Abstract
The origin of Plasmodium falciparum in South America is controversial. Some studies suggest a recent introduction during the European colonizations and the transatlantic slave trade. Other evidence—archeological and genetic—suggests a much older origin. We collected and analyzed P. falciparum isolates from different regions of the world, encompassing the distribution range of the parasite, including populations from sub-Saharan Africa, the Middle East, Southeast Asia, and South America. Analyses of microsatellite and SNP polymorphisms show that the populations of P. falciparum in South America are subdivided in two main genetic clusters (northern and southern). Phylogenetic analyses, as well as Approximate Bayesian Computation methods suggest independent introductions of the two clusters from African sources. Our estimates of divergence time between the South American populations and their likely sources favor a likely introduction from Africa during the transatlantic slave trade.
Keywords: malaria origin, New World, human migrations, genetic diversity
Malaria is an important human parasitic disease, responsible for 500 million clinical cases each year within its wide circumtropical range (1). This vector-borne disease is caused by a protozoan of the genus Plasmodium, with five species now known to infect humans, of which Plasmodium falciparum, widespread in tropical and subtropical regions, is the most malignant, accounting for the death of more than one million people every year (1).
An African origin of P. falciparum seems likely (2, 3), but the routes and times by which this pathogen dispersed out of Africa and colonized other parts of the world remain controversial (4). Regarding its expansion toward Asia, it has been proposed that African P. falciparum expanded rapidly about 6,000 y ago in association with the emergence of agricultural societies and increased mosquito transmission (5). An alternative hypothesis suggests that P. falciparum spread with humans as Homo sapiens populations migrated out of Africa, starting about 60,000 y ago (6, 7).
Concerning South America, the conventional hypothesis is that P. falciparum was introduced into the Americas by European immigrants and the transatlantic slave trade, a hypothesis supported by genetic (8–12) and archeological (13) evidence, although not quite conclusive. Mitochondrial DNA shows a clear link between South American and African haplotypes (11), consistent with a recent introduction of the pathogen through the slave trade. However, some time to the most-recent common ancestor estimates suggest a much more ancient origin (11). Some archaeological and historical studies are inconsistent with a recent introduction, and instead suggest that P. falciparum malaria was present in South America long before the European colonizations and the slave trade (13).
In this article, we seek to unravel the events surrounding the origin of P. falciparum in South America. We address the following questions: Where did the South American P. falciparum come from? Were there one or several events of invasion? Does the transatlantic slave trade from Africa account for the origin of this parasite in South America, or was it present there before the European colonizations?
Results
We obtained P. falciparum-infected human blood samples from 24 localities in 17 countries: nine from Africa, four from South America, one from the Middle East, and three from Asia (Fig. 1). The African localities were intended to represent the places of origin of the African slaves during the transatlantic slave trade. (See SI Appendix, Table S1 for study sites and sampling procedures.) We PCR-amplified 12 polymorphic microsatellites located on 8 of P. falciparum's 14 chromosomes (SI Appendix, Table S2). We thus obtained 577 monoinfected isolates from all 24 localities in 17 countries, which we designate as the “MS” dataset. In some analyses we combined our data with the data from ref. 8, using nine microsatellite markers common to both studies, resulting in a total of 1,047 isolates from 33 localities in 23 countries (hereafter referred to as the \x{201c}MS+” dataset) (Fig. 1). In addition, we obtained the genotypes of 210 isolates at 272 SNPs (SI Appendix, Table S3) from 17 populations in 13 countries, which we designate as the “SNP” dataset (SI Appendix, Table S1). See Materials and Methods for additional details.
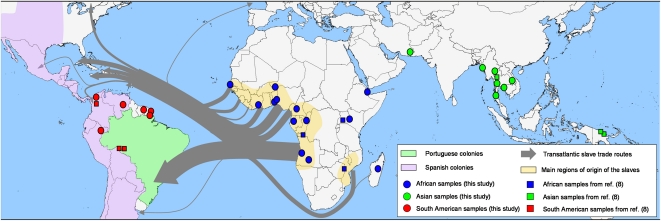
Fig. 1.
Sampling sites, main routes of the transatlantic slave trade, and major European Empires between the 16th and the 19th centuries in South America. Circles represent populations sampled for this study: the MS dataset. Squares represent populations sampled in ref. 8. The MS+ dataset includes all populations. Detailed information is in SI Appendix, Table S1.
FST-Based Test of Selection.
For microsatellites, the analysis of genetic differentiation over all populations is suggestive of positive selection at two loci: Pfg377 and TA109 (SI Appendix, Fig. S1 A and B). For the SNPs, the test of selection identified 46 SNPs that lie above the upper 95% boundary and 26 SNPs that are less genetically differentiated than expected by chance (SI Appendix, Fig. S1C). See SI Appendix, Table S4 for the SNPs identified as being under selection. In what follows, we only present the results obtained for datasets that exclude the loci under selection. (If the excluded loci are included in the analyses, the results obtained are similar to those presented below, except in the case when all microsatellite markers are included. These alternative results are presented in SI Appendix, Figs. S9–S12.)
Genetic Diversity.
SI Appendix, Table S5 displays the expected heterozygozities (Hs) for the separate populations, as well as the averages for each continent. A consistent pattern is apparent for both microsatellite and SNP markers. Asian populations exhibit slightly lower genetic diversity than African ones, but the difference is not statistically significant for SNPs. For South American populations, genetic diversity is significantly lower than in the Old World populations for all datasets (Student t test on logarithms) (SI Appendix, Table S5).
Geographic Differentiation.
There are striking differences in genetic differentiation among continents (SI Appendix, Fig. S2). In Asia and South America, there is a clear pattern of isolation by distance, with genetic differentiation increasing with geographic distance between populations, whereas there is little genetic differentiation between populations in Africa, with no significant correlation with geographic distance.
Worldwide Structure and Origin of the South American Populations of P. falciparum.
The principal component analysis (PCA) on SNP data (SI Appendix, Figs. S3C and S7C) shows clear differentiation between the three continents, with the African samples lying in the middle. The separation between African and Asian samples is far less marked for the PCA of microsatellite data (SI Appendix, Fig. S3C). Regarding the South American populations, the following distinctive features emerge from microsatellite and SNP data: (i) the French Guianan populations form a tight cluster (SI Appendix, Fig. S3C, all datasets); (ii) Brazilian and Bolivian populations form a tight cluster close to the French Guianan populations (SI Appendix, Fig. S3C, MS+ dataset); (iii) Colombian samples are close to one another, much closer to the Old World populations than are the previously mentioned populations (SI Appendix, Fig. S3C, all datasets); and (iv) the Peruvian and Venezuelan populations are (a) at an intermediate position between the group of populations that includes French Guiana, Brazil and Bolivia, and the Colombian populations; and (b) much more dispersed than the other South American populations (SI Appendix, Fig. S3C, all datasets). The dispersion of the Peruvian and Venezuelan populations suggests a recent history of admixture for these two populations.
Bayesian Clustering is consistent with the PCA analyses, in which the separation between Asia and Africa is far clearer in the SNP than in the microsatellite analysis (SI Appendix, Figs. S3B and S7B). In South America, the first and most significant split is between Colombia and the other populations. Under a model with K = 2 clusters, Colombia is assigned to the same group as the Old World populations (SI Appendix, Fig. S3B, all datasets). As the number of clusters increases, Peru and Venezuela separate from the set of the Brazilian, Bolivian (SI Appendix, Fig. S3B, MS+ dataset), and the three Guianan populations (SI Appendix, Fig. S3B, all datasets), which remain together in all analyses. Peru, and to a lesser extent Venezuela, seem to be a mixture between African or Colombian populations on the one side and the set of French Guiana, Brazil, and Bolivia on the other side. The optimal number of clusters as determined by the method proposed in ref. 14 is K = 2 for both the worldwide analysis and the one restricted to South American isolates (SI Appendix, Fig. S3B).
Phylogenetic trees reconstructed from the genetic distances between populations show that populations cluster according to their geographic origin when all American populations are included (SI Appendix, Figs. S3A and S7A). When South American populations are included one at a time in the tree, they all branch off the African cluster for the microsatellite data and closer to the African than to the Asian cluster for the SNP data (SI Appendix, Figs. S4–S6).
Fig. 2A shows the neighbor-joining tree built from shared allelic distances between pairs of isolates computed using SNPs. Again, the Peruvian and Venezuelan populations display a mixed composition, with some isolates being closer to the Colombian population and others closer to the French Guianan populations. The Colombian and the French Guianan populations branch off independently from the African clade, thus suggesting independent introductions from Africa of the Colombian and the French Guianan populations (Fig. 2A and SI Appendix, Fig. S8B).
Fig. 2.
Neighbor-joining trees of isolates obtained using: (A) the SNP dataset including all American populations; (B) the SNP dataset excluding the Peruvian and the Venezuelan populations. Genetic distances were computed without the loci suspected to be under selection. Ang, Angola; Ben, Benin; Cmp, Camopi (French Guiana); Col, Colombia; Con, Republic of the Congo; Gab, Gabon; Ira, Iran; Lao, Laos; Mad, Madagascar; Mar, Maripassoula (French Guiana); Mya, Myanmar; Per, Peru; Tai, Thailand; Trs, Trois Sauts (French Guiana); Ven, Venezuela.
Test of Multiple Introductions Based on Genetic Trees.
Based on all of the previous analyses, the South American populations are clearly subdivided in at least two main genetic clusters: a northern cluster that includes the populations from Colombia and a southern cluster comprising the populations from Brazil, Bolivia, and French Guiana. The individual-based SNP tree further suggests an independent introduction of these two clusters from Africa (Fig. 2A). In the following, we evaluate how strong the evidence is in favor of a scenario of independent introduction.
Two populations independently introduced from the same source would be expected to be genetically less closely related to each other than to the source population, and they should not cluster together in a population neighbor-joining tree (15). Such a scenario of independent introduction is congruent with the comparison of the genetic distances computed between the South American and the African populations (Table 1) but is at odds with the apparent monophyly of the South American populations observed in the population-based trees (SI Appendix, Fig. S3A). However, there are good reasons to believe that the grouping of the introduced populations in the neighbor-joining tree is an artifact inherent to the approach (see ref. 15 for an explanation of this phenomenon).
Table 1.
Comparison of the genetic distances computed between the northern cluster, the southern cluster and the African cluster for each dataset (MS, MS+, and SNP)
| Dataset | Mean genetic distance between clusters | P value | ||
| MS | Northern Africa | Northern–Southern | ||
| 0.574 | vs. | 0.669 | 0.005 | |
| Southern Africa | Northern–Southern | |||
| 0.629 | vs. | 0.669 | 0.038 | |
| MS+ | Northern Africa | Northern–Southern | ||
| 0.615 | vs. | 0.681 | 0.005 | |
| Southern Africa | Northern–Southern | |||
| 0.649 | vs. | 0.681 | 0.044 | |
| SNP | Northern Africa | Northern–Southern | ||
| 0.142 | vs. | 0.215 | 0.036 | |
| Southern Africa | Northern–Southern | |||
| 0.134 | vs. | 0.215 | 0.036 | |
For all datasets: The northern cluster includes the two populations from Colombia. The African cluster comprises the populations from Africa. For the MS and SNP datasets, the southern cluster includes the populations from French Guiana. For the MS+ dataset, the southern cluster comprises the populations from Brazil, Bolivia, and French Guiana. For all tests: Peru (Per) and Venezuela (Ven) were excluded because of clear genetic admixture. The genetic distances for the MS and MS+ datasets were Cavalli-Sforza and Edward's chord distances. For the SNP dataset, the genetic distances were shared allelic distances. For each pairwise comparison between pairs of clusters a P value was computed from a Wilcoxon test.
The paraphyly of the South American populations becomes clearly apparent in the individual-based SNP tree when the isolates from Peru and Venezuela (that are admixed) are excluded from the analysis: the populations from the northern and the southern clusters clearly branch off from the African lineage independently (Fig. 2B). The analysis of the alternative trees obtained after bootstrapping of the loci shows that this paraphyly of the two South American clusters is very robust: 96% of 50 bootstrapped trees show a clear paraphyly of the South American populations.
Test of Multiple Introductions Based on Approximate Bayesian Computation Analyses.
To further confirm the hypothesis of an independent introduction of the two genetic clusters in South America, we also used a Bayesian framework (Approximate Bayesian Computation or ABC methods) using the microsatellite datasets (MS and MS+). We considered three possible colonization scenarios (SI Appendix, Fig. S13): (i) “Independent Introduction” (SI Appendix, Fig. S13A); (ii) “Serial Introduction,” so that there was one introduction from Africa, with other South American populations derived from the first (SI Appendix, Fig. S13B); and (iii) “unsampled population scenario,” which assumes that all American populations derive from an unsampled population, which would have been the only one originating from Africa (SI Appendix, Fig. S13C). The results presented in SI Appendix, Table S6 consistently show that the “Independent Introduction Scenario (A)” is supported with the highest probabilities, with South American populations (including the Colombian populations) corresponding to one introduction event (northern cluster) and the French Guianan, Brazilian, and Bolivian populations reflecting a different introduction (southern cluster). The 95% confidence intervals for the probabilities associated with this scenario rarely overlap with those from alternative scenarios. The unsampled population scenario is supported with high probability in only a few cases. The only instance in which a “Serial Introduction Scenario (B)” is supported with the highest probability is the case in which the South American populations belong to the same cluster (northern or southern).We did not include in these analyses the Peruvian and the Venezuelan populations because they show a clear pattern of recent admixture.
Divergence time estimates based on these analyses are presented in SI Appendix, Table S6. Time estimates can be expressed in years by considering that P. falciparum experiences either 6 (8) or 12 (16) generations per year. Restricting ourselves to the scenarios of independent introductions, including populations from the two clusters, we estimate that, on average, P. falciparum reached South America 434–990 y ago (at 6 generations per year) or 217–495 y ago (at 12 generations per year). These dates are congruent with the transatlantic slave trade that occurred between the 16th and the mid-19th centuries.
Discussion
To ascertain the origin of P. falciparum in South America, we collected and analyzed 577 P. falciparum isolates from 24 populations encompassing the entire distribution range of the parasite: sub-Saharan Africa, Middle East, Southeast Asia, Oceania, and South America. In Africa, attention was given to collect samples from the localities of origin of the African slaves during the transatlantic slave trade. We have used an expanded dataset that includes data from ref. 8 (making a total of 1,047 isolates from 33 locations in 23 different countries), as well as an SNP dataset comprising 210 isolates from 17 locations in 13 different countries. Analyses of our data and of the combined datasets give very similar results.
Worldwide Population Structure.
The patterns of P. falciparum genetic structure observed at a worldwide scale are consistent with previous published results (7, 8, 12, 17). The African populations are the most genetically diverse (with the exception of a population from Djibouti), followed by the Asian populations and last, the South American populations. Genetic differentiation among populations is low in Africa, intermediate in Asia, and high in South America. In these two latter continents, genetic distance is largely explained by geography, with the closest populations displaying the lowest pairwise differentiation (SI Appendix, Fig. S2). The high levels of genetic differentiation observed among populations in South America could be due in part to the very low within-population genetic diversity, which is known to bias upward genetic distance estimates (18), but are also impacted by the epidemic nature and low effective population size of South American P. falciparum (8). Large genetic differentiation among the South American populations may also be attributed to independent introduction events of P. falciparum, as discussed below.
P. falciparum in South America: Several Introductions from Africa.
The most widely accepted scenario for the origin of P. falciparum in the Americas postulates introductions from Africa during the slave trade that took place within the 300 y following the early European colonizations (5). Several genetic studies support this scenario, although on the basis of limited evidence: either a limited number of isolates, or few sampled geographic areas in South America, or a small number of genetic markers (8–12).
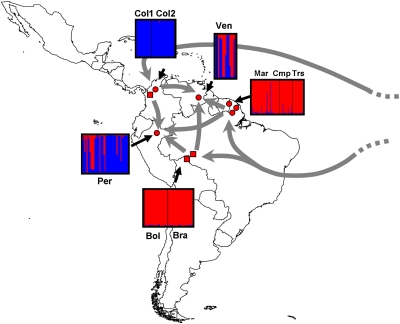
Our study confirms an African origin of P. falciparum in the New World. In addition, because our study now encompasses much of the distribution range of P. falciparum in South America, it provides a far more detailed picture of the genetic structure of the parasite in this continent, and hence of its colonization history. We show that P. falciparum is subdivided in South America in at least two main genetic clusters: a northern cluster that notably includes the Colombian populations, and a southern cluster that includes populations from French Guiana, Brazil, and Bolivia. Note that despite the fact that two populations of the northern cluster came from the same country, namely Colombia, they nevertheless were sampled independently in space and time and are genetically differentiated (FST = 0.0893, P value = 0.006). Hence, the genetic peculiarity of Colombia cannot be attributed to any sampling or genotyping artifacts. A transition zone between the two clusters is represented by populations from Venezuela and Peru, which exhibit typical genetic patterns of admixed populations (i.e., elevated genetic diversity, overdispersed isolates on PCA, as well as throughout the individual-based neighbor-joining tree). ABC analyses, individual-based phylogenetic trees (Fig. 2 and SI Appendix, Fig. S8B), as well as the comparison of genetic distances between introduced populations and their sources (Table 1), strongly suggest that the northern and southern clusters were introduced separately into South America (Fig. 3).
Fig. 3.
Proposed introduction and migration pathways of P. falciparum in South America. Boxes represent the assignment proportions to two clusters (K = 2), the optimal number of South American clusters inferred from STRUCTURE simulations, obtained using the MS+ dataset, excluding loci suspected to be under selection. Red circles: samples collected and genotyped in this study. Red squares: samples collected and genotyped in ref. 8.
Our estimates of divergence times between the South American and the African populations obtained using ABC methods suggest that the northern and southern clusters were both introduced during the transatlantic slave trade.
The European colonization history of South America and the characteristics of the transatlantic slave trade are central to understanding the origin of these independent introduction events of P. falciparum in South America. From the 16th to the 19th century, South America was largely divided into two main Empires. The Spanish Empire included much of the Caribbean and Central America, but in Continental South America it was mostly restricted to the north and west coast, stretching from present-day Venezuela to present-day Argentina and Chile. The Portuguese Empire was restricted to the east coast, from just north of the Amazon river to the south of present-day Brazil (Fig. 1) (19).
For three centuries, between 1560 and 1852, the Portuguese Empire was the principal destination for African slaves in South America (20). Over this period, almost five million African slaves were brought to Brazil and disembarked in two main ports: Rio de Janeiro and Bahia. Around 40% of all Africans forced into the slave trade ended their lives in Brazil.
The Spanish Empire ferried most slaves to the West Indies colonies (Jamaica, Cuba, and other islands), but some were also disembarked in Central and South America. The Spanish imported most enslaved Africans into Central America via Veracruz (Mexico) and in South America via Cartagena (Colombia) (20). From these ports of disembarkation, slaves were then dispatched into the other regions of the Central and South American Spanish Empire.
This subdivision of the Americas into two main empires and the fact that the two empires imported African slaves into different regions (using different ports of disembarkation) is very likely at the origin of the two independent introductions of P. falciparum shown by our genetic data (Fig. 3). It is further likely that the introduction of P. falciparum into South America was a recurrent process, lasting as long as African slaves were deported onto the continent by both the Spanish and Portuguese Empires. These recurrent introductions certainly played an important role in shaping the genetic diversity currently observed in the two South American clusters. The Andes, which form a natural geographic barrier separating Central and western South America from the rest of South America, may have played a role in preventing these two clusters to homogenize over the course of history. A recent human immigration history of people coming from north of the Andes and from the southern part of the continent into the region of Loreto in Peru, as well as into the regions of Bolivar and Amazonas in Venezuela (where our samples originate from), could account for the origin of the admixture of P. falciparum populations in these two regions. Indeed, in the Loreto region of Peru, malaria was almost eliminated by the end of the 1960s (21), but in the early 1990s it rose again, simultaneously with a strong immigration from the Andean region (22, 23). Concerning Venezuela, most of the samples collected from the provinces of Bolivar and Amazonas came from people infected in the gold mining regions, close to the border with Brazil, where many people from Colombia and other neighboring countries are known to have come to work as (largely illegal) miners.
Selection and Population History.
The basic premise behind the use of genetic markers to infer connectivity among populations or to reconstruct their demographic history is that the spatial distribution of allele frequencies within and between populations is a consequence of “neutral processes” alone (such as migration, genetic drift, and mutation), so that the distribution of the genetic markers deployed has not been shaped by natural selection. This assumption is rarely questioned, especially for microsatellites, which are generally assumed to be evolving neutrally.
The 12 microsatellites markers used in this study are no exception to this trend. The markers were described for the first time in 1999 (24) and were used multiple times without their neutrality being questioned since then (e.g., refs. 8 and 25–27). We have shown that two microsatellites (Pfg377 and TA109) have likely evolved under divergent selection. We accordingly excluded these two loci from further analyses. As shown in SI Appendix, Figs. S9–S12, including these loci could have led to spurious conclusions. Indeed, some analyses based on the whole set of loci suggest that some populations (French Guiana, Brazil, Bolivia, Venezuela) are mainly of African descent, and others (Peru and Colombia) would appear as primarily of Asian descent. A possible explanation for this misleading pattern could stem from the fact that the two microsatellite markers under apparent divergent selection are located within two genes (PFsXLX: gametocyte specific antigen, and PFF0930W: unknown function), which may be subject to similar selective pressures in the western part of South America and in Asia.
Conclusion
We present evidence for multiple introductions of P. falciparum from Africa into South America during the slave trade. Our results highlight the importance of human migrations on the current circum-tropical distribution of P. falciparum, be they natural migrations, such as the expansion out of Africa (7), or coerced, such as the transatlantic slave trade. Given the importance of human migrations in the dispersal of P. falciparum, this finding raises the question whether immigration during the colonial period from other P. falciparum endemic regions [e.g., southern Europe and Asia (19, 28)] might also have contributed to the current genetic makeup of South American P. falciparum populations.
Materials and Methods
Study Sites and Plasmodium falciparum Isolates.
We have studied P. falciparum-infected human blood samples from 24 localities in 17 countries: nine from Africa (Ang1 and Ang2: Angola; Ben1 and Ben2: Benin; Cam: Cameroon; Con1: Republic of the Congo; Dji: Djibouti; Gab: Gabon; Mad: Madagascar; Nig: Niger; and Sen: Senegal), four from South America (Col1: Colombia; Cmp, Trs, and Mar: French Guiana; Per: Peru; and Ven: Venezuela), one from the Middle East (Ira: Iran), and three from Asia (Lao: Laos; Mya: Myanmar; and Tai1–Tai4: Thailand) (Fig. 1). African sampling localities were selected to represent the places of origin of the African slaves during the transatlantic slave trade. Infected blood samples were collected either by venous puncture (∼500 μL) or by finger-prick (∼50 μL). All blood samples were collected after informed consent. Ethical clearance was obtained from local ethics committees in each country sampled. Additional information about the study sites and sampling procedures are given in SI Appendix, Table S1.
Microsatellite and SNP Genotyping.
Details regarding the genotyping of the 12 microsatellite markers and the 384 SNPs are given in SI Appendix, SI Materials and Methods.
Multiplicity of Infections.
Blood samples are frequently infected with two or more haploid clones of P. falciparum, resulting in the detection of two or more alleles at polymorphic loci. Isolates with more than one allele at any of the 12 microsatellite loci were removed from the analyses (SI Appendix, Table S1).
Analyzed Datasets.
Three datasets were analyzed. The first dataset corresponds to the microsatellite dataset (577 newly collected isolates from 24 locations in 17 countries, called MS). A second microsatellite dataset, called MS+, includes our data pooled with those from ref. 8 (Con2: Democratic Republic of the Congo; Uga: Uganda; Zim: Zimbabwe; Tai5: Thailand; PNG1 and PNG2: Papua New Guinea; Bol: Bolivia; Bra: Brazil; and Col2: Colombia). The number of loci in this second dataset is reduced to the nine markers in common between the two studies, but this increases the number of samples to 1,047 isolates from 33 localities in 23 countries (see Fig. 1). Third is the SNP dataset (210 isolates from 17 locations in 13 countries: Ang: Angola; Ben: Benin; Con1: Republic of the Congo; Gab: Gabon; Mad: Madagascar; Ira: Iran; Tai1: Thailand; Mya: Myanmar; Lao: Laos; Col1: Colombia; Cmp, Trs, and Mar: French Guiana; Per: Peru; and Ven: Venezuela). For this dataset, we pooled the two populations from Angola (Ang1 and Ang2), as well as the two populations from Benin (Ben1 and Ben2), to keep sample size large enough for genetic analyses.
Markers Under Natural Selection.
Natural selection, either divergent or convergent, may impact genetic frequencies differently in different populations. We searched for loci (microsatellites and SNPs) that could be under positive or balancing selection, using the method developed in ref. 29 and implemented in the program LOSITAN (30). A detailed description of the method is provided in SI Appendix, SI Materials and Methods.
Genetic Diversity and Genetic Differentiation.
We computed genetic diversity within each population using Nei's unbiased estimate of heterozygosity Hs (31). Weir and Cockerham's FST estimates (32) between pairs of populations were computed with the Genepop software (33). To explain patterns of isolation by distance within each continent, we studied the correlation between genetic differentiation (FST) and geographical distance between pairs of populations. Pairwise geographic distances were computed using MAPINFO (34). The significance of the relationship was assessed with a Mantel test using 10,000 permutations.
Statistical, Population Genetic, and Phylogenetic Data Analyses.
Worldwide population genetic structure and the origin of P. falciparum in South America were determined using: (i) population and individual-based genetic trees, (ii) multivariate analyses (PCA), (iii) clustering methods (STRUCTURE analyses), and (iv) model-based Bayesian methods (ABC). A detailed description of these methods is given in the SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the two anonymous reviewers for very helpful comments on a previous version of the manuscript; Dr. Frank Smithuis and the staff at Medecins Sans Frontieres-Holland (Myanmar) for providing parasite samples from Rakhine state, Myanmar (Samlane Phompida, Rattanaxay Phetsouvanh, Maniphone Khanthavone, Tiengkham Pongvongsa); Dr. Tepanata Pumpaibool and Dr. Pongchai Harnyuttanakorn for providing samples from Thailand; Susana Campino for her help in designing the SNPs; Dr. Laurence Lachaud for her help in performing quantitative PCR for SNPs at Centre Hospitalier Universitaire Carémeau, Nimes, France; and the program Centre National de la Recherche Scientifique–Amazonie Ecologie de la Santé. For microsatellite genotyping, we thank the “plateforme séquençage-génotypage de l'Institut Fédératif de Recherche 199”, Montpellier. The staff of the Centre for Malariology, Parasitology and Entomology in Vientiane and Savannakhet, Laos assisted in the collection of samples from Xekong. Samples from Peru were collected within a project funded by the Directorate General for Development Cooperation (DGCD) of the Belgian Government (Framework Agreement 02, 2003–2007, Project 95501) in collaboration with Dr. Alejandro Llanos-Cuentas. Samples from the Republic of Congo were collected thanks to the Laboratoire National de Santé Publique. This study was funded by Agence Nationale de Recherche Grant MGANE 07 SEST 012 (Programme Sante-Environnement et Sante-Travail–Malaria Genetic Adaptation to a New Environment), Centre National de la Recherche Scientifique, Institut de Recherche pour le Développement, Centre Régional des Oeuvres Universitaires et Scolaires, Ministère des Affaires Etrangères, and Institut Kurde de Paris. D.G. is supported by DGCD (Framework Agreement 03, 2008–2010, Project 910000) and US Public Health Service grants, National Institutes of Health/National Institute of Allergy and Infectious Diseases Grant R01 AI067727, and International Centers of Excellence for Malaria Research Grant 1U19AI089681-01.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119058109/-/DCSupplemental.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krief S, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartl DL. The origin of malaria: Mixed messages from genetic diversity. Nat Rev Microbiol. 2004;2:15–22. doi: 10.1038/nrmicro795. [DOI] [PubMed] [Google Scholar]
- 5.Hume JC, Lyons EJ, Day KP. Human migration, mosquitoes and the evolution of Plasmodium falciparum. Trends Parasitol. 2003;19:144–149. doi: 10.1016/s1471-4922(03)00008-4. [DOI] [PubMed] [Google Scholar]
- 6.Hughes AL, Verra F. Malaria parasite sequences from chimpanzee support the co-speciation hypothesis for the origin of virulent human malaria (Plasmodium falciparum) Mol Phylogenet Evol. 2010;57:135–143. doi: 10.1016/j.ympev.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe K, et al. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20:1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 8.Anderson TJC, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 9.Conway DJ. Tracing the dawn of Plasmodium falciparum with mitochondrial genome sequences. Trends Genet. 2003;19:671–674. doi: 10.1016/j.tig.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Conway DJ, et al. Origin of Plasmodium falciparum malaria is traced by mitochondrial DNA. Mol Biochem Parasitol. 2000;111:163–171. doi: 10.1016/s0166-6851(00)00313-3. [DOI] [PubMed] [Google Scholar]
- 11.Joy DA, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 12.Neafsey DE, et al. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 2008;9:R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Castro MC, Singer BH. Was malaria present in the Amazon before the European Conquest? Available evidence and future research agenda. J Archaeol Sci. 2005;32:334–340. [Google Scholar]
- 14.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 15.Estoup A, Guillemaud T. Reconstructing routes of invasion using genetic data: Why, how and so what? Mol Ecol. 2010;19:4113–4130. doi: 10.1111/j.1365-294X.2010.04773.x. [DOI] [PubMed] [Google Scholar]
- 16.Wéry M, Paskoff S. Protozoologie Médicale. Paris: Bruxelles; 1995. [Google Scholar]
- 17.Mu J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- 19.McAlister LN. Spain and Portugal in the New World, 1492–1700 (Europe and the World in the Age of Expansion) Minneapolis: University of Minnesota Press; 1984. [Google Scholar]
- 20.Voyages database 2009. Available at http://www.slavevoyages.org/tast/index.faces. Accessed June 2010.
- 21.Babione RW. Epidemiology of malaria eradication. II. Epidemiology of malaria eradication in Central America: A study of technical problems. Am J Public Health Nations Health. 1966;56:76–90. doi: 10.2105/ajph.56.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branch OH, et al. Plasmodium falciparum genetic diversity maintained and amplified over five years of low endemic malaria transmission in the Peruvian Amazon. Mol Biol Evol. 2011;28:1973–1986. doi: 10.1093/molbev/msq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vittor AY, et al. The effect of deforestation on the human-biting rate of Anopheles darlingi, the primary vector of Falciparum malaria in the Peruvian Amazon. Am J Trop Med Hyg. 2006;74:3–11. [PubMed] [Google Scholar]
- 24.Anderson TJC, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- 25.Iwagami M, et al. Phylogenetic relationship of Plasmodium falciparum populations in the Philippines. Southeast Asian J Trop Med Public Health. 2008;39:200–204. [PubMed] [Google Scholar]
- 26.Prugnolle F, et al. A comparison of Anopheles gambiae and Plasmodium falciparum genetic structure over space and time. Microbes Infect. 2008;10:269–275. doi: 10.1016/j.micinf.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Pumpaibool T, et al. Genetic diversity and population structure of Plasmodium falciparum in Thailand, a low transmission country. Malar J. 2009;8:96. doi: 10.1186/1475-2875-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meagher AJ. The Coolie Trade: The Traffic in Chinese Laborers to Latin America 1847–1874. Philadelphia: Xlibris Corporation; 2008. [Google Scholar]
- 29.Beaumont MA, Nichols RA. Evaluation loci for the use in the genetic analysis of population structure. Proc Biol Sci. 1996;263:1619–1626. [Google Scholar]
- 30.Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 32.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 33.Raymond M, Rousset F. GENEPOP Version 1.2: Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 34.Pitney Bowes. 2010. Mapinfo (Pitney Bowes Software Inc., Lanham, MD)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.