Abstract
Eutrophication and global climate change lead to expansion of hypoxia in the ocean, often accompanied by the production of hydrogen sulfide, which is toxic to higher organisms. Chemoautotrophic bacteria are thought to buffer against increased sulfide concentrations by oxidizing hydrogen sulfide before its diffusion to oxygenated surface waters. Model organisms from such environments have not been readily available, which has contributed to a poor understanding of these microbes. We present here a detailed study of “Sulfurimonas gotlandica” str. GD1, an Epsilonproteobacterium isolated from the Baltic Sea oxic-anoxic interface, where it plays a key role in nitrogen and sulfur cycling. Whole-genome analysis and laboratory experiments revealed a high metabolic flexibility, suggesting a considerable capacity for adaptation to variable redox conditions. S. gotlandica str. GD1 was shown to grow chemolithoautotrophically by coupling denitrification with oxidation of reduced sulfur compounds and dark CO2 fixation. Metabolic versatility was further suggested by the use of a range of different electron donors and acceptors and organic carbon sources. The number of genes involved in signal transduction and metabolic pathways exceeds those of other Epsilonproteobacteria. Oxygen tolerance and environmental-sensing systems combined with chemotactic responses enable this organism to thrive successfully in marine oxygen-depletion zones. We propose that S. gotlandica str. GD1 will serve as a model organism in investigations that will lead to a better understanding how members of the Epsilonproteobacteria are able to cope with water column anoxia and the role these microorganisms play in the detoxification of sulfidic waters.
Keywords: marine bacteria, sulfide oxidation, isolation, genomics
Oxygen-deficient (“hypoxic,” typically defined as dissolved oxygen < 2 mg/L−1) conditions in marine water columns occur seasonally near productive upwelling areas (1) and permanently in enclosed, stratified seas (Black Sea, Baltic Sea) or isolated basins (e.g., Cariaco) (2, 3). Severe hypoxia and total lack of oxygen (anoxia) can lead to the production of hydrogen sulfide, which results from anaerobic mineralization of organic matter by sulfate reducing bacteria. Climate change, which causes higher water temperatures and stronger water column stratification combined with eutrophication, results in the expansion of oxygen-depleted areas (4) and more frequent occurrence of sulfidic conditions (5). Hydrogen sulfide toxicity has been shown to be responsible for the severe mortality of marine organisms (6), the deterioration of coastal ecosystems, and drastic reductions in secondary production (5, 7), leading to “dead zones” in the ocean (5, 6). Indeed, coastal dead zones have become an important environmental issue, as they pose a serious threat to benthic fauna and fishery-based economies (5, 7).
Diverse groups of heterotrophic, chemolithoautotrophic, and phototrophic bacteria are known to oxidize hydrogen sulfide and other reduced sulfur species (8), thereby lowering the concentration of these harmful compounds. There is also evidence to suggest that some microbes remove sulfide before it reaches the oxygenated zone (2). Recently, it has been demonstrated that blooming chemolithoautotrophic Gamma- and Epsilonproteobacteria counteract the expansion of sulfidic waters, for example in African shelf subsurface waters (9), or in a seasonally anoxic, sulfidic fjord in Canada (10). Chemolithoautotrophic Epsilonproteobacteria are known mainly from deep-sea vent ecosystems, where they constitute a dominant and diverse bacterial group (11) with important roles in the cycling of carbon, nitrogen, and sulfur (12, 13). However, Epsilonproteobacteria have also been detected in other marine sulfidic environments (9, 14–16), and are thought to be associated with chemoautotrophy and sulfur cycling.
In the central Baltic Sea, a stable halocline separates deep anoxic, sulfidic water from oxygenated surface water. Epsilonproteobacteria in the transition zone of suboxic and anoxic/sulfidic waters (the redoxline) are mainly comprised of a single phylogenetic cluster, Sulfurimonas subgroup GD17, which dominates chemoautotrophic production in the redoxcline (17). It has been proposed that these bacteria combine the oxidation of reduced sulfur compounds with denitrification (14). This chemoautotrophic denitrification has been shown to constitute a major nitrogen loss process in the water column of marine systems with a sulfide-nitrate interface (18, 19).
Still, the physiology of these pelagic Epsilonproteobacteria and their ecological role in counteracting expanding sulfidic waters are not well understood because of missing representative isolates. In this study we present autecological, genomic, and physiological analyses of “Sulfurimonas gotlandica” str. GD1, a strain recently isolated from a Baltic Sea redoxcline where close phylogenetic relatives occur in considerable abundance (20). The unique physiological and genomic features of this microorganism help to illustrate how these bacteria adapt and thrive in hypoxic waters, and how their activities contribute to hydrogen sulfide removal in marine environments.
Results and Discussion
Isolation and in Situ Occurrence.
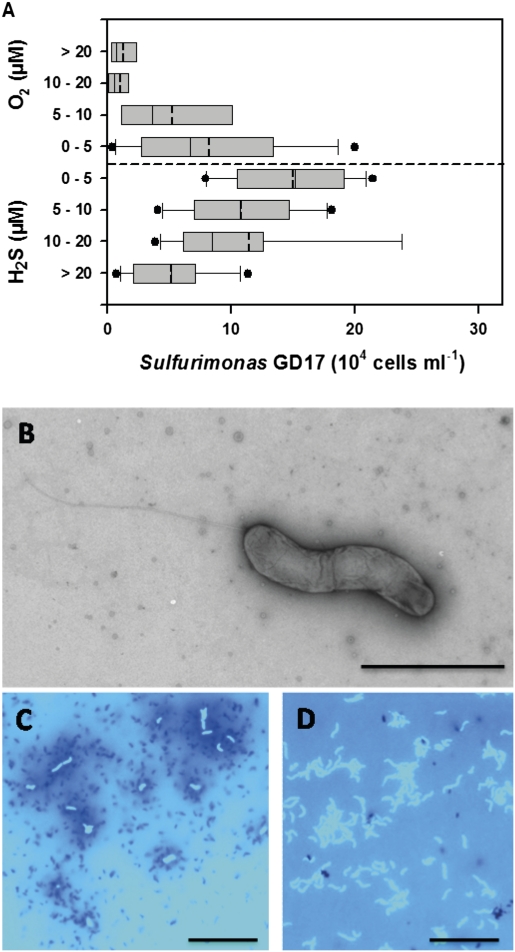
A compilation of samples taken during different seasons and years and quantification by a catalyzed reporter deposition (CARD)-FISH probe (SUL90) specific for the Sulfurimonas GD17 cluster (20) showed that this cluster is consistently present in hypoxic waters of the central Baltic Sea (Fig. 1A). Sulfurimonas GD17 is most abundant directly below the oxic-anoxic interface, where the water is slightly sulfidic (< 5 μM H2S) and can account for up to 25% of total prokaryotic abundance. Sulfide generally overlaps with nitrate, but not with oxygen, in this zone (18). Cell numbers quickly decline in the oxic part of the water column, but relatively high concentrations (104 to 105 cells mL−1) extend deeper into the sulfidic zone.
Fig. 1.
Sulfurimonas GD17 in Baltic Sea redoxclines. (A) Abundance of the Sulfurimonas subgroup GD17 as determined by CARD-FISH vs. grouped concentrations of sulfide and oxygen in Baltic Sea redoxcline samples. Values for sulfide are expressed as negative oxygen concentrations. Box-and-whisker plots with outliers include median (solid) and mean (dashed) values within the boxes. Data from 89 samples collected from redoxclines in the central Baltic Sea during cruises between 2003 and 2009, including 35 datapoints from refs 17 and 20, is displayed. (B) Electron micrograph of a phosphotungstic acid negatively stained S. gotlandica str. GD1 cell cultivated on nitrate and thiosulfate demonstrating spirilla-like morphology and monopolar flagella. (Scale bar, 2 μm.) (C) Fluorescence microscopy of DAPI-stained S. gotlandica str. GD1 cells grown on 14C-bicarbonate, followed by microautoradiographic incubation (17). Silver granules surrounding GD1 cells indicate 14C-bicarbonate, suggesting chemolithoautotrophic activity. (Scale bar, 10 μm.) (D) Fluorescence microscopy of DAPI-stained S. gotlandica str. GD1 cells. (Scale bar, 10 μm.)
A bacterial strain closely related to the Sulfurimonas GD17 cluster was isolated from the oxic-anoxic interface of the central Baltic Sea and grown anaerobically on a liquid mineral medium containing thiosulfate and nitrate (SI Materials and Methods). This strain, S. gotlandica str. GD1, is a spiral-shaped motile rod, 0.5–1.0 μm in size (Fig. 1 B and D). Cell numbers reached around 5 × 107 cells mL−1 at the experimental growth conditions used in this study. Inorganic carbon (carbon dioxide, carbonate, bicarbonate) was the sole carbon source, and dark CO2 fixation was confirmed microautoradiographically using 14C-bicarbonate (Fig. 1C). In contrast to most cultivated relatives within the genus Sulfurimonas, which are hyperthermophilic, S. gotlandica str. GD1 showed optimal growth within a temperature range of 10–20 °C, with doubling times of about 13 h in an anoxic synthetic brackish water medium containing thiosulfate and nitrate.
Genomic Analysis of S. gotlandica str. GD1.
Genomic sequencing of S. gotlandica str. GD1 yielded a single contig representing almost the complete but not the full circular chromosome, as a single small gap prevailed. The 2.95-Mb sequence is the largest epsilonproteobacterial genome sequenced thus far. A total of 2,879 coding sequences were identified, along with four copies of the ribosomal RNA (rRNA) operon and 47 transfer RNAs (tRNA) (Table S1).
S. gotlandica str. GD1 and group GD17 exhibit 95.7% 16S rRNA sequence identity, and 93.6% or 93.9%, respectively, to Sulfurimonas denitrificans DSM 1251T, the closest cultivated relative, isolated from a tidal mud flat (21). Phylogenetically, these bacteria are members of the Helicobacteraceae family of Epsilonproteobacteria. Epsilonproteobacteria occupy habitats ranging from human blood to hydrothermal vents to mesophilic marine systems, and as such display extremely different physiologies (Fig. S1). S. gotlandica str. GD1 is the only known cultivated representative from pelagic marine systems.
S. gotlandica str. GD1 Displays an Enhanced Metabolic Versatility.
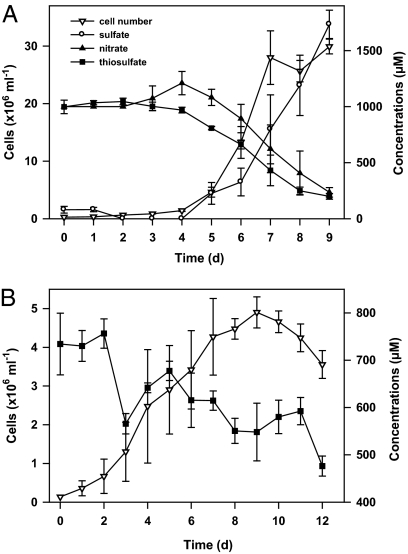
In vitro experiments and genome analysis revealed that S gotlandica str. GD1 has a considerable metabolic versatility, capable of both chemoautotrophy (fueled by different electron donor and acceptor combinations) and utilization of organic substrates. Experiments confirmed the consumption of nitrate and hydrogen sulfide or thiosulfate and production of sulfate, suggesting that chemolithoautotrophic denitrification is an important respiratory pathway in this organism (Fig. 2A). S gotlandica str. GD1 showed a pronounced ability to exploit a wide range of reduced sulfur compounds as electron donors, including elemental sulfur, sulfite, and hydrogen sulfide (Table S2). Growth in the presence of hydrogen sulfide was observed at low concentrations (10 μM), similar to concentrations found at the oxic-anoxic interfaces in the Baltic Sea, indicating that hydrogen sulfide is toxic at higher concentrations (Fig. S2B). Although S. gotlandica str. GD1 was tolerant of high oxygen concentrations and growth was not inhibited until about 10% oxygen saturation (Fig. S2A), it could not be demonstrated that oxygen was used as an electron acceptor.
Fig. 2.
Anaerobic, chemoautotrophic growth of S. gotlandica str. GD1 in batch cultures. (A) Growth on thiosulfate (1 mM) and nitrate (1 mM): a time course measuring nitrate and thiosulfate used and sulfate produced. Results clearly indicate chemolithoautotrophic denitrification. (B) Cells grow well on thiosulfate (1 mM) as the sole substrate, but do not develop products typical for thiosulfate disproportionation. All datapoints shown in A and B are the means of three replicates; error bars are SEs.
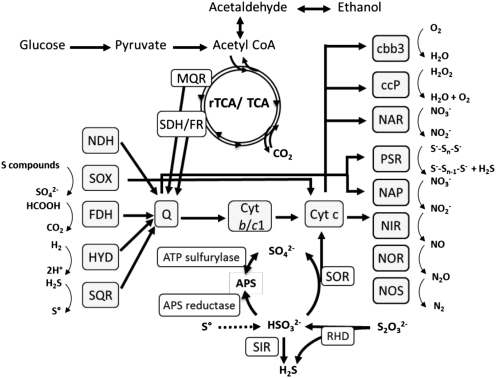
Genome analysis revealed that the S. gotlandica str. GD1 chromosome contains a complete set of genes required for chemolithoautotrophy and encodes the components of the reductive citric acid cycle for inorganic carbon fixation, all reductases necessary to reduce nitrate to nitrogen gas, including NAP and NAR, and different enzyme systems for the oxidation of reduced sulfur compounds (Fig. 3 and Table S3). In addition to the sulfur reducing Sox multienzyme complex, several enzymes known to oxidize sulfite to sulfate via the intermediate adenosine-5′-phosphosulfate in a Sox-independent pathway (22) were also encoded in the genome. The simultaneous presence of both sulfite-oxidizing pathways has also been observed in several other sulfur oxidizing bacteria, and is hypothesized to provide the organisms with a higher flexibility in their dissimilatory sulfur metabolism (22).
Fig. 3.
Theoretical model of the metabolic pathways and electron transport in S. gotlandica str. GD1. Enzymes catalyzing key reactions are indicated; reactions for which no enzyme was annotated in S. gotlandica str. GD1 but are predicted to occur are indicated by dotted lines. cbb3, cytochrome c oxidase; ccP, cytochrome c peroxidase; Cyt b/c1: quinone cytochrome oxidoreductase; FDH, formate dehydrogenase; HYD, hydrogenase; MQR, malate-quinone-reductase; NAP/NAR, nitrate reductase; NIR, nitrite reductase; NOR, nitric oxide reductase; NOS, nitrous oxide reductase; PSR, polysulfide reductase; RHD, rhodanese-related sulfurtransferase; Q, quinine; SDH/FR, succinate dehydrogenase/ fumarate reductase; SIR, sulfite reductase; SOR, sulfite oxidoreductase; SOX, sulfur oxidation multienzyme complex; SQR, sulfide quinone reductase.
Both adenylyl sulfate reductase and ATP sulfurylase, as well as sulfite oxidoreductase, could be potentially used for inorganic disproportionation of reduced sulfur compounds, as has been shown for Deltaproteobacteria (23). This use would provide a small energy gain and might explain the growth of GD1 on thiosulfate as the sole substrate (Fig. 2B). However, although we could show the conversion of thiosulfate to sulfate alongside dark carbon dioxide fixation, hydrogen sulfide, zero-valent sulfur, and polymeric sulfur compounds were not detected in significant amounts, suggesting that disproportionation of thiosulfate may not occur. Hepes, the organic buffer used in the media, may potentially be used as an alternative electron acceptor. Similar chemical groups (sulfonic acid-, hydroxy-, and carboxyamide-residues) are found in humic acids, which are known to function as electron acceptors (24), and may potentially serve as electron acceptors for these bacteria in the absence of nitrate (e.g., in the upper sulfidic zone of the redoxcline). Nevertheless, the genomic features of S. gotlandica str. GD1 (Fig. 3 and Table S3) suggest this strain may be capable of disproportionation of reduced sulfur compounds, potentially induced only in very specific redox conditions. Furthermore, the genome encodes a membrane-bound polysulfide reductase, which may enable the strain to use polysulfide as an alternative electron acceptor (e.g., for the oxidation of H2 or formate, as described for Nautilia profundicola and Wolinella succinogenes) (25, 26). Polysulfides have been shown to be important sulfur species in the oxic-anoxic interfaces of diverse systems (27).
These results, together with the observed dominance of Epsilonproteobacteria in the upper sulfidic zone, suggest that the oxidation of reduced sulfur compounds is likely the primary energy source of S. gotlandica str. GD1 and the cluster GD17. Interestingly, we detected a gene encoding a putative cbb3-type cytochrome c oxidase with the potential to mediate aerobic respiration or to act as an electron acceptor in oxygen scavenging and thus in the prevention of oxidative stress (21). The latter function is consistent with our experimental observations that, with oxygen as a terminal electron acceptor, no growth was observed under the conditions tested (organic and inorganic substrates in different concentrations). However, we cannot exclude the possibility that the experimental design of the oxygen utilization experiments (e.g., concentrations of electron donors, carbon sources) was not optimal to detect growth under the described conditions. Therefore, the precise function of the proton-pumping cbb3-type cytochrome c oxidase remains unknown.
By using nitrate as an electron acceptor instead of oxygen, this bacterial group can extend its ecological niche in the water column to below the oxic-anoxic interface. Sulfide oxidation rates (Fig. S2B), in situ chemical profiles, and the in situ abundance of the Sulfurimonas GD17 cluster (Fig. 1A) indicate that these bacteria are capable of oxidizing the upwardly diffusing sulfide completely before it reaches the oxygenated water layer in the central Baltic Sea.
In the absence of nitrate unknown electron acceptors, such as organic compounds and polysulfides, are likely relevant in the anoxic, upper sulfidic layer, where high rates of chemoautotrophic activity have been measured (28) and attributed to the Sulfurimonas GD17 cluster (17). In addition to the sulfur-oxidizing activities observed in S. gotlandica str. GD1, this organism was also capable of using hydrogen as an electron donor with nitrate as electron acceptor (Table S2); this has also been observed in microbes of deep-sea hydrothermal habitats (12). The presence of multiple hydrogenases in the genome (Table S3) suggests adaptations to different concentrations of hydrogen, as an important alternative electron donor, and a variety of electron acceptors (29). S. gotlandica str. GD1 was also able to grow on nitrate combined with several different organic compounds, such as formate, acetate, yeast, pyruvate, and peptone, indicating its capability for heterotrophic denitrification (Table S2). The genetic basis for utilization of organic carbon compounds (e.g., homologs for glycolysis and proteolysis, were also present in the genome) (Fig. 3).
Physiological, Behavioral, and Genetic Adaptations to Pelagic Redoxclines.
Although the pelagic hypoxia that is found in the Baltic or Black Sea can be relatively stable and long-lasting, the fine-scale structure of the redoxcline is subject to frequent disturbances because of lateral intrusions and small-scale mixing events (30). Thus, organisms thriving in this layer must cope with vertical shifts in the oxic-anoxic interface, changes in electron acceptor and donor concentrations, and intermittent exposure to unfavorable concentrations of oxygen or hydrogen sulfide. The presence of genes that encode enzymes that protect against oxygen stress (Table S3) and the high oxygen tolerance of S. gotlandica str. GD1 observed experimentally (Fig. S2A), suggests that this strain is well equipped to cope with fluctuating oxygen concentrations.
Directed motility is advantageous in environments with fluctuating nutrient availabilities, such as the suboxic/sulfidic transition zones. Genes involved in environmental sensing and chemotaxis were found in the genome and a positive chemotactic response to nitrate was shown experimentally in capillary assays (enrichment factor of 2.6–2.9) (Fig. S3). Chemotaxis enables S. gotlandica str. GD1 to locate zones within redox gradients favorable for growth.
Comparative Genomics of S. gotlandica str. GD1 and Related Members of the Epsilonproteobacteria.
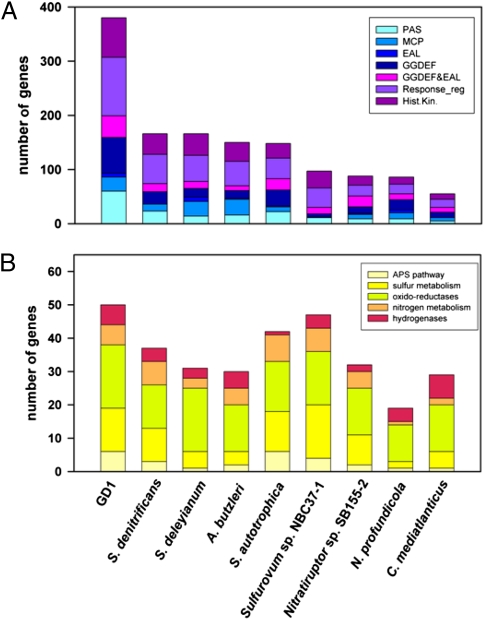
Members of the Epsilonproteobacteria are known for their high metabolic versatility (12, 13), and this feature is particularly pronounced in S. gotlandica str. GD1. Hydrothermal vent Epsilonproteobacteria have numerous signal sensing and transduction mechanisms, essential for free-living bacteria in highly variable environments (29, 31). Compared with previously described Epsilonproteobacteria, however, the total number of protein domains involved in environmental sensing and signal transduction is notably increased in the genome of S. gotlandica str. GD1 (Fig. 4A). Higher numbers of functional domains involved in energy metabolism were also observed in S. gotlandica str. GD1 (Fig. 4B), compared with the epsilonproteobacterial genomes from hydrothermal vents or from vent-associated metazoans, such as Caminibacter mediatlanticus, N. profundicola, and Nitratiruptor sp., for which a relatively simple electron transport chain has been described (29).
Fig. 4.
Quantitative comparative genomic analysis of S. gotlandica str. GD1 with other members of the Epsilonproteobacteria. (A) Number of sensing and signal transduction domains. EAL, EAL domain; GGDEF, GGDEF domain; Hist.Kin., Histidine kinase; MCP, methyl-accepting chemotaxis protein signaling domains; PAS, PAS fold; Response_reg, Response regulator receiver domain. (B) Number of genes related to energy generation by redox reactions, grouped in different functional categories. The complete set of genes considered for the different categories and information on the respective bacterial taxa are given in Table S4.
These observations suggest that S. gotlandica str. GD1 has evolved to cope with a particularly challenging and dynamic environment. The fact that multiple homologs of genes involved in metabolic processes are present in the GD1 genome (e.g., hydrogenases and sulfide-quinone oxidoreductases) (Table S3) suggests that different genes with similar functions are expressed under different environmental conditions, enabling a varied array of different electron donor/acceptor combinations in fluctuating redox-environments. Other genes predicted to be involved in energy metabolism (e.g., molybdopterin oxidoreductases) could not be determined. Thus, S. gotlandica str. GD1 may harbor additional metabolic capabilities not yet identified that play a critical role in helping it to adapt to life in a highly dynamic, fluctuating environment, such as pelagic redoxclines.
Global Distribution of Sulfur Oxidizing Bacteria in Hypoxic Marine Water Columns.
Epsilonproteobacterial sequences related to the Sulfurimonas group, but also from the genus Arcobacter, have been detected in other stratified hypoxic marine systems with permanent or seasonal sulfidic waters and a nitrate or oxygen/sulfide interface, such as the Black Sea, the Cariaco Basin, and Canadian fjords (Table S5). Furthermore, representatives of this group have been retrieved from the water column of lakes with an oxic-anoxic interface (32). The other globally distributed but still uncultivated bacterial group related to sulfur oxidation in marine hypoxic systems comprises the γ-sulfur oxidizers, and particularly the SUP05 cluster, closely affiliated with sulfur-oxidizing symbionts (10). These bacteria seem to have an even wider distribution and are widespread also in marine oxygen minimum zones (OMZ), in which sulfidic conditions occur only irregularely, such as in the Namibian upwelling (9), or are hardly detectable because of low sulfide concentrations and a high turnover, as in the Chilean OMZ (33). This finding clearly shows that we have two major sulfur oxidizing bacterial clades in hypoxic marine water columns, within the Gamma- and Epsilonproteobacteria, which have an overlapping but slightly different distribution, potentially related to different redox potentials. Both chemoautotrophic sulfur-oxidizing bacterial groups can use nitrate to respire reduced sulfur compounds. However, these groups also exhibit substantial differences in their metabolic pathways (10), and their niche differentiation in hypoxic water columns remains to be resolved.
Conclusion
S. gotlandica str. GD1 is a cultivated Epsilonproteobacterium isolated from a marine hypoxic water column and a unique isolate from this phylum with proven high abundances in nature. Results from this study suggest that this strain is highly adaptable to the fluctuating redox conditions and varying oxygen, nitrate, and sulfide concentrations in suboxic and sulfidic waters and is exceptionally versatile, even for members of the Epsilonproteobacteria, which are known for their adaptability. In Baltic Sea redoxclines, these organisms are capable of oxidizing the upwardly diffusing sulfide completely. Related Epsilonproteobacteria are globally distributed in marine dead zones with sulfide accumulation in stratified water columns, where they play an important role in the detoxification of sulfide before it enters oxic waters and impacts higher organisms. The natural abundance and cultivability of S. gotlandica str. GD1 make it a suitable model organism for further studies of Epsilonproteobacteria biology and for understanding the ecological impact of these organisms in expanding hypoxic/sulfidic areas in the oceans worldwide.
Materials and Methods
Microbial Isolation and Growth.
S. gotlandica str. GD1 was isolated from a water sample collected from the Baltic Sea in May 2005 (215 m depth, 5 °C, Gotland Deep, 5719.2′N, 2003′E). Strain isolation was carried out in an anoxic artificial brackish water medium (ABW) (34) supplemented with 100 μM KNO3 and 100 μM Na2S2O3. A pure culture was obtained using the dilution to extinction method, and purity was finally confirmed by molecular methods. Growth experiments were conducted in anoxic ABW amended with nitrate and thiosulfate at 15 °C in the dark. Growth rate, temperature, sea salt requirements, and oxygen sensitivity were investigated, and alternative electron donors (sulfur compounds, hydrogen, organic substances) and electron acceptors (nitrite, fumarate, Hepes) were tested. Growth rates were calculated by measuring cell numbers during a 7-d incubation, as assessed by flow cytometry (35). Chemical analyses were performed as described previously (36–40). Dark CO2 fixation rates were determined as described by Jost et al. (28). Chemotactic responses toward nitrate were examined using a modified capillary assay for chemotaxis (41). Capillary assays including background controls were performed in three to four parallel experiments, and repeated three times. After a 1.5- to 2-h incubation, the capillaries were removed, DAPI staining was performed and the relative response was determined (42).
Genome Sequencing, Annotation, and Analysis.
DNA extraction, shotgun cloning, and sequencing was done by the J. Craig Venter Institute as a part of the Gordon and Betty Moore Foundation Marine Microbiology Initiative. Gene prediction and automatic annotation was done using Glimmer (43) and Michanhti (44), as well as the RAST server (45). Manual curation was done using GenDB v2.2 (46) and JCoast (47). Alignment and phylogenetic analyses of the S. gotlandica str. GD1 16S rRNA sequence were performed using the ARB software package (48) and the SILVA database (49).
Supplementary Material
Acknowledgments
We thank the captain and the crew of the RV Alkor; Bärbel Buuk, Christian Meeske, Katja Becker, Kerstin Mammitzsch, and Annett Grüttmüller for their excellent technical assistance; Michael Hannig for help during isolation procedures; and Michael Laue for providing electron micrographs. Genome sequencing was done by the J. Craig Venter Institute as a part of the Gordon and Betty Moore Foundation Marine Microbiology Initiative. M.L. was supported by Deutsche Forschungsgemeinschaft Projects LA 1466/4-1 and LA 1466/4-2, and C.G.B. by BMBF project “MIMAS” (03F0480E).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Whole Genome Shotgun project has been deposited in the DNA Databank of Japan/European Molecular Biology Laboratory/GenBank (accession no. AFRZ00000000; the version reported in this paper is the first version, AFRZ01000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111262109/-/DCSupplemental.
References
- 1.Paulmier A, Ruiz-Pino D. Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr. 2009;80:113–128. [Google Scholar]
- 2.Jørgensen BB, Fossing H, Wirsen CO, Jannasch HW. Sulfide oxidation in the anoxic Black Sea chemocline. Deep-Sea Res. 1991;38:S1083–S1103. [Google Scholar]
- 3.Taylor GT, et al. Chemoautotrophy in the redox transition zone of the Cariaco Basin: A significant midwater source of organic carbon production. Limnol Oceanogr. 2001;46:148–163. [Google Scholar]
- 4.Stramma L, Johnson GC, Sprintall J, Mohrholz V. Expanding oxygen-minimum zones in the tropical oceans. Science. 2008;320:655–658. doi: 10.1126/science.1153847. [DOI] [PubMed] [Google Scholar]
- 5.Diaz RJ, Rosenberg R. Spreading dead zones and consequences for marine ecosystems. Science. 2008;321:926–929. doi: 10.1126/science.1156401. [DOI] [PubMed] [Google Scholar]
- 6.Vaquer-Sunyer R, Duarte CM. Thresholds of hypoxia for marine biodiversity. Proc Natl Acad Sci USA. 2008;105:15452–15457. doi: 10.1073/pnas.0803833105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin LA, et al. Effects of natural and human-induced hypoxia on coastal benthos. Biogeosciences. 2009;6:2063–2098. [Google Scholar]
- 8.Kelly DP, Shergill JK, Lu WP, Wood AP. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie van Leeuwenhoek. 1997;71:95–107. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 9.Lavik G, et al. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature. 2009;457:581–584. doi: 10.1038/nature07588. [DOI] [PubMed] [Google Scholar]
- 10.Walsh DA, et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science. 2009;326:578–582. doi: 10.1126/science.1175309. [DOI] [PubMed] [Google Scholar]
- 11.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: Diversity, biochemistry and ecological significance. FEMS Microbiol Ecol. 2008;65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell BJ, Engel AS, Porter ML, Takai K. The versatile ε-proteobacteria: Key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 14.Labrenz M, et al. Impact of varying in vitro electron donator-/acceptor conditions on potential chemolithoautotrophic communities from pelagic redoxclines. Appl Environ Microbiol. 2005;71:6664–6672. doi: 10.1128/AEM.71.11.6664-6672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, et al. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl Environ Microbiol. 2006;72:2679–2690. doi: 10.1128/AEM.72.4.2679-2690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakimov MM, et al. Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L'Atalante, Eastern Mediterranean Sea. ISME J. 2007;1:743–755. doi: 10.1038/ismej.2007.83. [DOI] [PubMed] [Google Scholar]
- 17.Grote J, Jost G, Labrenz M, Herndl GJ, Jürgens K. Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl Environ Microbiol. 2008;74:7546–7551. doi: 10.1128/AEM.01186-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannig M, et al. Shift from denitrification to anammox after inflow events in the central Baltic Sea. Limnol Oceanogr. 2007;52:1336–1345. [Google Scholar]
- 19.Jensen MM, Petersen J, Dalsgaard T, Thamdrup B. Pathways, rates, and regulation of N2 production in the chemocline of an anoxic basin, Mariager Fjord, Denmark. Mar Chem. 2009;113:102–113. [Google Scholar]
- 20.Grote J, Labrenz M, Pfeiffer B, Jost G, Jürgens K. Quantitative distributions of Epsilonproteobacteria and a Sulfurimonas subgroup in pelagic redoxclines of the central Baltic Sea. Appl Environ Microbiol. 2007;73:7155–7161. doi: 10.1128/AEM.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievert SM, et al. USF Genomics Class. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol. 2008;74:1145–1156. doi: 10.1128/AEM.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kappler U, Dahl C. Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol Lett. 2001;203:1–9. doi: 10.1111/j.1574-6968.2001.tb10813.x. [DOI] [PubMed] [Google Scholar]
- 23.Finster K. Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem. 2008;29:281–292. [Google Scholar]
- 24.Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. Humic substances as electron acceptors for microbial respiration. Nature. 1996;382:445–448. [Google Scholar]
- 25.Ringel M, Gross R, Krafft T, Kröger A, Schauder R. Growth of Wolinella succinogenes with elemental sulfur in the absence of polysulfide. Arch Microbiol. 1996;165:62–64. [Google Scholar]
- 26.Hedderich R, et al. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev. 1998;22:353–381. [Google Scholar]
- 27.Luther GW, 3rd, et al. Sulfur speciation monitored in situ with solid state gold amalgam voltammetric microelectrodes: Polysulfides as a special case in sediments, microbial mats and hydrothermal vent waters. J Environ Monit. 2001;3:61–66. doi: 10.1039/b006499h. [DOI] [PubMed] [Google Scholar]
- 28.Jost G, Martens-Habbena W, Pollehne F, Schnetger B, Labrenz M. Anaerobic sulfur oxidation in the absence of nitrate dominates microbial chemoautotrophy beneath the pelagic chemocline of the eastern Gotland Basin, Baltic Sea. FEMS Microbiol Ecol. 2010;71:226–236. doi: 10.1111/j.1574-6941.2009.00798.x. [DOI] [PubMed] [Google Scholar]
- 29.Campbell BJ, et al. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 2009;5:e1000362. doi: 10.1371/journal.pgen.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reissmann JH, et al. Vertical mixing in the Baltic Sea and consequences for eutrophication—A review. Prog Oceanogr. 2009;82:47–80. [Google Scholar]
- 31.Nakagawa S, et al. Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA. 2007;104:12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehours A-C, Evans P, Bardot C, Joblin K, Gérard F. Phylogenetic diversity of archaea and bacteria in the anoxic zone of a meromictic lake (Lake Pavin, France) Appl Environ Microbiol. 2007;73:2016–2019. doi: 10.1128/AEM.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canfield DE, et al. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- 34.Bruns A, Cypionka H, Overmann J. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl Environ Microbiol. 2002;68:3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasol JM, et al. Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol. 2004;34:193–206. [Google Scholar]
- 36.Schneider B, Sadkowiak B, Wachholz F. A new method for continuous measurements of O2 in surface water in combination with pCO2 measurements: Implications for gas phase equilibration. Mar Chem. 2007;103:163–171. [Google Scholar]
- 37.Zopfi J, Ferdelman TG, Fossing H. In: Sulfur Biogeochemistry—Past and Present. Amend JP, Edwards KJ, Lyons TW, editors. Boulder, Colorado: Geological Society of America; 2004. pp. 97–116. [Google Scholar]
- 38.Chan CW, Suzuki I. Quantitative extraction and determination of elemental sulfur and stoichiometric oxidation of sulfide to elemental sulfur by Thiobacillus thiooxidans. Can J Microbiol. 1993;39:1166–1168. [Google Scholar]
- 39.Jones MN. Nitrate reduction by shaking with cadmium: Alternative to cadmium columns. Water Res. 1984;18:643–646. [Google Scholar]
- 40.Tabatabai MA. Determination of sulfate in water samples. Sulphur Inst J. 1974;10:11–13. [Google Scholar]
- 41.Overmann J. Chemotaxis and behavioral physiology of not-yet-cultivated microbes. Methods Enzymol. 2005;397:133–147. doi: 10.1016/S0076-6879(05)97008-0. [DOI] [PubMed] [Google Scholar]
- 42.Barak R, Nur I, Okon Y. Detection of chemotaxis in Azospirillum brasilense. J Appl Bacteriol. 1983;53:399–403. [Google Scholar]
- 43.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quast C. Bremen, Germany: Univ Bremen; 2006. MicHanThi - design and implementation of a system for the prediction of gene functions in genome annotation projects. PhD thesis; p. 120. [Google Scholar]
- 45.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer F, et al. GenDB—An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter M, et al. JCoast—A biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes. BMC Bioinformatics. 2008;9:177. doi: 10.1186/1471-2105-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.