Abstract
Calorie restriction delays brain senescence and prevents neurodegeneration, but critical regulators of these beneficial responses other than the NAD+-dependent histone deacetylase Sirtuin-1 (Sirt-1) are unknown. We report that effects of calorie restriction on neuronal plasticity, memory and social behavior are abolished in mice lacking cAMP responsive-element binding (CREB)-1 in the forebrain. Moreover, CREB deficiency drastically reduces the expression of Sirt-1 and the induction of genes relevant to neuronal metabolism and survival in the cortex and hippocampus of dietary-restricted animals. Biochemical studies reveal a complex interplay between CREB and Sirt-1: CREB directly regulates the transcription of the sirtuin in neuronal cells by binding to Sirt-1 chromatin; Sirt-1, in turn, is recruited by CREB to DNA and promotes CREB-dependent expression of target gene peroxisome proliferator-activated receptor-γ coactivator-1α and neuronal NO Synthase. Accordingly, expression of these CREB targets is markedly reduced in the brain of Sirt KO mice that are, like CREB-deficient mice, poorly responsive to calorie restriction. Thus, the above circuitry, modulated by nutrient availability, links energy metabolism with neurotrophin signaling, participates in brain adaptation to nutrient restriction, and is potentially relevant to accelerated brain aging by overnutrition and diabetes.
Progressive decline of cognitive functions and an increased risk of developing neurodegenerative disorders, such as Alzheimer's and Parkinson diseases, are severe and debilitating consequences on brain senescence. A moderate reduction of food intake (calorie restriction, CR) delays aging and improves resistance to disease in a fashion that is evolutionarily conserved from yeast to primates and humans (1), and these beneficial effects include in mammals the prevention of age-associated cognitive impairment and neurodegeneration (2). Conversely, obesity and type 2 diabetes increase the risk of developing Alzheimer's disease (AD) (3), and reduced insulin signaling in the brain may contribute to neurodegeneration (4). Elucidation of the molecular mechanisms whereby nutritional/metabolic cues impinge on neuronal survival and health may be an avenue to new pharmacological strategies that exploit nutrient-sensitive protective circuitries to prevent the catastrophic impact of aging and dysmetabolism on the brain.
Most of the current molecular knowledge on the beneficial effects of CR involves Sirtuins, the mammalian homologs of the yeast Silent Information Regulator (Sir2.1) NAD+-dependent histone deacetylase (5, 6). This class of enzymes plays an evolutionarily conserved role in promoting genomic stability, cell survival, and stress resistance in response to limited nutrient availability. Such action results in extended longevity in lower organisms, and prevents in mammals a variety of age-associated pathologic changes from cardiovascular disease and diabetes, to cancer, to neurologic disorders. In particular, neuronal Sirt-1 has been shown to regulate endocrine and behavioral responses to CR (7) and to prevent AD amyloid neuropathology in dietary-restricted mice (8, 9). To date, however, the nutrient-sensitive signaling cascades upstream of sirtuins, as well as their downstream molecular effectors, have been incompletely characterized, especially in the brain.
Another class of neuroprotective factors, neurotrophins, promotes neuronal cell health by triggering genetic programs that are largely dependent on the cAMP responsive-element binding (CREB) factor (10, 11); accordingly, neurotrophin and CREB activities are critically reduced in the context of aging and of age-associated brain diseases (12–14). Of note, CREB also serves as a metabolic sensor in several tissues, including the brain (15), and a functional interplay between CREB and Sirt-1 in neurons through miR134 has been recently reported (16). It is therefore conceivable that CREB integrates neurotrophic and metabolic signals in the orchestration of complex neuroprotective responses that oppose brain aging.
This attractive hypothesis prompted us to investigate whether: (i) neuronal CREB has a role in brain response to CR and, by extension, in the metabolic regulation of brain function and health in mice; and (ii) this unique CREB function involves molecular interactions with Sirt-1.
Results
Effects of CR on Long-Term Potentiation, Memory, and Behavior Are Impaired in Mice Lacking Neuronal CREB.
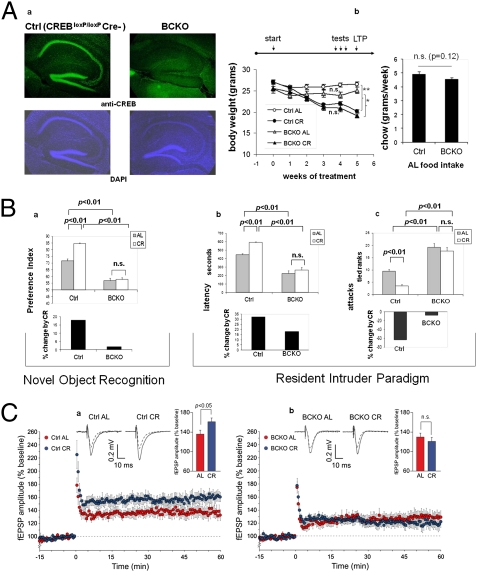
Mice subject to moderate CR display behavioral (increase in physical and exploratory activity) and cognitive (improved object memory and spatial learning) changes largely related to increased wakefulness and food-seeking (17, 7). To explore the potential role of CREB in brain response to dietary restriction, we took advantage of a “floxed” conditional mutant mouse strain (CREB1loxP/loxP) in which postnatal inactivation of the gene in the forebrain is driven by a Cre recombinase transgene under the transcriptional control of the calcium/calmodulin-dependent protein kinase II-α gene (Camk2a) promoter (18). Brain immunostaining of 6-mo-old CREBloxP/loxPCamkCre+ (henceforth indicated as BCKO, Brain CREB KO) male mice confirmed a dramatic reduction of CREB1 signal in cortex and hippocampus compared with control littermates (Fig. 1Aa). These mice appear phenotypically normal, although slightly smaller than their recombinase-negative littermates (CREBloxP/loxP CamkCre negative, henceforth “control” mice) (Fig. 1Ab). BCKO mice and their CREB-proficient controls were left with free access to food (Ad libitum regimen, AL) or subject to a CR regimen (70% of the AL food intake of the corresponding genotype) for a period of 5 wk; food consumption, and effect of food restriction on body weight, both monitored weekly, were comparable in the two strains (Fig. 1Ab), indicating that neither feeding behavior nor gross energy metabolism were affected in mutant mice.
Fig. 1.
Impaired brain response to calorie restriction in mice lacking neuronal CREB. (Aa) Immunofluorescence analysis reveals a near complete deletion of CREB1 in the cortex and hippocampus of 6-mo-old BCKO mice. Nuclear staining with DAPI confirms normal cellularity in the same areas. (b) Weight loss under calorie restriction (Left) and food consumption ad libitum (Right) in control (Ctrl) and BCKO mice (n = 5–6 per group, one of two independent experiments); (Left) **P < 0.01; *P < 0.05; n.s. nonsignificant by two-way ANOVA (4 wk time-point); (Right) P by two-tailed t test. (B) Cognitive and behavioral effects of CR. (a) (Upper) Preference toward the novel object in a novel object recognition paradigm. Values (seconds) are mean ± SEM. (Lower) Percent change by CR. (b and c) (Upper) Latency of the first attack (Left, values in seconds, mean ± SEM) and number of attacks in 10 min (Right, rank in an ordinal scale ± SEM) were scored in a resident-intruder paradigm. (Lower) Percentage changes of the corresponding parameter by calorie restriction in control and BCKO mice datasets (n = 6 animals per group) were analyzed by two-way ANOVA; P values are indicated. Experiments were performed twice with similar results. (C) Time course of Schaffer collateral-CA1 LTP induced by tetanic stimulation in control (a) and BCKO (b) mice. Values are percentages of baseline fEPSP amplitude (100%). (Insets) Representative fEPSPs at baseline (dashed line) and during the last 10 min of LTP recording (solid line). Traces are averages of 10 consecutive responses at the time points selected. Bar graphs compare average LTP magnitudes observed during the last 10 min of recording (percentage of baseline fEPSP amplitude). (Student t test, *P < 0.05).
We first asked whether CREB deletion affected the beneficial effect of CR on the cognitive performance of our mice. To this end, a standard novel object recognition paradigm (19) was used to assess object memory 24 h after a training section. We found that preference for the novel object (expressed as preference index) was significantly more pronounced in control than in BCKO mice (Fig. 1Ba), consistent with a reported role for CREB in memory consolidation (20). Importantly, memory performance was clearly improved by CR in control mice (P < 0.01 by bifactorial ANOVA) but not in the BCKO strain (Fig. 1Ba), suggesting a major role for CREB in the effect of nutrient restriction on this cognitive task.
CR reduces male aggressiveness in mice, most likely by acting on the brain serotoninergic system (21). Prompted by the observation that BCKO male mice appeared more aggressive than their control littermates, we decided to investigate this aspect in detail in the context of a resident-intruder paradigm (22). BCKO mice fed AL attacked the intruder with shorter latency (Fig. 1Bb) and higher frequency (Fig. 1Bc) compared with controls, confirming a more aggressive behavior of these animals. This behavior pheno-copies that of mice lacking neuronal nitric oxide synthase (nNOS) (22), an enzyme transcriptionally regulated by CREB (23). Aggressiveness was markedly diminished in CR control mice compared with littermates fed AL (P < 0.01, two-way ANOVA); conversely, BCKO mice showed only a marginal decrease of aggressive behavior in response to food restriction (Fig. 1B, b and c). Thus, neuronal CREB appears to be necessary for at least some of the cognitive and behavioral changes induced by CR in mice. Importantly, additional controls ruled out the possibility that the small difference in food intake between control and BCKO mice or the expression of the Cre recombinase per se in the latter strain may contribute to the above CREB-related phenotypic changes (Fig. S1).
Long-term potentiation (LTP) in the hippocampus underlies higher-order brain functions, including object and spatial memory. This indicator of neuronal plasticity declines in rodents with age, in a fashion that is attenuated by calorie restriction (24). To extend our analysis of CR effects on the brain of CREB-deficient mice, we tested LTP at CA3-CA1 synapses in hippocampal brain slices obtained from animals euthanized after 5 wk of either CR or AL feeding regimen. As previously shown (25), LTP was not affected by CREB status in the two AL-fed groups. Strikingly, however, LTP increased in control mice under CR [AL 136.6 ± 8.0% (n = 10 slices) vs. CR 161.3 ± 8.2% (n = 9 slices); P < 0.05] but not in the BCKO strain [AL 130.3 ± 8.6% (n = 9 slices) vs. CR 121.0 ± 8.5% (n = 10 slices), n.s.] (Fig. 1C), indicating that beneficial effect of CR on LTP is also abrogated by brain-specific CREB deletion. The increased LTP induced by CR in control mice was independent on changes in basal synaptic transmission, because input/output curves obtained plotting field excitatory postsynaptic potential (fEPSP) amplitudes vs. stimulus intensity in hippocampal brain slices of CR and AL animals were superimposable.
Brain CREB Is Activated by CR and Increases the Expression of Sirt-1.
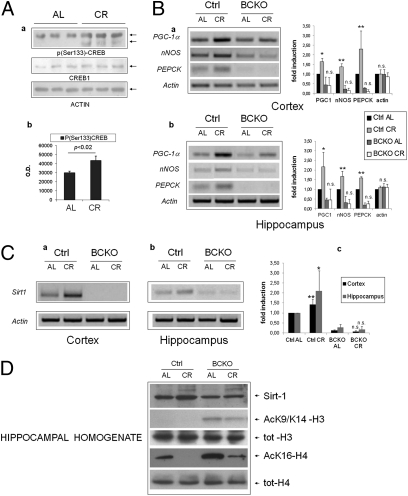
Because CREB appeared to participate in brain response to CR, we assessed whether diet affected the total amount/phosphorylation of CREB protein, and the expression level of a number of mRNAs known to be regulated by CREB (23). CREB1 protein expression in control mice was not changed by the dietary regimen; instead, an increase in CREB phosphorylation on Serine 133 in the hippocampi of the CR group was revealed by phospho-specific immunoblotting (Fig. 2A), suggesting that CR activates CREB in this brain area. Accordingly, mRNAs of “canonical” CREB targets peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), nNOS, and phosphoenolpyruvate carboxykinase (PEPCK) were induced by CR in the cortex and hippocampus of CREB-proficient mice (Fig. 2B). Of note, both PGC-1α and NO, the product of NOS enzymes, are known to promote mitochondrial biogenesis and to participate in organismal response to CR (26). The three mRNAs were overall down-regulated in BCKO mice fed AL compared with controls, and their induction by CR, observed in control mice, was nearly abolished (Fig. 2B). Interestingly, the expression of other putative CREB targets like bcl-2, NGF, and c-fos were unaffected by CR and CREB deletion (Fig. S2). Thus, collectively, these findings demonstrate that nutrient availability selectively regulates CREB-dependent gene expression in the forebrain.
Fig. 2.
Brain CREB is activated by calorie restriction and increases the expression of Sirt-1. (Aa) Western blot analysis of hippocampal homogenates from individual mice revealing increased phosphorylation of CREB on Ser-133 by calorie restriction. (b) Band densitometry normalized to actin; statistics by two-tailed t test. (B) RT-PCR analysis of three canonical CREB target mRNAs in the cortex (a) and hippocampus (b) of control and BCKO mice after 4 wk of AL or CR feeding. Actin was used as loading control. Histograms report fold-induction values compared with Ctrl AL (mean ± SD of three to five mice). *P < 0.05; **P < 0.01; n.s., nonsignificant by two-way ANOVA (CR vs. AL). (C) RT-PCR analysis of Sirt-1 mRNA expression and up-regulation by CR in the cortex (a) and hippocampus (b) of control and BCKO mice. Histogram in c displays fold-induction values relative to Ctrl AL (mean ± SD of three to five mice). Statistics as in B. (D) Western blot analysis of hippocampal homogenates showing impaired up-regulation of Sirt-1 by CR and increased acetylation of histones H3 and H4 at Sirt-1-sensitive sites in BCKO mice. Anti-total H3 and H4 histones confirms equal protein input throughout the lanes. Each lane is the pool of hippocampi from two different mice.
We next asked how CR may affect CREB activity. Because Sirt-1 is a metabolic sensor involved in several biological consequences of nutrient deprivation (27, 6), and animals lacking Sirt-1 in the brain show defective behavioral and hormonal responses to CR (7), we investigated the sirtuin as a potential CREB interactor. Sirt-1 mRNA was drastically reduced both in the cortex and hippocampus of BCKO mice, irrespective of the dietary regimen. Moreover, the moderate induction by CR that we observed in control brains was completely lost in the corresponding CREB-deficient tissues (Fig. 2C). Finally, acetylation of Histones H3 (AcK9) and H4 (AcK16), an inverse correlate of Sirt-1 activity (28), was abnormally high in BCKO hippocampal homogenates (Fig. 2D), confirming an overall reduction of Sirt-1 activity in this area of the brain.
Transcriptional Regulation of Sirt-1 by CREB in Neurons.
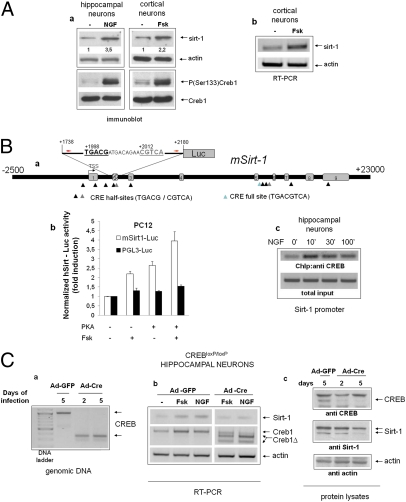
These observations suggested that Sirt-1 may represent a direct transcriptional target of CREB. Accordingly, CREB activators NGF and Forskolin (Fsk) (29) raised the level of immunoreactive Sirt-1 and of the corresponding mRNA in cultured primary cortical and hippocampal neurons (Fig. 3A). Bioinformatic analysis of the mouse Sirt-1 locus (NC_000076) revealed the presence of several putative cAMP Responsive Elements (CRE) both upstream and downstream of the transcription start site (TSS) (30), and an ∼300-bp segment encompassing two of those elements (TGACG at +1,998 and CGTCA at +2,012 from the annotated TSS) drove transcriptional response to Fsk and PKA in a standard luciferase reporter assay performed in PC12 pheochromocytoma cells (Fig. 3B, a and b). Moreover, ChIP from hippocampal neurons revealed that CREB binds the same genomic region in a fashion inducible by NGF (Fig. 3Bc). Finally, in primary neurons isolated from CREBloxP/loxP mice, deletion of CREB by adenoviral delivery of the Cre recombinase (Fig. 3C, a and b) led to a reduction of Sirt-1 immunoreactivity (Fig. 3Cc), and inhibited Sirt-1 mRNA induction by NGF and Fsk (Fig. 3Cb). Thus, we conclude that CREB directly regulates Sirt-1 mRNA and protein expression in neurons.
Fig. 3.
Transcriptional regulation of Sirt-1 by CREB in neuronal cells. (Aa) Immunoblot analysis of whole-cell lysates from primary neurons (hippocampal or cortical) exposed to CREB-activating stimuli. NGF, 50 ng/mL; Fsk, 10 μM. Relevant bands are indicated by arrows. Relative densitometric values for the Sirt-1 band are indicated. CREB phosphorylation and Sirt-1 expression were assayed at different times (30 min and 16 h, respectively). (b) RT PCR analysis of Sirt-1 mRNA in cortical neurons treated with Fsk for 6 h. Actin was amplified as an internal loading control. Panels representative of several independent experiments. (Ba) Scheme displaying several putative CRE elements within the mouse Sirt-1 gene (MGSCv37 C57BL/6J, locus NC_000076). The segment inserted in the Sirt-1-Luc reporter gene, and primers used in ChIP studies (red arrows) are indicated. Two CRE half-sites internal to the segment are also highlighted; numbers are positions relative to the annotated TSS. Exons refer to transcript variant 1. The exon 2 box is shaded because this exon is absent in transcript variants 2 and 3. (b) Luciferase reporter assay confirming responsiveness of the 1738–2180 genomic fragment to Fsk and PKA in PC12 cells. Bars are fold-induction ± SD of triplicate samples; picture representative of two independent experiments. (c) ChIP assay showing NGF-induced binding of CREB to the 1824–2090 Sirt-1 region in hippocampal neurons. Minutes of stimulation are indicated. Sirt-1 promoter was amplified from the total chromatin input as quantitative control (Lower). (Ca) Deletion of CREB exon 10 in cultured CREBloxP/loxP hippocampal neurons 2 or 5 d after adenoviral delivery of Cre recombinase (Ad-Cre). Genomic DNA was amplified with two primers external to the recombination sites. Bands corresponding to undeleted (Upper) and deleted (Lower) alleles are indicated by arrows. (b) RT-PCR analysis revealing defective induction of Sirt-1 mRNA by NGF and Fsk (6 h) in CREB-deleted hippocampal neurons. Actin was amplified as loading control. Bands corresponding to deleted (ΔCREB) and residual undeleted CREB mRNA are indicated by arrows. Picture is representative of several independent experiments. (c) Western blot analysis of whole cell lysates from mock (Ad-GFP) and Cre-infected cells indicating reduced expression of Sirt-1 in the latter cell population. Actin band confirms equal protein loading.
Sirt-1 Binds CREB and Promotes CREB-Dependent Gene Expression.
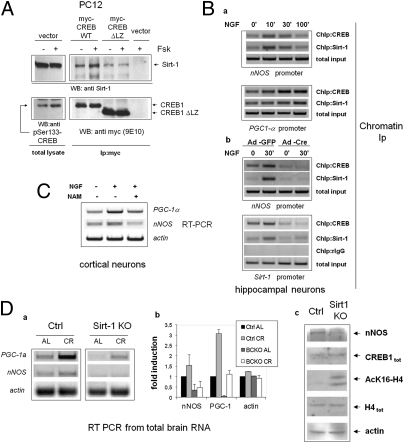
Known transcriptional regulators of Sirt-1, such as p53 and Foxo3a (31), also interact with, and are either positively or negatively modulated by the deacetylase (32, 33); similarly, a myc-tagged form of human CREB1 and Sirt-1 could be easily coimmunoprecipated in naive PC12 cells (Fig. 4A), indicating physical association between the two proteins; the association was rapidly induced by the PKA agonist Fsk, in parallel with phosphorylation of CREB on Serine 133. Short-time (30 min) stimulation with Fsk did not affect the total amount of cellular Sirt-1, but rather increased the stoichiometry of the binding to CREB, suggesting either an increased affinity between the two proteins, or the enhanced interaction with intermediate molecular partners co-recruited at target gene promoters. In keeping with the latter possibility, a mutant CREB that retains the phosphorylation site but is unable to bind DNA because of the deletion of the C-terminal transactivation domain (ΔLZ CREB), displayed marginal physical interaction with Sirt-1, nor was the binding induced by Fsk (Fig. 4A).
Fig. 4.
Sirt-1 promotes CREB-dependent gene expression. (A) Forskolin-inducible physical association of CREB with Sirt-1 in PC12 cells. mycCREB or mycCREB-ΔLZ were transfected in PC12 cells and immunoprecipitated from protein lysates untreated or stimulated with Fsk for 30 min; the presence of Sirt-1 in immunocomplexes was verified by immunoblotting (Upper); anti-Ser133 CREB and anti-myc immunoblotting were used to confirm CREB phosphorylation by Fsk and assess the expression level of transfected CREB isoforms, respectively (Lower). (Ba) ChIP assays showing parallel interaction of CREB and Sirt-1 with the CRE-containing promoter regions of nNOS and PGC-1α in hippocampal neurons treated with NGF. Promoter amplification from total chromatin input is also reported as control. (b) CREB mediates Sirt-1 interaction with CRE-containing promoter regions. NGF-inducible binding of Sirt-1 to nNOS (Upper) and Sirt-1 promoter (Lower) are drastically reduced in hippocampal neurons lacking CREB. Binding of CREB to the same promoter regions confirms severe reduction of CREB binding in Cre-infected cells. rIgG (rabbit IgG) is a negative control for ChIP. Total chromatin input was equal throughout the lanes. (C) RT-PCR analysis showing reduced induction of nNOS and PGC-1α mRNA by NGF in cortical neurons treated with the Sirt-1 inhibitor Nicotinamide (NAM). (D) (a and b) Representative RT-PCR analysis of PGC-1α and nNOS mRNA expression in whole brains from WT and Sirt-1-deficient mice under both AL and CR (6 mo) feeding. Each lane represents a pool of two mice. Actin was used as loading control. (c) Western blot analysis of whole brain protein homogenates from WT and Sirt-1 KO mice (fed AL) indicating reduced expression of nNOS but normal levels of immunoreactive CREB and total histone H4. Anti-AcH4K16 and antiactin immunostaining confirm, respectively, reduced deacetylase activity in the SirtKO sample and equal protein loading in the two lanes.
To confirm that SIRT1 and CREB colocalize on CREB-responsive chromatin regions in cortical neurons, cross-linked chromatin was immunoprecipitated with an anti-Sirt-1 antiserum and Sirt-1 binding to the regions of nNOS and PGC-1α promoters surrounding the CRE elements determined by semiquantitative PCR. Sirt-1 was found to interact with both genes, in a fashion inducible by NGF and with a binding kinetic similar to that of CREB (Fig. 4Ba). Sirt-1 also bound the same Sirt-1 chromatin region surrounding the +1,998/+2,012 half-CRE sites that copre-cipitates with CREB, suggesting that Sirt-1 may self-regulate its CRE-dependent transactivation (Fig. 4Bb). Of note, Sirt-1 interaction with nNOS and Sirt-1 chromatin was indeed mediated by CREB, as revealed by its drastic reduction in CREB loxP/loxP neurons following the recombinase-mediated deletion of the factor (Fig. 4Bb) and by the failure of forced Sirt-1 overexpression to restore Sirt-1 chromatin binding in CREB-deficient cells (Fig. S3).
To determine whether Sirt-1 modulates CREB transcriptional activity, cortical neurons were stimulated with NGF in the presence of sirtuin inhibitor Nicotinamide (34); induction of nNOS and PGC-1α mRNA by NGF was blunted upon inhibition of Sirt-1 deacetylase activity (Fig. 4C). A similar result was obtained in naive PC12 cells by siRNA-mediated knock-down of Sirt-1 (Fig. S4A). In addition, Sirt-1–deficient PC12 displayed impaired differentiation by NGF (as assessed by quantification of neurite outgrowth), a response largely dependent on CREB and nNOS in this cell model (35, 36) (Fig. S4B). In a complementary set of experiments, lentivirus-mediated overexpression of Sirt-1 increased the expression of nNOS and PGC-1α mRNA in hippocampal neurons treated with NGF. However, Sirt-1 did not fully restore the expression of these genes in CREB-deficient cells, nor NGF protection from hydrogen peroxide-dependent cell death, which is largely lost in these cells (Fig. S5), was recovered upon transduction of the Sirt-1 cDNA. Collectively, the above data suggest that Sirt-1 and CREB are, at least to some extent, reciprocally dependent, and act in concert in the context of neurotrophin signaling.
Unlike previously reported findings (16), manipulations aimed at modulating Sirt-1 expression and activity in the above contexts did not affect the level of immunoreactive CREB1 (Fig. S4Aa).
Defective Expression of CREB Target Genes in the Brain of CR Sirt-1 KO Mice.
Finally, to verify whether Sirt-1 actually regulates CREB-dependent transcription in vivo and this is relevant for response to CR, we analyzed the expression of CREB targets nNOS and PGC-1α mRNAs in whole brains of Sirt-1 KO mice fed AL or CR for 25–28 wk (37). This analysis revealed that both genes are markedly hypoexpressed in Sirt-deficient mice compared with littermate controls under both dietary regimens (Fig. 4D), and their induction by CR, that was more pronounced for PGC-1α, was also attenuated (Fig. 4D, a and b). The amount of immunoreactive CREB was comparable in brain homogenates of the two strains (Fig. 4Dc), nor CREB acetylation on lysine residues was affected by either Sirt-1 deletion or CR (Fig. S6), indicating that, at least in these experimental settings, Sirt-1 regulates CREB transcriptional activity independent of the acetylation and expression level of the factor.
Taken together, the above findings strongly suggest that Sirt-1 modulates the expression of CREB-dependent genes in mouse brain, and by extension, identify in the decrease of CREB-dependent transcription a molecular signature for a defective brain response to CR, shared by CREB and Sirt-1 mutant mice (7).
Discussion
The CREB transcription factor has been widely investigated as a metabolic sensor and regulator of glucose homeostasis in liver and fat tissue (15), and as a master switch of calcium and neurotrophin-triggered transcriptional programs regulating neuronal differentiation, survival, and plasticity in the brain and peripheral nervous system (10, 11, 18). Evidence also exist for CREB roles in the control of appetite and food intake in the hypothalamus (38), but whether neuronal plasticity and high-order cognitive functions may be influenced by nutrient cues and energy metabolism through CREB, an issue relevant to major diseases like AD and type 2 diabetes (3, 4), remains to be established. Our demonstration of impaired electrophysiological, cognitive, and emotional response to CR in brain CREB KO mice, clearly suggests that this may indeed be the case. Interestingly, these differences emerge in the context of an overall comparable feeding behavior and metabolic response to CR between control and BCKO mice (Fig. 1Ab), indicating that potential effects of hypothalamic CREB signaling on energy balance and appetite regulation unlikely account for the observed phenotypes (38).
Data reported in Fig. 2, showing CREB phosphorylation and transcriptional activation by CR, indicate that CREB is metabolically regulated in the cortex and hippocampus, although the mechanism of such regulation needs to be further investigated. Because plasma from CR rodents has been previously shown to induce Sirt-1 expression in several organs/tissues, a humoral/hormonal mechanism for CREB activation could be envisaged (39); as an alternative, NO (40) or oxygen species (41) may mediate, cell-autonomously, this effect. Further research in this direction is warranted.
Our data identify in the complex interplay with the nutrient-sensitive histone deacetylase Sirt-1 a molecular connection between CREB and neuronal response to CR. Previous work from others has compellingly involved neuronal Sirt-1 in hormonal and metabolic adaptation to dietary restriction in mice (7). Results presented here clearly suggest that the two molecules are part of the same CR-sensitive signaling cascade. However, other genes potentially relevant to mitochondrial biogenesis and neuronal response to calorie restriction, namely nNOS and PGC-1α (40), and some PGC-1α targets—including CPT1, CoxIV, and UCP-2—are also critically down-regulated, together with Sirt-1, in the cortex and hippocampus of BCKO mice (Fig. 2B and Fig. S2b). This finding, and evidence from Fig. S3 that overexpression of Sirt-1 does not rescue NGF signaling in CREB-deficient hippocampal neurons in vitro, suggests that up-regulation of Sirt-1 is not the only mechanism whereby CREB participates in neuronal response to nutrients. Instead, cooperation with CREB is likely critical for the action of neuronal Sirt-1 in CR. This view is supported by the unexpected finding that CREB-dependent genes involved in neuronal plasticity, survival, and stress resistance (42, 43), and induced by calorie restriction (Fig. 2), are markedly down-regulated in Sirt-1–deficient cultured neurons (PC12) and in the brain of Sirt-1 KO mice. Because the latter strain is, like BCKO mice, impaired in brain response to reduced food intake (7), the above evidence further support the notion that CREB-dependent transcription has a pivotal role in the neuronal effects of calorie restriction, and identify in the CREB–Sirt-1 axis a major component of the nutrient sensitive molecular network that connects caloric intake and energy metabolism to brain health.
Biochemical details of how Sirt-1 affects CREB activity remains to be clarified. Unlike other transcription factors, CREB does not appear to be deacetylated by Sirt-1 (Fig. S6); moreover, we could not detect consistent effects of Sirt-1 on the expression of CREB (Fig. 4Dc, and Figs. S4Aa and S6), unlike that described by Gao et al. (16). Those authors, however, made their important observations on Sirt-1 Δex4 mice that, unlike Sirt-1KO mice used in our experiments, do express an inactive form of Sirt-1 potentially acting in a dominant negative fashion against other molecules (deacetylases?) capable of regulating CREB expression.
On the other hand, Sirt-1 dependent modulation of nutrient sensitive cofactors of CREB, like TORC/Crtc (38), has been described in the liver and may occur in neuronal cells. Alternatively, Sirt-1 may be recruited by CREB on target promoters and act either on histones (Fig. 2D) or, in trans, on other (maybe competing) transcription factors.
In conclusion, CREB1, an effector of neurotrophins involved in several age-associated neurodegenerative diseases, mediates at least some brain responses to dietary restriction. This action involves the up-regulation of Sirt-1 that in turn bolsters the CREB-dependent expression of genes involved in neuronal metabolism, survival and plasticity (Fig. S7). Although molecular details on how Sirt-1 regulates gene transcription by CREB need to be further investigated, the CREB-Sirt-1 axis outlines a unique molecular network at the crossroad of energy metabolism, metabolic diseases, and brain aging.
Materials and Methods
Mice.
BCKO (CREB1loxP/loxP- Camk2aCRE) mice, Sirt-1 KO mice, and the relative control strains have been previously described elsewhere (18, 37).
Calorie Restriction.
For CR studies, the amount of food consumed by AL mice was determined weekly, and CR mice were fed daily 80% of that value for the first week and 60% for the following 4 wk. Absolute food consumption AL was slightly (about 10%) higher in control than in BCKO mice, but the amount of chow per gram body weight comparable, because of the smaller size of the latter strain. CR in BCKO mice was calculated either on the AL feeding of mice of the same genotype, or on the AL consumption of control mice, with similar results. Body weight was monitored weekly.
Behavioral Tests.
All behavioral tests were conducted on male, 6-mo-old mice, during the dark cycle (the animals’ active phase). Resident-intruder and novel object recognition paradigms were performed according to refs. 11 and 13, respectively, with minimal changes.
Long-Term Potentiation.
Coronal hippocampal slices (400-μm thick) were obtained from adult male C57BL/6 mice. fEPSP evoked by Schaffer collateral stimulation were recorded from the CA1 subfield of the hippocampus. The stimulation intensity that produced one-third of the maximal response was used for the test pulses and LTP induction protocol consisting of four trains of 50 stimuli at 100 Hz repeated every 20 s (referred to as tetanus). The magnitude of LTP was measured 60 min after tetanus and expressed as a percentage of baseline fEPSP peak amplitude.
Statistics.
Datasets were compared by bifactorial (2 × 2) ANOVA or two-tailed Student t test where appropriate, using either raw or tied-ranked values. Threshold for significance was set at P < 0.05.
Additional methods and the associated references are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Drs. M. Greenberg and D. Ginty for DNA constructs; Drs. Giuseppe Maulucci and Vito Vetrugno for critically reading the manuscript; and the laboratory members for experimental contributions and valuable suggestions. This study was funded by Grant EASD/Glaxo Smith Kline from the European Association for the Study of Diabetes and Grant IG8634/2009 from the Italian Association for Cancer Research (to G.P.), and by Catholic University intramural Grants Linea D.1 (to G.P.), D3.2 (to C.G.), and D3.2 (to T.G.). S.F. is the recipient of a Research Doctorate Fellowship from the Italian Ministry of University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109237109/-/DCSupplemental.
References
- 1.Fontana L, Partridge L, Longo VD. Extending healthy life span—From yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maalouf M, Rho JM, Mattson MP. The neuroprotective properties of calorie restriction, the ketogenic diet, and ketone bodies. Brain Res Brain Res Rev. 2009;59:293–315. doi: 10.1016/j.brainresrev.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda S, et al. Diabetes-accelerated memory dysfunction via cerebrovascular inflammation and Abeta deposition in an Alzheimer mouse model with diabetes. Proc Natl Acad Sci USA. 2010;107:7036–7041. doi: 10.1073/pnas.1000645107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Monte SM. Insulin resistance and Alzheimer's disease. BMB Rep. 2009;42:475–481. doi: 10.5483/bmbrep.2009.42.8.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haigis MC, Guarente LP. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 6.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin W, et al. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 9.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 11.Finkbeiner S. CREB couples neurotrophin signals to survival messages. Neuron. 2000;25:11–14. doi: 10.1016/s0896-6273(00)80866-1. [DOI] [PubMed] [Google Scholar]
- 12.Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 13.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA. 2010;107:22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halagappa VK, et al. Intermittent fasting and caloric restriction ameliorate age-related behavioral deficits in the triple-transgenic mouse model of Alzheimer's disease. Neurobiol Dis. 2007;26:212–220. doi: 10.1016/j.nbd.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Mantamadiotis T, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 19.Bevins RA, Besheer J. Object recognition in rats and mice: A one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc. 2006;1:1306–1311. doi: 10.1038/nprot.2006.205. [DOI] [PubMed] [Google Scholar]
- 20.Bourtchuladze R, et al. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 21.Koizumi H, Hashimoto K, Iyo M. Dietary restriction changes behaviours in brain-derived neurotrophic factor heterozygous mice: Role of serotonergic system. Eur J Neurosci. 2006;24:2335–2344. doi: 10.1111/j.1460-9568.2006.05094.x. [DOI] [PubMed] [Google Scholar]
- 22.Nelson RJ, et al. Behavioural abnormalities in male mice lacking neuronal nitric oxide synthase. Nature. 1995;378:383–386. doi: 10.1038/378383a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hori N, Hirotsu I, Davis PJ, Carpenter DO. Long-term potentiation is lost in aged rats but preserved by calorie restriction. Neuroreport. 1992;3:1085–1088. doi: 10.1097/00001756-199212000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Balschun D, et al. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nisoli E, Clementi E, Moncada S, Carruba MO. Mitochondrial biogenesis as a cellular signaling framework. Biochem Pharmacol. 2004;67:1–15. doi: 10.1016/j.bcp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 28.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 29.Riccio A, et al. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Impey S, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 31.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 32.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 33.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 34.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 35.Du K, Asahara H, Jhala US, Wagner BL, Montminy M. Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo. Mol Cell Biol. 2000;20:4320–4327. doi: 10.1128/mcb.20.12.4320-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peunova N, Enikolopov G. Nitric oxide triggers a switch to growth arrest during differentiation of neuronal cells. Nature. 1995;375:68–73. doi: 10.1038/375068a0. [DOI] [PubMed] [Google Scholar]
- 37.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altarejos JY, et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat Med. 2008;14:1112–1117. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 40.Nisoli E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 41.Bedogni B, et al. Redox regulation of cAMP-responsive element-binding protein and induction of manganous superoxide dismutase in nerve growth factor-dependent cell survival. J Biol Chem. 2003;278:16510–16519. doi: 10.1074/jbc.M301089200. [DOI] [PubMed] [Google Scholar]
- 42.Dawson VL, Dawson TM. Nitric oxide in neurodegeneration. Prog Brain Res. 1998;118:215–229. doi: 10.1016/s0079-6123(08)63210-0. [DOI] [PubMed] [Google Scholar]
- 43.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.