Abstract
Synthesis of many proteins is tightly controlled at the level of translation, and plays an essential role in fundamental processes such as cell growth and proliferation, signaling, differentiation, or death. Methods that allow imaging and identification of nascent proteins are critical for dissecting regulation of translation, both spatially and temporally, particularly in whole organisms. We introduce a simple and robust chemical method to image and affinity-purify nascent proteins in cells and in animals, based on an alkyne analog of puromycin, O-propargyl-puromycin (OP-puro). OP-puro forms covalent conjugates with nascent polypeptide chains, which are rapidly turned over by the proteasome and can be visualized or captured by copper(I)-catalyzed azide-alkyne cycloaddition. Unlike methionine analogs, OP-puro does not require methionine-free conditions and, uniquely, can be used to label and assay nascent proteins in whole organisms. This strategy should have broad applicability for imaging protein synthesis and for identifying proteins synthesized under various physiological and pathological conditions in vivo.
Keywords: click chemistry, microscopy, ribosome
The entire set of cellular proteins is generated through translation of mRNAs by ribosomes. The identity and amount of the proteins that a cell synthesizes are critical parameters in determining the physiological state of the cell. Protein synthesis is frequently not proportional to mRNA levels, primarily because translation is often tightly regulated, and many critical controls in gene expression occur at the level of translation (1–3). Under specific conditions (such as heat shock, starvation, availability of iron, etc.), translational controls ensure that synthesis of specific cellular proteins is quickly turned on or off. Translational controls are particularly prominent in systems in which transcription is inhibited, such as in early embryonic development before the onset of zygotic transcription. Furthermore, translation of many proteins is spatially localized, as underscored by the finding that the majority of mRNAs in Drosophila embryos display distinct subcellular patterns (4).
Understanding how gene expression is regulated at the level of translation, spatially and temporally, requires tools for visualizing and identifying nascent polypeptide chains. One of the major methods used for this purpose relies on the biosynthetic incorporation of azide- or alkyne-bearing methionine (Met) analogs such as azidohomoalanine (Aha) (5–7) or homopropargylglycine (Hpg) (7, 8). The resulting azide- or alkyne-labeled proteins can be detected by copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (9–11) with reagents for fluorescent detection (8) or for affinity purification and identification by mass spectrometry (5). Though simple and robust, this method has a number of drawbacks. Cells prefer Met over Aha or Hpg by a factor of about 500 (8), which means cultured cells need to be labeled with Aha or Hpg in Met-free media, a limitation that precludes the use of Aha and Hpg to study protein synthesis in whole animals. To be incorporated into proteins, Aha and Hpg need to be activated as aminoacyl-tRNAs, a step which limits the temporal resolution of this method. Finally, this method generates full-length Aha- or Hpg-labeled proteins, not nascent polypeptide chains.
Other methods to assay translation rely on the incorporation of puromycin (puro) into nascent proteins. Puro (Fig. 1A) is an aminonucleoside antibiotic that blocks protein synthesis in both prokaryotes and eukaryotes, by causing premature termination of nascent polypeptide chains. Puro mimics an aminoacyl-tRNA molecule and binds to the acceptor site of translating ribosomes, which leads to the formation of an amide bond between the C terminus of the nascent polypeptide chain and the primary amine group of puro (12, 13). The translation inhibition mechanism of puro has been exploited in the past to assay the rate of synthesis of specific proteins, by metabolic labeling with radioactive puro followed by immunoprecipitation of the protein of interest (14). More recently, fluorescent derivatives of puro have been used to label newly synthesized proteins in cells (15, 16) and cell-free extracts (17), although imaging nascent proteins in cells by this method has had limited applicability due to low signal-to-noise ratios (15, 16). Finally, another strategy relies on detecting nascent polypeptide-puro conjugates with anti-puro antibodies (18). Although this method works robustly on immunoblots, the subcellular pattern of nascent proteins detected by anti-puro immunofluorescence (18, 19) differs significantly from that obtained with Hpg labeling (8) and is inconsistent with the expected subcellular localization of newly synthesized proteins identified by Aha labeling and mass spectrometry (5).
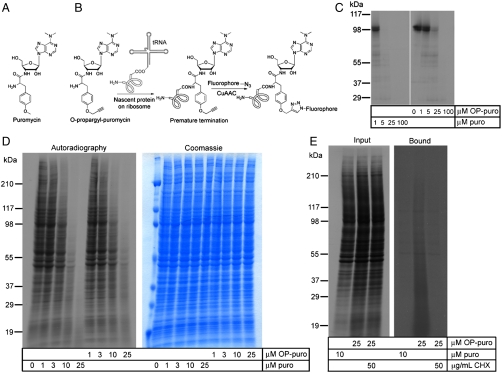
Fig. 1.
OP-puro, an alkyne puroanalog, is a potent protein synthesis inhibitor. (A) Structure of puro and the analog OP-puro, which bears a terminal alkyne group. (B) Schematic of OP-puro incorporation into nascent polypeptide chains on translating ribosomes. The prematurely terminated polypeptides are subsequently detected by CuAAC using a fluorescent azide. (C) Inhibition of protein translation in vitro by puro and OP-puro. A 35S-Met-labeled protein (a GFP fusion of mouse SuFu) was generated by translation in rabbit reticulocyte lysates, in the absence or presence of varying concentrations of puro and OP-puro. The translation reactions were separated by SDS-PAGE, and the translated protein was visualized by autoradiography. OP-puro inhibits protein synthesis in a dose-dependent manner. (D) Inhibition of protein translation in cultured cells by puro and OP-puro. Human embryonic kidney 293T cells were incubated in methionine-free media supplemented with 35S-Met, in the absence or presence of varying concentrations of puro and OP-puro. Total cell lysates were analyzed by SDS-PAGE, followed by autoradiography, to measure bulk protein translation. The gel was also stained with Coomassie blue, to demonstrate equal protein loading. (E) Formation of conjugates between OP-puro and nascent polypeptide chains. Cultured 293T cells were labeled with 35S-Met as in D), in the presence of OP-puro, puro, or OP-puro and the protein synthesis inhibitor CHX. Cellular lysates were reacted with biotin-azide under conditions for CuAAC, after which biotinylated molecules were purified on streptavidin beads. Bound proteins were eluted, separated by SDS-PAGE, and followed by autoradiography to detect nascent proteins.
From the discussion above it is clear that improved labeling techniques are still needed for the study of nascent proteins in vivo, in particular methods that are rapid, sensitive, and work well in whole organisms. To circumvent some of the limitations of current methodologies, we have developed a simple and robust method to detect nascent proteins in cultured cells and in animals, based on a puro analog that bears a terminal alkyne group, O-propargyl-puromycin (OP-puro). OP-puro inhibits protein synthesis in cells similar to puro, acts rapidly, and forms covalent polypeptide-OP-puro conjugates, which can be reacted by CuAAC with either fluorescent azides for visualization by fluorescence microscopy, or with biotin-azide for affinity isolation. Unlike Met analogues, OP-puro labels nascent proteins in whole animals, allowing the visualization of protein synthesis patterns in large explants from tissues and organs. We anticipate this method will be useful for microscopic imaging of nascent proteins in cells and in tissues, and will allow identification of proteins synthesized in vivo under various conditions.
Results
O-Propargyl-Puromycin, a Puromycin Analog That Retains Translation Inhibition Activity.
We wanted to develop a chemically-tagged puro to label newly synthesized proteins, for subsequent imaging by fluorescence microscopy and for isolation by affinity chromatography. Structure-activity studies of puro (20–24) indicated that the molecule tolerates modifications of the O-Me phenyl ring, without significant loss of activity. We synthesized O-propargyl-puromycin (OP-puro, Fig. 1B), a puro analog that bears a terminal alkyne group, which should allow detection of nascent polypeptide chains by CuAAC (11). OP-puro inhibits protein synthesis, both in reticulocyte lysates (Fig. 1C) and in cultured cells (Fig. 1D), displaying a potency two- to threefold lower than that of unmodified puro. Thus OP-puro should allow the covalent labeling of nascent polypeptide chains with a terminal alkyne group.
We next asked if OP-puro forms conjugates with nascent polypeptide chains. Cultured cells were preincubated with 35S-methionine (35S-Met), followed by incubation with 35S-Met in media supplemented with OP-puro, puro, or OP-puro and the protein synthesis inhibitor, cycloheximide (CHX). Cellular extracts were subjected to CuAAC with biotin-azide, and biotinylated molecules were purified on streptavidin beads, eluted and analyzed by SDS-PAGE, followed by autoradiography. OP-puro forms conjugates with 35S-Met-labeled nascent polypeptide chains (Fig. 1E), which can be specifically retrieved on streptavidin beads. Importantly, the OP-puro conjugates do not form if protein synthesis is inhibited by CHX, which blocks translational initiation, demonstrating that translating ribosomes are required for OP-puro incorporation into nascent polypeptide chains, at the level of polypeptide chain elongation. These results demonstrate that OP-puro is a translation inhibitor that forms covalent adducts with elongating polypeptide chains on the ribosome. A concentration of 25 μM OP-puro blocked protein synthesis almost completely, defining a concentration that should capture almost quantitatively the proteins synthesized by a given cell in the form of OP-puro-polypeptide conjugates.
Imaging Nascent Polypeptide Chains in Cells with O-Propargyl-Puromycin.
We next tested if OP-puro-labeled nascent proteins can be visualized by fluorescence microscopy using a CuAAC reaction with a fluorescent azide. When cultured cells were treated for 1 h with varying concentrations of OP-puro followed by fixation and staining with Alexa568-azide (25), a specific fluorescent signal proportional to the concentration of OP-puro was detected (Fig. 2A). At a concentration of 50 μM OP-puro, the fluorescent staining reached a signal-to-noise ratio of 24 (see graph in Fig. 2A), which is significantly higher than the signal-to-noise ratio of 2–4 reported for fluorescent puro conjugates (15, 16). Importantly, OP-puro incorporation required functional ribosomes and was abolished if cells were treated with CHX (right-most panel of Fig. 2A), as seen before by autoradiography (Fig. 1E). The intensity of the OP-puro stain increased with incubation time (Fig. 2B) and reached saturation after about 1 h. With OP-puro, a strong signal was seen after as little as 15 min of incubation. Although the OP-puro staining pattern was mostly cytoplasmic, many cells also showed a punctate nuclear stain, suggesting that the truncated protein-OP-puro conjugates released from ribosomes can localize to various subcellular compartments. The OP-puro staining pattern is very similar to the one observed in cells labeled with the methionine analog Hpg (8) and is consistent with the finding that nuclear and nucleolar proteins are among the most abundant newly synthesized proteins in cells (5). At later time points (see the 3 h image of Fig. 2B), the cytoplasmic OP-puro signal is significantly decreased, suggesting that the polypeptide-OP-puro conjugates are turned over.
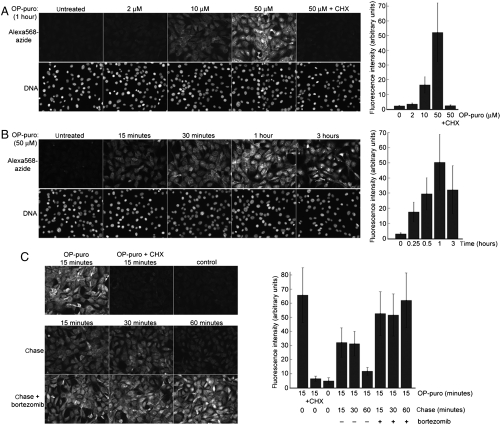
Fig. 2.
Imaging nascent proteins in cultured cells with OP-puro. (A) Cultured NIH 3T3 cells were incubated for 1 h in complete media supplemented with increasing concentrations of OP-puro, OP-puro, and CHX, or control vehicle. The cells were then fixed, stained by CuAAC with Alexa568-azide, and imaged by fluorescence microscopy. A specific signal is observed in cells treated with OP-puro, which is proportional to the concentration of added OP-puro. This signal is abolished if protein translation is blocked with CHX (50 μg/mL), which blocks translation elongation and thus prevents the formation of conjugates between nascent polypeptide chains and OP-puro. The graph on the right shows the quantification of Alexa568 fluorescence intensity in this experiment. (B) Time course of OP-puro incorporation into nascent proteins. NIH 3T3 cells were incubated with OP-puro (50 μM, which is sufficient to completely block protein synthesis) for varying amounts of time, after which OP-puro incorporation was imaged as in A. The intensity of the OP-puro signal reaches a maximum after about 1 h. The graph on the right shows the quantification of Alexa568 fluorescence intensity in this experiment. (C) The nascent protein-OP-puro conjugates are unstable and are cleared from cells in a proteasome-dependent manner. NIH 3T3 cells were treated with 50 μM OP-puro for 15 min, followed by incubation in media without OP-puro, in the absence or presence of 5 μM of the proteasome inhibitor bortezomib. Parallel cultures were fixed at the indicated times after removal of OP-puro, and nascent protein-OP-puro conjugates were imaged by CuAAC with Alexa568-azide. The OP-puro conjugates have largely disappeared after 1 h but are completely stabilized by proteasome inhibition. Untreated cells and cells incubated for 15 min with OP-puro (50 μM) and CHX (50 μg/mL) served as negative controls. The graph on the right shows the quantification of Alexa568 fluorescence intensity in this experiment.
After puro is covalently attached to the C terminus of nascent polypeptide chains, the polypeptide chain-puro conjugate is released from the ribosome, which is followed by its quick, ubiquitin-dependent proteolysis (26, 27). We asked if polypeptide-OP-puro conjugates are similarly unstable. Cultured cells were labeled with a short pulse of OP-puro, after which the cells were washed and chased in the absence or presence of the proteasome inhibitor bortezomib. In the absence of bortezomib, OP-puro conjugates are unstable and disappear from cells in under 1 h (Fig. 2C, Middle). If the proteasome is inhibited with bortezomib, the OP-puro conjugates become stable (Fig. 2C, Bottom), demonstrating that the rapid disappearance of OP-puro conjugates is due to degradation by the proteasome. We conclude that the majority of conjugates of OP-puro with nascent polypeptide chains are short-lived, which suggests that the OP-puro signal is a good reflection of instant protein synthesis in cells.
Visualizing Patterns of Protein Translation in Animals with O-Propargyl-Puromycin.
Finally we tested if OP-puro can be used to visualize nascent proteins in animals. Mice were injected intraperitoneally with OP-puro, and tissues were harvested 1 h later, fixed and stained with fluorescent azide, either after sectioning or in whole mount. As shown in Fig. 3 and in Fig. S1, tissues from uninjected mice showed low nonspecific staining, whereas tissues from OP-puro-injected mice displayed specific patterns of OP-puro incorporation into nascent proteins. In the small intestine, translation was strongest in cells in the crypts and at the base of intestinal villi (Fig. 3A), consistent with the high proliferative and secretory activity of these cells. The stain was particularly strong in Paneth cells, which are located close to the base of the crypts and are filled with secretory vesicles. The intense OP-puro labeling of vesicles in Paneth cells (Fig. 3A, Bottom) suggests that prematurely terminated, OP-puro-conjugated secretory proteins are translocated into the endoplasmic reticulum (ER) lumen, as described before for puro (28). The same pattern of OP-puro labeling was observed in whole-mount stains of the small intestine (Fig. 3B), suggesting that OP-puro is uniquely suited for whole-mount visualization of protein synthesis in tissues and organs, with high sensitivity.
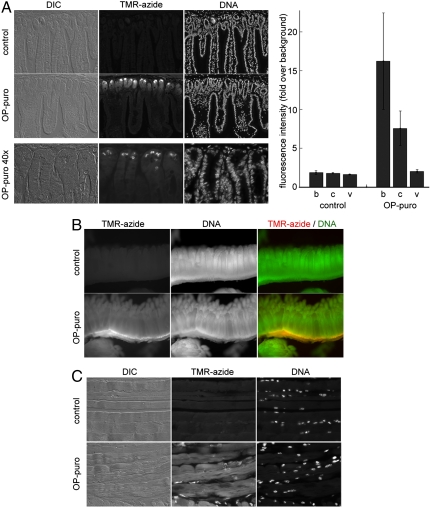
Fig. 3.
Using OP-puro to image protein synthesis in whole animals. One hundred microliters of a 20 mM OP-puro solution in PBS or PBS alone (negative control) were injected intraperitoneally into mice. Organs were harvested 1 h later, fixed in formalin, and stained by CuAAC with TMR-azide, either after paraffin sectioning or whole mount. (A) Section through mouse small intestine showing intestinal vili sectioned longitudinally. OP-puro stains strongly the cells in the crypts (particularly Paneth cells) and the cells at the base of the villi. Bottom panels show a higher magnification (40× objective) view of the intestinal crypts in an OP-puro-injected mouse. Note the intense staining of the secretory granules characteristic of Paneth cells. The graph on the right shows the quantification of TMR fluorescence in three different regions of the intestinal epithelium: the bottom of the crypts (b), the crypts proper (c), and the epithelium covering the vili (v). The rate of protein translation is significantly higher in crypts compared to vili. (B) Whole-mount staining of mouse small intestine, showing the localization of the OP-puro stain in the crypts. Protein-OP-puro conjugates were detected with TMR-azide (red), and nuclear DNA was stained with OliGreen (green). (C) OP-puro incorporation into striated muscle fibers. Paraffin sections of muscle were stained as in A. Sarcomeres are strongly stained with OP-puro, likely because some protein-OP-puro conjugates are functional and are properly assembled into sarcomeres. Images of OP-puro staining of other mouse tissues (spleen, kidney, liver) are shown in Fig. S1.
Patterns of protein synthesis in other mouse tissues surveyed (liver, kidney, and spleen) are shown in Fig. S1. The level of protein synthesis varies between tissues, being the strongest in hepatocytes (Fig. S1A), consistent with the high levels of protein synthesis in the liver. Protein synthesis levels are uniformly high in hepatocytes but can vary significantly within other tissues, such as spleen (Fig. S1C), in which the strongest OP-puro signal is found at the organ’s periphery, under the capsule. The functional significance of such patterns of protein synthesis is currently unclear and deserves further study. Interestingly, in muscle the OP-puro stain shows a striking striated pattern (Fig. 3C), suggesting that some muscle OP-puro-protein conjugates are properly incorporated into sarcomeres. Thus OP-puro might be suitable for imaging the assembly and turnover of subcellular structures such as sarcomeres.
Discussion
Regulation of mRNA translation is a critical mechanism in controlling the expression of a large number of genes, both temporally and spatially. Methods that allow imaging and biochemical analysis of protein synthesis are essential for our understanding of the role played by mRNA translation in numerous biological processes. Although powerful methods are available to detect translation of recombinant proteins in cells with high spatial and temporal resolution (29, 30), current strategies for labeling and imaging endogenous proteins have several limitations (see below). With the goal of developing a bioorthogonal method for assaying endogenous nascent proteins in whole organisms, we synthesized the alkyne-bearing analog of (OP-puro, which incorporates cotranslationally at the C terminus of nascent polypeptide chains, forming covalent conjugates that can be detected by a CuAAC reaction. We used OP-puro to label and visualize nascent proteins in cells, with high sensitivity. Uniquely, due to the small size of the fluorescent azides used for detection by CuAAC, OP-puro labeling allowed the visualization of protein synthesis patterns by whole-mount staining of large fragments of organs and tissues. Our analysis revealed that the rate of protein synthesis varies greatly between tissues and organs, as well as within tissues. We envision that this simple method will be useful for microscopic studies of protein synthesis and for the affinity isolation and mass spectrometric identification of proteins synthesized in vivo under various conditions. In particular, OP-puro should allow the identification of proteins regulated at the level of translation, such as targets of specific micro-RNAs (2, 3), targets of other RNA-binding proteins that control translation of specific mRNAs, and targets of signaling pathways that regulate translation.
The major method currently used to label newly synthesized proteins for detection by CuAAC is based on bioorthogonal Met analogs such as the azido analog Aha and the alkyne analog Hpg (5–8). Compared to these Met analogs, OP-puro has a number of important advantages: (i) Metabolic labeling with Met analogs requires Met-free media, which prevents the use of this method in animals, whereas OP-puro works well in complete media and in animals; (ii) Met analog incorporation is proportional to the number of Met residues in a protein, whereas OP-puro incorporates at exactly one molecule per nascent polypeptide chain, generating a “normal” representation of protein translation; (iii) Met analogs will not label proteins that do not start with or contain a Met residue, whereas OP-puro labeling does not depend on amino acid content; (iv) Met analogs need to be first activated and converted to amino acyl-tRNAs, before incorporation into proteins, whereas OP-puro generates covalent conjugates with nascent polypeptide chains directly, without any prior modification, and could conceivably be used when higher temporal resolution is needed.
Two other strategies to assay protein synthesis rely on the mechanism of translation inhibition by puro. In one approach, fluorescent puro conjugates have been used to image newly synthesized proteins in cells (15, 16). Although these conjugates are potent inhibitors of translation in vitro, their signal-to-noise ratio in cells is only 2–4, indicating the low sensitivity of this method for imaging protein translation. We speculate that this low signal-to-noise ratio might be due to the poor cellular permeability of the fluorescent puro derivatives, which contain 2–3 phosphate ester groups as well as cell-impermeable fluorophores such as fluorescein or Cy5. In contrast, OP-puro is cell permeable and generates a signal-to-noise ratio of 24 in cells, allowing a significantly more sensitive microscopic detection of protein synthesis.
In another approach, polypeptide-puro conjugates are detected with anti-puro antibodies (18). Although this approach is simple and allows the sensitive detection of nascent proteins by imunoblot, the subcellular pattern seen by anti-puro immunofluorescence (18, 19) differs significantly from that obtained using Hpg labeling (8) and is inconsistent with the expected subcellular localization of newly synthesized proteins identified by Aha labeling and mass spectrometry (5). Specifically, anti-puro staining shows the strongest signal at the plasma membrane, although the majority of newly synthesized proteins are nuclear and cytoplasmic (5). In contrast, OP-puro labeling has a subcellular pattern very similar to that obtained with Hpg and is consistent with the identity of newly synthesized proteins. We speculate that the discrepancy between OP-puro detection by CuAAC and anti-puro immunofluorescence might be because many of the polypeptide-OP-puro conjugates are inaccessible to anti-puro antibodies, but are readily accessible to the small fluorescent azides used to stain OP-puro-labeled cells.
Aside from imaging proteins synthesis in vivo, we anticipate that an important application of OP-puro will be the identification of proteins synthesized under specific conditions and in various organs and tissues. Recently, ribosome profiling has been used to characterize mRNA translation in cells (31, 32). This approach relies on sequencing mRNA fragments protected against RNase digestion by the ribosome footprint as a way to identify actively translated mRNAs. Although this strategy generates a detailed view of protein synthesis, it is considerably more labor intensive and experimentally demanding than the OP-puro method. Ribosome profiling requires the biochemical isolation of RNase-protected monosomes, followed by the generation and sequencing of a cDNA library. In contrast, the OP-puro method does not require complicated or delicate biochemical purifications, which can be problematic when analyzing animal tissues and organs. Because OP-puro is covalently attached to nascent proteins, tissue and organ samples can be harvested and quickly denatured, followed by affinity purification of OP-puro-labeled polypeptides under denaturing conditions, and finally by protein identification by mass spectrometry. Thus the OP-puro method should allow accurate and reproducible analysis of protein synthesis in vivo.
Materials and Methods
Op-Puro Labeling of Cultured Cells and Detection by Fluorescence Microscopy.
NIH 3T3 cells were grown on glass coverslips in DMEM supplemented with 10% bovine calf serum, penicillin, and streptomycin. OP-puro was added to cells in complete culture medium. After incubation for the desired amount of time, the cells were washed with PBS and then fixed with cold methanol for 2 min at -20 °C. The cells were washed with Tris buffered saline (TBS: 10 mM Tris, pH 7.5, 150 mM NaCl), permeabilized with TBST (TBS with 0.2% Triton X-100), and then washed with TBS again. CuAAC detection of OP-puro incorporated into nascent protein was performed by reacting the fixed cells for 30 min at room temperature with 20 μM Alexa568-azide, as described (33). After staining, the coverslips were washed several times with TBST, counterstained with Hoechst, and mounted in standard mounting media. The stained cells were imaged by differential interference contrast (DIC) and by epifluorescence microscopy on a Nikon TE2000U microscope equipped with an OrcaER camera (Hammamatsu), and 20× PlanApo 0.75NA and 40× PlanApo 0.95NA air objectives (Nikon). Images were collected using Metamorph image acquisition software (Applied Precision).
To determine if protein synthesis is required for OP-puro incorporation into nascent polypeptides, cells were incubated with OP-puro, in the presence or absence of 50 μg/mL CHX, and added to the cells 15 min before OP-puro. This concentration of CHX was determined to completely block protein translation in cells.
To determine the effect of inhibiting the proteasome on the stability of OP-puro-conjugated polypeptide chains, cells were pulse-labeled with 50 μM OP-puro for 15 min and were then incubated in complete media supplemented with 50 μg/mL CHX (to block further incorporation of OP-puro into nascent proteins), in the presence or absence of 5 μM of the proteasome inhibitor bortezomib. Cells were fixed at the indicated times and were stained in parallel with Alexa568-azide. Two negative controls were used: (i) untreated cells, and (ii) cells preincubated with 50 μg/mL CHX for 15 min, followed by incubation with 50 μM OP-puro and 50 μg/mL CHX for another 15 min.
Quantification of OP-Puro Labeling.
The intensity of the fluorescent OP-puro stain in single cells was quantified by automated analysis of microscopic images. Briefly, the centers of nuclei were first identified by local adaptive thresholding of Hoechst images. Cell boundaries were then determined by the watershed method. The fluorescence intensity in each cell was corrected by subtracting the local background, defined as the median intensity of background pixels surrounding the cell. The fluorescence intensity of each cell was finally computed as the mean of the corrected pixel values. In the bar plots, the height of each bar represents the mean cellular intensity for a given condition, and the error bar represents the standard deviation of the mean. Between 63 and 145 cells were quantified per condition.
Inhibition of Translation by OP-Puro.
A plasmid carrying a GFP fusion of the mouse Suppressor of Fused (SuFu) gene under the control of an SP6 RNA polymerase promoter was used to generate 35S-Met-labeled GFP-SuFu by in vitro translation in rabbit reticulocyte lysates (TNT SP6 quick coupled transcription/translation kit; Promega), according to the manufacturer’s instructions. Translation reactions were performed for 1 h at 30°C, in the absence or presence of varying concentrations of puro and OP-puro. The reactions were stopped by addition of SDS-PAGE sample buffer with 50 mM DTT and boiling. Equal amounts of lysate were separated by SDS-PAGE and the amount of in vitro translated GFP-SuFu was determined by autoradiography.
To measure translation inhibition in cells, human embryonic kidney 293T cells were preincubated for 30 min with varying concentrations of puro or OP-puro, in complete media. The cells were then incubated for 3 h in Met-free media supplemented with 35S-Met (from Perkin-Elmer, at 100 μCi/mL final concentration), in the continued presence of varying concentrations of puro or OP-puro. The cells were harvested, lyzed in TBS with 1% Triton X-100 and protease inhibitors (complete tablets; Roche), and centrifuged for 15 min at 20,000 × g in a refrigerated centrifuge. The clarified cells lysates were analyzed by SDS-PAGE, followed by autoradiography, to measure bulk protein translation.
Affinity Purification of Nascent Polypeptide-Op-Puro Conjugates.
Human 293T cells were labeled for 1 h in Met-free DMEM supplemented with 10% dialyzed fetal bovine serum and 35S-Met (100 μCi/mL final). The cells were then incubated in the same media, in the presence of 10 μM puro, 25 μM OP-puro, or 25 μM OP-puro and 50 μg/mL CHX, for an additional 1 h. The cells were harvested, washed with ice-cold PBS, and then lyzed on ice in 100 mM Tris, pH 8.5, with 1% Triton-X10 and protease inhibitors. The lysates were clarified by centrifugation for 15 min at 20,000 × g and 4 °C, and were then subjected to CuAAC with biotin-azide (25) for 30 min at room temperature. The final concentrations in the CuAAC reaction were 100 μM biotin-azide, 1 mM CuSO4, and 50 mM ascorbic acid (added last to the lysates). Biotinylated proteins were diluted in binding buffer (20 mM Tris, pH 8, 500 mM NaCl, 1% Triton-X100) and were bound to Neutravidin beads (Pierce). After extensive washes with binding buffer, bound proteins were eluted, separated by SDS-PAGE, and detected by autoradiography.
Use of Op-Puro to Image Protein Synthesis in Animals.
One hundred microliters of a 20 mM solution of OP-puro in PBS were injected intraperitoneally into a 3-wk-old mouse, and a mouse injected with 100 μL of PBS was used as negative control. Various organs were harvested after 1 h and were fixed in formalin overnight. Organ fragments were embedded in paraffin, sectioned, and washed with xylene to remove the paraffin. After washing with ethanol and rehydration in TBS, the tissue sections were stained with 20 μM tetramethylrhodamine(TMR)-azide, as described (33). The tissue sections were counterstained with Hoechst, mounted in standard mounting media, and were then imaged by fluorescence microscopy and DIC. Whole-mount staining of intestine fragments was performed as described (33). The fluorescent images of sections through mouse small intestine were quantified manually using Image J. For both control and OP-puro-injected specimens, the image was divided into three zones: the bottom of the crypts, the crypts proper, and the epithelium lining the vili. Circular regions of interest (ROI, n = 6–15) were randomly defined in each zone, and the mean fluorescence intensity was calculated for each ROI. The mean ROI intensity was plotted as the ratio to background, defined as fluorescence intensity in a region devoid of cells. The error bars represent the standard deviation of the mean ROI intensity.
Chemicals.
The detailed synthesis of OP-puro is shown in SI Text. TMR-azide and Alexa568-azide were described previously (33, 34).
Supplementary Material
Acknowledgments.
A.S. acknowledges support from the Harvard-Armenise Foundation. D.S. is supported by a Postdoctoral Fellowship from the Charles King Trust of the Medical Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111561108/-/DCSupplemental.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 3.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lecuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Link AJ, Tirrell DA. Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 7.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beatty KE, et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 9.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 10.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, et al. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 12.Nathans D. Puromycin inhibition of protein synthesis: Incorporation of puromycin into peptide chains. Proc Natl Acad Sci USA. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathans D. Inhibition of protein synthesis by puromycin. Fed Proc. 1964;23:984–989. [PubMed] [Google Scholar]
- 14.Isaacs WB, Fulton AB. Cotranslational assembly of myosin heavy chain in developing cultured skeletal muscle. Proc Natl Acad Sci USA. 1987;84:6174–6178. doi: 10.1073/pnas.84.17.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starck SR, Green HM, Alberola-Ila J, Roberts RW. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chem Biol. 2004;11:999–1008. doi: 10.1016/j.chembiol.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 19.Goodman CA, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nathans D, Neidle A. Structural requirements for puromycin inhibition of protein synthesis. Nature. 1963;197:1076–1077. doi: 10.1038/1971076a0. [DOI] [PubMed] [Google Scholar]
- 21.Pestka S, Vince R, Daluge S, Harris R. Effect of puromycin analogues and other agents on peptidyl-puromycin synthesis on polyribosomes. Antimicrob Agents Chemother. 1973;4:37–43. doi: 10.1128/aac.4.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckermann DJ, Greenwell P, Symons RH. Peptide-bond formation on the ribosome. A comparison of the acceptor-substrate specificity of peptidyl transferase in bacterial and mammalian ribosomes using puromycin analogues. Eur J Biochem. 1974;41:547–554. doi: 10.1111/j.1432-1033.1974.tb03296.x. [DOI] [PubMed] [Google Scholar]
- 23.Vanin EF, Greenwell P, Symons RH. Structure-activity relationships of puromycin analogues on Escherichia coli polysomes. FEBS Lett. 1974;40:124–126. doi: 10.1016/0014-5793(74)80908-7. [DOI] [PubMed] [Google Scholar]
- 24.Lee H, Fong KL, Vince R. Puromycin analogues. Effect of aryl-substituted puromycin analogues on the ribosomal peptidyltransferase reaction. J Med Chem. 1981;24:304–308. doi: 10.1021/jm00135a013. [DOI] [PubMed] [Google Scholar]
- 25.Jao CY, Roth M, Welti R, Salic A. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc Natl Acad Sci USA. 2009;106:15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg AL. Degradation of abnormal proteins in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wharton SA, Hipkiss AR. Abnormal proteins of shortened length are preferentially degraded in the cytosol of cultured MRC5 fibroblasts. FEBS Lett. 1984;168:134–138. doi: 10.1016/0014-5793(84)80222-7. [DOI] [PubMed] [Google Scholar]
- 28.Andrews TM, Tata JR. Protein synthesis by membrane-bound and free ribosomes of secretory and non-secretory tissues. Biochem J. 1971;121:683–694. doi: 10.1042/bj1210683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin MZ, Glenn JS, Tsien RY. A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proc Natl Acad Sci USA. 2008;105:7744–7749. doi: 10.1073/pnas.0803060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arava Y, Boas FE, Brown PO, Herschlag D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 2005;33:2421–2432. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.