated, placed within a jacket of limb bud ectoderm, and grafted onto a host embryo (13). After several days of development in the host egg, recombinant limbs are patterned by endogenous signals and form recognizable skeletal structures (14, 15).
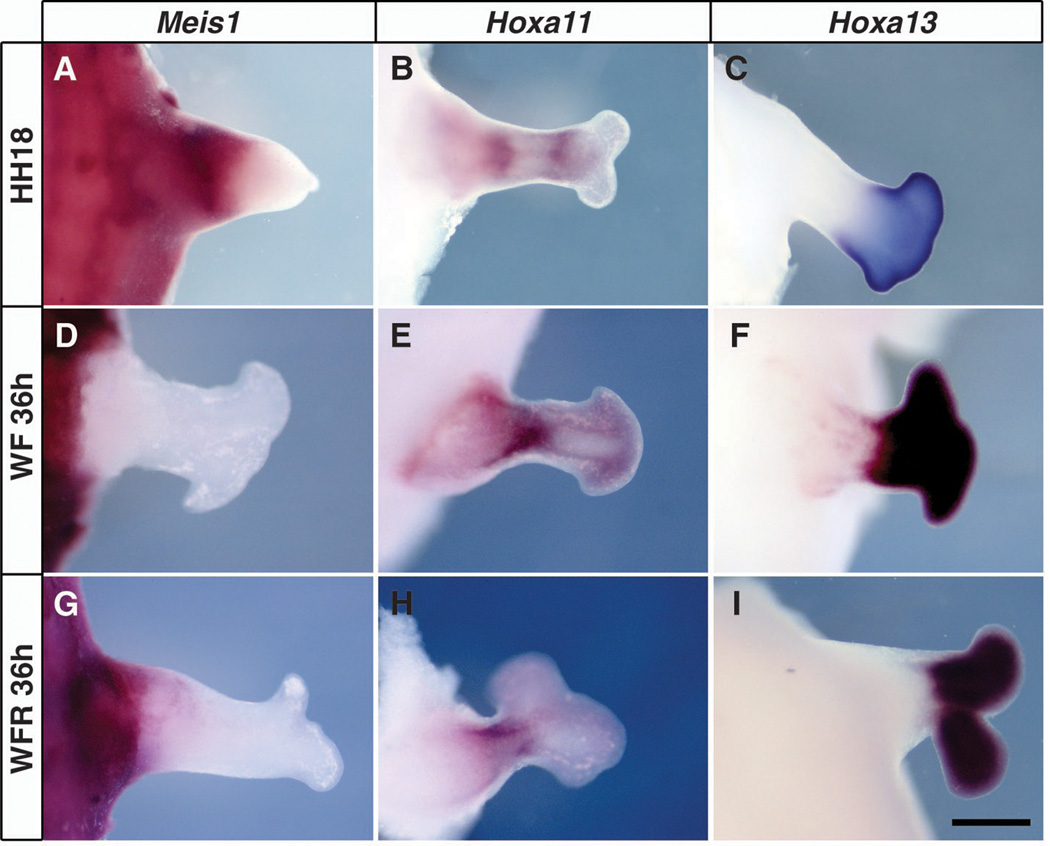
Recombinant limbs, made from limb mesenchyme cultured under various conditions, were first assessed 3 days after grafting to determine how expression of the segmental markers resolved in this in vivo setting. As in normal limb development, these markers are expressed in a segment-specific manner in recombinants generated from freshly dissociated HH18 limb mesenchyme (Fig. 2, A to C). In contrast, recombinants made from mesenchymal cells cultured for 36 hours in Wnt3a and FGF8 lacked proximal Meis1 expression but did express Hoxa11 in a middle domain and Hoxa13 distally (Fig. 2, D to F). Thus, limb mesenchyme cultured without RA shuts off Meis1 expression in vitro and does not reactivate its expression when reexposed to flank signals in vivo. However, recombinants made from cells exposed to Wnt3a, FGF8, and RA in culture continued to express Meis1 proximally, Hoxa11 centrally, and Hoxa13 distally (Fig. 2, G to I), comparable to fresh recombinants and normal limb buds.
Fig. 2.
Expression of Meis1, Hoxa11, and Hoxa13 delineate segmental domains in recombinant limbs. Whole-mount in situ hybridization with Meis1, Hoxa11, and Hoxa13 probes 72 hours after grafting. (A to C) Recombinants using freshly dissociated HH18 hindlimb cells. (D to F) Recombinants using HH18 hindlimb cells cultured for 36 hours in Wnt3a and FGF8. (G to I) Recombinants using HH18 hindlimb cells cultured for 36 hours in the presence of Wnt3a, FGF8, and RA. Scale bars: 500 µm in (A) to (C) and 800 µm in (D) to (I).
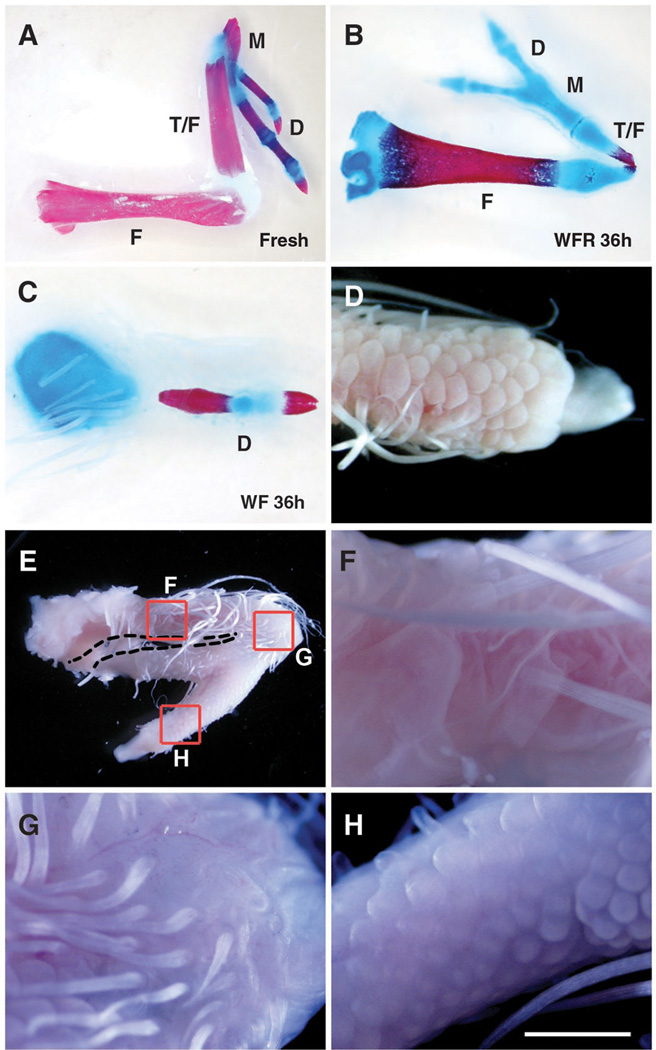
Similar to previous studies, recombinant limbs made from fresh HH18 leg bud mesenchyme that were allowed to develop for 14 days after grafting formed segmented skeletons approximating the PD organization of normal limbs including apparent femur, fused tibia and fibula, and digits (Fig. 3A) When made from cells cultured for as little as 12 hours in medium with serum alone, which (as noted above) rapidly initiate chondrogenic differentiation in vitro, they lost the ability to form more than a small cartilaginous nodule in ovo (fig. S2A). When similar recombinants were made from limb mesenchyme that was cultured with Wnt3a and FGF8 for 18, 24, or 36 hours, there was a progressive loss of proximal structures (Figs. 4, D and F, and 3C, respectively; fig. S2; and table S1), such that those cultured for 36 hours were reduced to a cartilage nodule embedded in the flank and a single digit. This loss of ability of cells cultured in Wnt3a and FGF8 to form proximal skeletal elements was not due to detectable decrease in proliferation, increase in apoptosis, or inhibition of chondrogenesis within the recombinants (fig. S3). In contrast, primary limb cells cultured for 36 hours with Wnt3a, FGF8, and RA and then assayed in recombinant limbs gave rise to multiple well-formed segments, similar to those produced in recombinants made with fresh HH18 mesenchyme, although typically smaller in size and often exhibiting a bend or break at the thinnest point in the middle of the second skeletal segment (Fig. 3B, and table S1). Our best interpretation of these skeletons is one of three segments—stylopod, zeugopod, and autopod, which is also consistent with the three distinct domains of segmental gene expression at earlier stages, discussed above.
Fig. 3.
Wnt3a, FGF8, and RA together maintain the potential of cells to form the complete PD axis. (A) Freshly dissociated HH18 hindlimb cells formed three distinct limb segments 14 days after grafting. Although not shown, proximal and middle segments were covered with feathers, and digits with terminal claws were covered by scales. (C and D) Cells cultured for 36 hours in the presence of Wnt3a and FGF8 (WF) lost the ability to form all but a single scale-covered digit extending from a cartilage nodule embedded in the flank (n = 25 out of 28 with one segment). (B and E to H) Cells cultured for 36 hours in Wnt3a, FGF8, and RA (WFR) formed an elongated feather-covered proximal segment, a short feather-covered middle segment, and a scale-covered digit with a terminal claw (n = 13 out of 24 with three segments). F, femur; T/F, tibia and fibula; M, metatarsal; D, digits. Scale bars: 5 mm in (A) and (E), 2.5 mm in (B) and (C), 1.6 mm in (D), and 1 mm in (F) to (H).
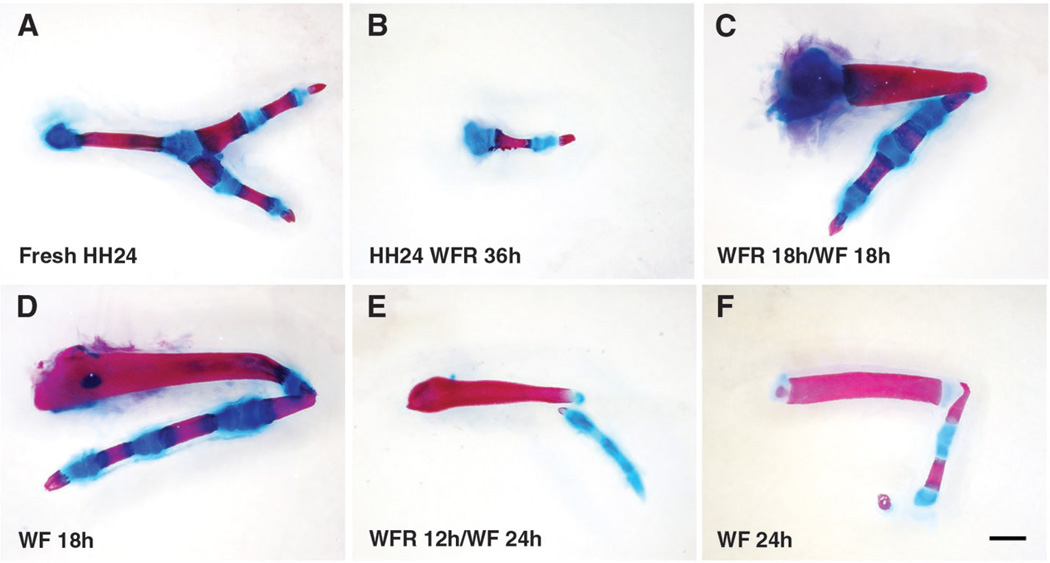
Fig. 4.
PD potential is restricted by time spent out of the influence of RA. (A) Recombinants made from freshly dissociated distal HH24 hindlimb cells. (B) Recombinants of distal HH24 cells cultured for 36 hours in Wnt3a, FGF8, and RA (WFR) (n = 8). (C and D) Recombinants of HH18 cells cultured for 18 hours in the presence of all three factors followed by 18 hours with Wnt3a and FGF8 (WF) resembled recombinants grafted immediately after culture in Wnt3a and FGF8 for 18 hours (C) n = 28; (D) n = 12. (E and F) HH18 cells cultured for 12 hours in all three factors followed by 24 hours in Wnt3a and FGF8 formed recombinants that resembled those cultured for 24 hours in the two factors alone (E) n = 12; (F) n = 9. Scale bars: 1.5 mm in (A) and (B) and 1 mm in (C) to (F).
This interpretation critically depends on our ability to correctly identify the skeletal structures resulting from recombinant limbs. Most problematic is the identification as a digit of the small rod-like ossified element that forms in recombinants made from mesenchyme cultured for 36 hours with Wnt3a and FGF8, but in the absence of RA (Fig. 3C). We therefore used a second criterion for establishing the identity of these structures using cultured leg bud mesenchyme repackaged in wing bud ectoderm. The identity of ectodermal appendages, feathers and scales is induced by the underlying mesenchyme late in embryonic development (16). The proximal part of the chick leg is covered with feathers, whereas scales cover the feet (including metatarsals and digits). Similarly, recombinant limbs generated from freshly dissociated HH18 leg mesenchyme formed feather-covered proximal elements and scale-covered feet with claws, another digit-specific structure. The elements we identified as digits in the recombinants made after culture with Wnt3a and FGF8 were invariably covered by scales and ended in claws (Fig. 3D). Similarly, the multisegmented recombinants produced by cells cultured in Wnt3a, FGF8, and RA displayed scales only over the distal elements we identified as digits, also terminating in claws (Fig. 3, E to H). Although this approach identifies the distal-most element as digit, both of the proximal segments of recombinants made with cells grown in all three factors are exclusively covered in feathers. Section in situ detection of Meis1 surrounding the proximal-most cartilage of recombinants harvested after 4 days in ovo clearly delineates this element as stylopod (fig. S4).
Thus, mesenchymal cells cultured in the combination of all three signaling molecules to which early limb cells are normally exposed maintain the capacity to form both proximal and distal structures despite the passage of time and continued proliferation. Indeed cells cultured in either Wnt3a and FGF8 or Wnt3a, FGF8, and RA divide with a cell cycle time of ~11 hours (11.43 ± 1.4 hours and 10.82 ± 1.25 hours, respectively) (movie S1), comparable to what has been reported for early limb mesenchyme in vivo (17, 18). This strongly argues against a mechanism linking PD specification to a cell cycle–based internal clock (1, 19, 20).
Freshly harvested HH18 limb bud cells give rise to multiple segments of the PD axis in a recombinant limb. Previous studies using a different experimental approach have also indicated plasticity of proximal HH20 limb bud cells in response to their environment (21). However, undifferentiated cells from the distal HH24 limb bud are committed to forming only autopod structures [Fig. 4A and Dudley et al. (22)], in spite of exposure to endogenous proximal signals after grafting to a host embryo. To determine whether this fate restriction is irreversible under culture conditions that maintain the ability of HH18 limb cells to form all three segments, we cultured dissociated distal HH24 limb mesenchyme in the presence of Wnt3a, FGF8, and RA for 36 hours. We found that when placed in a recombinant limb, these cells were at most capable of forming a single digit with terminal claw (Fig. 4B and table S1).
Although the combination of Wnt3a, FGF8, and RA cannot reverse digit specification once it starts, our data indicate that these factors are sufficient to maintain early limb mesenchyme in a state capable of giving rise to the full PD pattern. We propose that the trigger for initiating the process of specification of the zeugopod and autopod is the cessation (due to displacement) of RA exposure. If this model is correct, then cells initially cultured with Wnt3a, FGF8, and RA, and hence held in an early limb mesenchyme-like state, should start to lose the ability to form proximal structures in vitro as soon as RA is removed from the media. Indeed, we found that primary HH18 leg bud mesenchyme cultured for 18 hours in all three factors and then for 18 hours in only Wnt3a and FGF8 developed two segments, comparable to primary cells assayed immediately after culture for 18 hours in Wnt3a and FGF8 (Fig. 4, C and D; fig. S2B; and table S1). Similarly, HH18 limb cells cultured for 12 hours in all three factors followed by 24 hours in Wnt3a and FGF8 alone formed a digit with a shorter proximal element similar to those assayed after culture in Wnt3a and FGF8 for 24 hours (Fig. 4, E and F; fig. S2C; and table S1).
These data strongly suggest that exposure to the combined activities of Wnt3a, FGF8, and RA in the early limb bud or in culture maintains the potential to form both proximal and distal structures. As the limb bud grows, the proximal cells fall out of range of distal signals that act, in part, to keep the cells undifferentiated (7). Cells closer to the flank therefore differentiate and form proximal structures under the influence of proximal signals. Meanwhile, the potential of distal mesenchymal cells becomes restricted over time to zeugopod and autopod fates by virtue of their growing beyond the range of proximally produced RA. Similar conclusions were reached independently by Roselló-Díez et al. (11), as discussed in the accompanying paper.
Supplementary Material
Acknowledgements
We thank R. Nusse for generously providing Wnt3a protein, G. Martin for helpful discussions, M. Torres for sharing data before publication, and J. Gros for assistance with confocal live imaging. This work was supported by an NIH grant, R37HD032443, to C.T. and by BFU2008-00397, from the Spanish Ministry of Science and Innovation to M.R.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/VOL/ISSUE/PAGE/DC1 Materials and Methods
References and Notes
- 1.Summerbell D, Lewis JH, Wolpert L. Nature. 1973;244:492. doi: 10.1038/244492a0. [DOI] [PubMed] [Google Scholar]
- 2.Mercader N, et al. Development. 2000;127:3961. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- 3.Tabin C, Wolpert L. Genes Dev. 2007;21:1433. doi: 10.1101/gad.1547407. [DOI] [PubMed] [Google Scholar]
- 4.Mariani FV, Ahn CP, Martin GR. Nature. 2008;453:401. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamburger H, Hamilton V. J. Morphol. 1951;88:49. [PubMed] [Google Scholar]
- 6.Ahrens PB, Solursh M, Reiter RS. Dev. Biol. 1977;60:69. doi: 10.1016/0012-1606(77)90110-5. [DOI] [PubMed] [Google Scholar]
- 7.ten Berge D, Brugmann SA, Helms JA, Nusse R. Development. 2008;135:3247. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Materials and methods are available as supporting material on Science Online.
- 9.Zhao X, et al. Curr. Biol. 2009;19:1050. doi: 10.1016/j.cub.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham TJ, Chatzi C, Sandell LL, Trainor PA, Duester G. Dev. Dyn. 2011;240:1142. doi: 10.1002/dvdy.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roselló-Díez A, Ros M, Torres M. Diffusible signals, not autonomous mechanisms, determine the main proximodistal limb subdivision. Science. 2011;332 doi: 10.1126/science.1199489. XXX. [DOI] [PubMed] [Google Scholar]
- 12.Mic FA, Molotkov A, Benbrook DM, Duester G. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7135. doi: 10.1073/pnas.1231422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwilling E. Dev. Biol. 1964;9:20. doi: 10.1016/0012-1606(64)90012-0. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Teran M, Piedra ME, Ros MA, Fallon JF. Cell Tissue Res. 1999;296:121. doi: 10.1007/s004410051273. [DOI] [PubMed] [Google Scholar]
- 15.Ros MA, Simandl BK, Clark AW, Fallon JF. Methods Mol. Biol. 2000;137:245. doi: 10.1385/1-59259-066-7:245. [DOI] [PubMed] [Google Scholar]
- 16.Rawles ME. J. Embryol. Exp. Morphol. 1963;11:765. [PubMed] [Google Scholar]
- 17.Herrmann H, Marchok AC, Baril EF. Natl. Cancer Inst. Monogr. 1967;26:303. [PubMed] [Google Scholar]
- 18.Searls RL, Janners MY. Dev. Biol. 1971;24:198. doi: 10.1016/0012-1606(71)90095-9. [DOI] [PubMed] [Google Scholar]
- 19.Lewis JH. J. Embryol. Exp. Morphol. 1975;33:419. [PubMed] [Google Scholar]
- 20.Pascoal S, et al. J. Mol. Biol. 2007;368:303. doi: 10.1016/j.jmb.2007.01.089. [DOI] [PubMed] [Google Scholar]
- 21.Krabbenhoft KM, Fallon JF. Dev. Biol. 1989;131:373. doi: 10.1016/s0012-1606(89)80011-9. [DOI] [PubMed] [Google Scholar]
- 22.Dudley AT, Ros MA, Tabin CJ. Nature. 2002;418:539. doi: 10.1038/nature00945. [DOI] [PubMed] [Google Scholar]
- 23.Willert K, et al. Nature. 2003;423:448. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 24.Sato K, Koizumi Y, Takahashi M, Kuroiwa A, Tamura K. Development. 2007;134:1397. doi: 10.1242/dev.02822. [DOI] [PubMed] [Google Scholar]
- 25.Galloway JL, Delgado I, Ros MA, Tabin CJ. Nature. 2009;460:400. doi: 10.1038/nature08117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson CE, et al. Development. 1996;122:1449. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.